Abstract
The compound GW678248 is a novel benzophenone nonnucleoside reverse transcriptase inhibitor (NNRTI). Preclinical assessment of GW678248 indicates that this compound potently inhibits wild-type (WT) and mutant human immunodeficiency virus type 1 (HIV-1) reverse transcriptase in biochemical assays, with 50% inhibitory concentrations (IC50s) between 0.8 and 6.8 nM. In HeLa CD4 MAGI cell culture virus replication assays, GW678248 has an IC50 of ≤21 nM against HIV-1 isogenic strains with single or double mutations known to be associated with NNRTI resistance, including L100I, K101E, K103N, V106A/I/M, V108I, E138K, Y181C, Y188C, Y188L, G190A/E, P225H, and P236L and various combinations. An IC50 of 86 nM was obtained with a mutant virus having V106I, E138K, and P236L mutations that resulted from serial passage of WT virus in the presence of GW678248. The presence of 45 mg/ml human serum albumin plus 1 mg/ml α-1 acid glycoprotein increased the IC50 approximately sevenfold. Cytotoxicity studies with GW678248 indicate that the 50% cytotoxicity concentration is greater than the level of compound solubility and provides a selectivity index of >2,500-fold for WT, Y181C, or K103N HIV-1. This compound exhibits excellent preclinical antiviral properties and, as a prodrug designated GW695634, is being developed as a new generation of NNRTI for the treatment of HIV-1 in combination with other antiretroviral agents.
New antiretroviral drugs are currently needed, and more will be required in the future, to treat drug-resistant strains emerging from current therapies (8). The nonnucleoside reverse transcriptase inhibitor (NNRTI) compound class is a key component of effective combination regimens. However, in the absence of complete suppression of human immunodeficiency virus type 1 (HIV-1) replication, resistance to NNRTIs emerges rapidly due to a low genetic barrier.
We have recently described results obtained with analogs in a benzophenone compound series that were synthesized and screened for anti-HIV-1 activity, with particular emphasis on potency against key NNRTI-resistant HIV-1 strains emerging from current treatments (6). The most promising candidate emerging from this screen was compound GW678248 (K. R. Romines, G. A. Freeman, L. T. Schaller, J. R. Cowan, S. S. Gonzales, J. H. Tidwell, C. W. Andrews III, D. K. Stammers, R. J. Hazen, R. G. Ferris, S. A. Short, J. H. Chan, and L. R. Boone, submitted for publication). An N-propionyl sulfonamide derivative amide prodrug of GW678248, designated GW695634 (Fig. 1), was developed to improve solubility and bioavailability (L. Schaller, T. Burnette, J. Cowan, P. Feldman, G. Freeman, H. Marr, B. Owens, K. Romines, J. Shepard, L. Boone, and J. Chan, Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother., poster H-872, 2003). GW678248 is released from GW695634 by endogenous proteinases, which may include, but may not be limited to, the vitamin K-dependent serine proteinases. Studies of interspecies pharmacokinetics and biotransformation of GW695634 which predicted acceptable human bioavailability of GW678248 (T. Burnette, H. Marr, B. Owens, P. Wheelan, and K. Moore, Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother., poster F-1837, 2003) have been confirmed by clinical phase I studies. The safety, tolerability, and pharmacokinetics of GW695634 have been studied in a double-blind, parallel, randomized, placebo-controlled, single-ascending-dose study with healthy volunteers administered doses from 10 mg to 800 mg (J. Denning, J. Kim, B. Sanderson, L. Edwards, K. Moore, and W. Symonds, XV Int. AIDS Conf., poster TuPeB 4480, 2004) and following repeat oral administration of doses from 100 to 400 mg q12h for 10 days (Y. Kim, W. Symonds, H. Steel, J. Ng-Cashin, and K. Moore, Abstr. 44th Intersci. Conf. Antimicrob. Agents Chemother., poster A-23, 2004). GW695634 was generally well tolerated in healthy volunteers and reached trough exposure levels of GW678248 exceeding the in vitro 50% inhibitory concentration (IC50) against wild-type (WT), Y181C, and K103N strains.
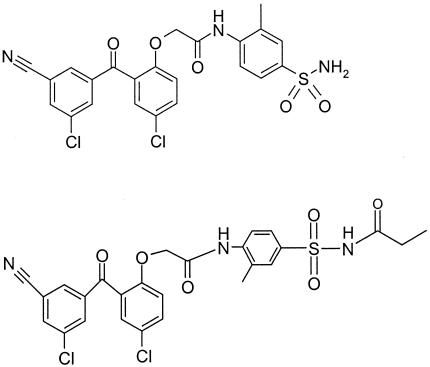
FIG. 1.
Structure of the benzophenone nonnucleoside reverse transcriptase inhibitor GW678248 and N-propionyl sulfonamide prodrug GW695634.
Herein we report initial preclinical studies describing the antiviral activity of GW678248. Additional studies of GW678248 resistance selection by in vitro passage, demonstration of the lack of susceptibility of HIV-2, combination with other antiretroviral drugs, and activity against clinical isolates from NNRTI-experienced patients are reported separately (9a). (The results of this study were presented in part at the 2nd IAS Conference on HIV Pathogenesis and Treatment, 13 to 16 July 2003, Paris, France [K. Romines et al., poster 535, and G. Freeman et al., poster 538].)
MATERIALS AND METHODS
Compounds.
GW678248 (structure shown in Fig. 1), GW695634 (structure shown in Fig. 1), efavirenz (EFV), nevirapine (NVP), zidovudine (AZT), and amprenavir (APV) were synthesized by GlaxoSmithKline, Research Triangle Park, NC.
Reverse transcriptase plasmids.
DNA encoding the HIV-1 reverse transcriptase (RT) was subcloned from an M13 phage into a general shuttle vector, pBCSK. Specific amino acid replacements were made using QuickChange reagents (Stratagene, La Jolla, CA) and mutagenic oligonucleotides (Oligos Etc., Wilsonville, OR). Following mutagenesis, the entire mutant RT coding sequence was verified by sequencing both DNA strands. RT plasmids containing the following mutations were constructed: L100I, K101E, K103N, V106A, V106I, V106M, V108I, E138K, Y181C, Y188C, Y188L, G190A, G190E, P225H, P236L, K103N/L100I, K103N/V108I, K103N/Y181C, K103N/G190A, K103N/P225H, V106A/Y181C, V106I/Y181C, V106I/P236L, V108I/Y181C, and V106I/E138K/P236L.
RT expression and purification.
For expression, each RT coding sequence was positioned 3′ to the TAC promoter of vector pKK233-2. Each RT mutant protein was purified from bacterial lysates by sequential column chromatography utilizing Toyopearl Super Q-650S, Toyopearl DEAE-650S, ceramic hydroxyapatite (Bio-Rad, Hercules, CA), Toyopearl SP-650S, and Superdex-200 resins. Fractions obtained following gel filtration that contained p66/p51 heterodimeric RT were pooled, concentrated to 10 mg/ml, and stored at −80°C.
RT enzyme assay.
Catalysis by WT and mutant RT enzymes was determined by measuring Cy5-dUMP incorporation into an Eu-labeled primer template by time-resolved fluorescence. The assays were initiated by the addition of enzyme and were linear over the 40-min assay period. In the absence of other nucleotides, only one Cy5-dUMP molecule is incorporated into each primer template, resulting in a distributive polymerization reaction. Each compound to be evaluated was serially diluted in 100% dimethyl sulfoxide (DMSO). Reactions contained 100 nM Cy5-dUTP, 40 nM Eu-labeled template primer, 1 nM purified recombinant RT, 47 mM Tris-HCl (pH 8.0), 75 mM KCl, 9.3 mM MgCl2, 0.003% Nonidet P-40 (Pierce Chemical Co., Rockford, IL), 9.3 mM dithiothreitol, test compound, and 2% DMSO in a total volume of 50 μl. Incorporation of Cy5-dUTP into the Eu-labeled template primer was monitored with a Victor2 1420 multilabel counter (Wallac-PerkinElmer Life and Analytical Sciences Inc., Boston, MA).
Construction of isogenic HIV-1 RT mutant virus.
Cultures of 1 × 107 Jurkat or MT-4 cells (maintained in RPMI medium 1640 containing 10% [vol/vol] fetal bovine serum [FBS] [HyClone, Logan, UT]; split 1:5 every 5 to 6 days) were cotransfected using DMRIE-C (Invitrogen, Carslbad, CA) with linearized mutant RT plasmid and BstEII-digested HXB2ΔRT DNA (an RT-deleted molecular clone) (11). The recombinant progeny virus was either sequenced across the mutation or verified using PCR. All confirmed mutants were expanded in either Jurkat or MT-4 cells and harvested, and titers were determined in the HeLa-CD4 MAGI assay. Recombinant DNA-derived isogenic strains consisted of virus containing the individual RT mutation L100I, K101E, K103N, V106A, V106I, V106M, V108I, E138K, Y181C, Y188C, Y188L, G190A, G190E, P225H, or P236L or the double or triple mutation combination K103N and L100I; K103N and V108I; K103N and Y181C; K103N and G190A; K103N and P225H; V106A and Y181C; V106I and Y181C; V106I and P236L; V108I and Y181C; or V106I, E138K, and P236L.
Viruses used in the nucleoside inhibitor-resistant virus panel.
Nucleoside-resistant HIV-1 strains: AZT-resistant strains RTMN (RT genotype = M41L T215Y K219Q) and RTMCY (RT genotype = D67N K70R T215Y), multinucleoside-resistant strain MDR151 (RT genotype = A62V V75I F77L Q151M), lamivudine (3TC)-resistant strain M184V (RT genotype = M184V), AZT- and 3TC-resistant strain DRSM34 (RT genotype = M41L D67N K70R M184V H208Y L210W T215Y K219E), abacavir (ABC)-, 3TC-, and didanosine (DDI)-resistant strain ABC4 (RT genotype = K65R LV4V M184V Y115F), and DDI-, zalcitabine (DDC)-, 3TC-, and ABC-resistant strain 74V/184V (RT genotype = L74V M184V) were used.
Viruses used in protease inhibitor-resistant virus panel.
Virus isolates and molecular clones were provided by Laurence Robinson (14): HxB2, Pro genotype, WT; isolate designated 468, derived from a clone from a subject receiving APV; Pro genotype, I15V E34G M36I S37E I50V L63P; Rp4K and Kp4′R in p7/p1 CS, Lp1′F in p1/p6 CS; protease inhibitor (PI) phenotype, SAQ and IDV resistant; isolate designated Triple, APV-resistant triple mutant; Pro genotype, M46I I47V I50V; PI phenotype, APV and LPV resistant; isolate designated 14330, derived from a heterogeneous plasma RT-PCR product; Pro genotype, D30N S37D I62V L63P I64M N88N/D; PI phenotype, NFV resistant; isolate designated 30813, virus derived from a plasmid clone from a subject receiving IDV; Pro genotype, I15V S37N I54V R57K I62V L63P H69Y A71T I72E V82A I85V; Ap2V in p7/p1 cleavage site; PI phenotype, IDV and LPV resistant; isolate designated D545701, this sample is not clonal and is derived directly from plasma RT-PCR products; Pro genotype, L10I L19Q K20R E35D M36I S37N M46I I50V I54V I62V L63P A71V V82A L90M; PI phenotype, multidrug resistant; isolate designated 31246, Pro genotype, L10I I54V L63P A71V I72V V77I V82A I84V L90M Q92K; PI phenotype, SAQ, RTV, IDV, LPV, and ATZ resistant; isolate designated EP13, Pro genotype, M46I L63P A71V V82F I84V; PI phenotype, RTV, IDV, LPV, and ATZ resistant; isolate designated I54V, Pro genotype, I54V; PI phenotype, WT; I50V, Pro genotype, I50V; PI phenotype, RTV and LPV resistant; Isolate designated EP14, Pro genotype, L10R M46I L63P V82T I84V; PI phenotype, IDV and LPV resistant.
Growth and maintenance of cell cultures.
Jurkat, MT-4, Molt-4, IM-9, and U-937 cells were propagated in RPMI medium 1640 containing 10% (vol/vol) FBS and 10 μg/ml gentamicin. MRC-5 cells were propagated in Dulbecco's modified Eagle medium (DMEM) containing 10% (vol/vol) FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. HepG2 cells were propagated in Eagle minimal essential medium containing 10% (vol/vol) FBS, 2 mM l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. HeLa-CD4-LTR-β-gal cells were propagated in DMEM containing 10% (vol/vol) FBS, 0.2 mg/ml G-418 sulfate (Invitrogen, Carlsbad, CA), and 0.1 mg/ml hygromycin B. All cells were maintained in log-phase growth. Peripheral blood mononuclear cells (PBMCs) were acquired as buffy coats from the American Red Cross (Durham region, NC) and separated from whole blood by density gradient centrifugation with lymphocyte separation medium. The cells were resuspended in 150 ml of RPMI medium 1640 containing 20% (vol/vol) FBS, 10% (vol/vol) CELLKINES TCGF (natural human T-cell growth factor) (Zeptometrix Corporation, buffalo, NY), and 50 μg/ml gentamicin, stimulated by the addition of 5 μg/ml phytohemagglutinin (PHA), and incubated at 37°C, 5% CO2, for 3 days. Following stimulation, PBMCs were centrifuged, washed once with phosphate-buffered saline, and then resuspended in medium without PHA.
HeLa-CD4 MAGI antiviral assay.
Multiwell (Costar 3904) plates were seeded with HeLa-CD4-LTR-β-gal (12) (obtained from Michael Emerman through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH) at 3 × 103 cells per well in 100 μl DMEM containing 10% (vol/vol) FBS, and placed in a humidified incubator at 37°C, 5% CO2, overnight. The following day, virus stocks were diluted in DMEM containing 10% (vol/vol) FBS and 25 μg/ml DEAE-dextran to a multiplicity of infection of 1,500 to 2,000 relative light units/ml. Thirty-five microliters (75 to 100 total relative light units) of diluted virus was added to each well, and the plates were placed in a humidified incubator at 37°C, 5% CO2, for 2 h. Test compounds were titrated in a fivefold stepwise manner at 1.35 times the final assay concentration in DMEM containing 10% (vol/vol) FBS and 0.135% DMSO. The final volume in each well was 150 μl, and DMSO was held constant at 0.1% in all wells, including those containing the no-compound controls. For assays involving protein binding assessments, human serum albumin (HSA), α-1 acid glycoprotein (AAG), or a combination of HSA and AAG was added to the compound mixture at appropriate concentrations. One hundred microliters of titrated compound was overlaid onto the HeLa-CD4 cells containing 35 μl of virus, and the plates were placed in a humidified incubator at 37°C, 5% CO2, for 3 days. Supernatants were aspirated, cells washed once with phosphate-buffered saline, and 15 μl of lysis buffer added followed by processing for β-galactosidase activity measurement according to kit instructions (Applied Biosystems, Foster City, CA). Plates were incubated at room temperature for at least 15 min and then read in a Topcount luminometer (Packard-PerkinElmer Life and Analytical Sciences Inc., Boston, MA) at 1 s/well.
MT-4 cell antiviral assay.
Anti-HIV-1 activity and compound-induced cytotoxicity were measured in parallel by means of a tetrazolium-based colorimetric procedure in the human T-cell lymphotropic virus-transformed cell line MT-4 (2, 13). Aliquots of the anti-HIV-1 test compounds were serially diluted in medium (RPMI 1640, 10% [vol/vol] FBS, and 10 μg/ml gentamicin) in 96-well plates (Costar 3598) using a Biomek automated dilution station (Beckman Coulter, Fullerton, CA). Test compounds, GW678248, AZT, or APV, were diluted to yield nine serial dilutions, resulting in a maximum final concentration of 50 nM, 1 μM, 4 μM, or 2 μM, respectively. Tests were performed in quadruplicate. Exponentially growing MT-4 cells were harvested and centrifuged at 192 × g in a Jouan centrifuge (model CR412) with an M4 swing-out rotor for 10 min. Cell pellets were resuspended in fresh medium (RPMI 1640, 20% [vol/vol] FBS, 20% [vol/vol] CELLKINES TCGF [natural human T-cell growth factor]; [Zeptometrix Corporation, buffalo, NY], and 10 μg/ml gentamicin) to a density of 6 × 105 cells/ml. Cell aliquots were infected by the addition of HIV-1 (WT or PI-resistant strains) diluted to give a viral multiplicity of infection of 0.001. A similar cell aliquot was diluted with medium to provide a mock-infected control. Cell infection was allowed to proceed for 1 h at 37°C in a tissue culture incubator with a humidified 5% CO2 atmosphere. After the 1-h incubation, the virus/cell suspensions were diluted sixfold with fresh medium, and 100 μl of the cell suspension was added to each well of the plate containing prediluted compounds. Plates were then placed in a tissue culture incubator with humidified 5% CO2 for 5 days. At the end of the incubation period, 20 μl of CellTiter 96 methanethiosulfonate (MTS) reagent (Promega, Madison, WI) was added to each well of the incubation plate. Plates were incubated at 37°C for 2.5 h to allow for color development. Optical density was measured at 492 nM using a microplate absorbance reader (Tecan US Inc., Durham, NC).
MTS cytotoxicity assay.
For the IM-9, Molt-4, MT-4, U-937, and PBMC cytotoxicity assessments, GW678248 and appropriate controls were dissolved and titrated by half-log increments into neat DMSO. Two microliters of titrated compound was transferred to 96-well plates containing 50 μl of appropriate medium. The cells were counted and diluted in appropriate medium, and 50 μl was dispensed onto compound plates. The final density for IM-9 and Molt-4 was 2 × 104 cells per well. The final densities for MT-4, U-937, and PBMC were 4 × 104, 5 × 103, and 1 × 105 cells per well, respectively. Following 3 days of incubation at 37°C, 5% CO2, the plates were viewed under a microscope and any visible compound precipitation was recorded. Twenty microliters of MTS reagent was added directly to the cells, and the plates were incubated at 37°C, 5% CO2, for 4 h, allowing color development of the formazan product. The absorbance of each well was then read at 490 nM in a Victor2 1420 Multilabel Counter (Wallac-PerkinElmer Life and Analytical Sciences Inc., Boston, MA).
For the MRC-5 and HepG2 cytotoxicity assessments, GW678248 was dissolved and titrated by twofold increments into neat DMSO. Culture medium was added to all wells to further dilute the compound fivefold. Five microliters of titrated GW678248 was transferred to a 96-well tissue culture plate prefilled with 95 μl of medium. A suspension of MRC-5 or HepG2 cells was then added to the GW678248 titrations at 3.3 × 104 cells per well. Following 3 days of incubation at 37°C, 5% CO2, the plates were viewed under a microscope and any visible compound precipitation was recorded. Medium was removed, and 120 μl of fresh medium containing MTS reagent was added. The plates were placed in an incubator at 37°C, 5% CO2, and periodically read as described above until the absorbance reached a value between 0.8 and 1.2 optical density units (usually after 45 to 90 min).
RESULTS
Inhibition of HIV-1 RT by GW678248.
The inhibitory activity of GW678248 against the molecular target was measured in a continuous time-resolved fluorescence enzyme assay using purified, recombinant WT RT and a collection of NNRTI-resistant RT mutant enzymes (Table 1). Against WT HIV-1 RT, GW678248 had an IC50 of 1.8 nM compared with 0.7 nM and 190 nM determined with EFV and NVP, respectively. The IC50s recorded for GW678248 with 11 RT mutants in the enzyme assay ranged from 0.8 nM to 6.8 nM, indicating a favorable resistance profile, since these values did not exceed that of WT RT by more than fivefold. The IC50 of NVP was increased more than fivefold for 8 of the 10 mutant enzymes tested, whereas the IC50 of EFV was within fivefold of the WT value for all but 4 of the 11 enzymes tested. The two highest IC50s for EFV were against K103N and L100I RTs, consistent with the reported antiviral data, which indicate a reduced sensitivity to EFV for viruses harboring these mutations in their RT (3, 4).
TABLE 1.
Inhibition of RT catalysis by GW678248 in a continuous time-resolved fluorescence assay
| HIV-I RT genotype | IC50 (nM)a of:
|
||
|---|---|---|---|
| GW678248b | Nevirapinec | Efavirenzd | |
| WT | 1.8 ± 0.6e | 190 | 0.7 ± 0.4 |
| L100I | 0.84 ± 0.05 | 1,500 | 18 ± 3 |
| K103N | 1.9 ± 0.2 | 11,000 | 28 ± 4 |
| V106A | 4.0 ± 0.6 | 50,000 | 2.6 ± 0.2 |
| V106I | 2.7 ± 0.2 | NT | 1.1 ± 0.2 |
| V108I | 2.0 ± 0.4 | 6,800 | 1.6 ± 0.5 |
| E138K | 4.2 ± 0.6 | 410 | 2.0 ± 0.3 |
| Y181C | 2.3 ± 0.4 | 35,000 | 1.9 ± 1.5 |
| Y188C | 3.1 ± 0.2 | >25,000 | 8.0 ± 0.8 |
| P236L | 6.8 ± 0.9 | 350 | 1.3 ± 0.6 |
| V106A Y181C | 3.5 ± 0.4 | >2,000 | 5.6 ± 0.8 |
| V108I Y181C | 4.8 ± 0.4 | >2,000 | 3.6 ± 0.1 |
IC50s are means ± standard deviations. NT, not tested.
n = 3.
n = 1.
n = 4.
n = 5.
Anti-HIV-1 activity and resistance profile of GW678248 in cell culture systems.
The activities of GW678248 against the WT and 25 genetically engineered HIV-1 strains with mutations known to be associated with NNRTI resistance (7) were determined in the HeLa-CD4 MAGI assay. GW678248 had an IC50 of <10 nM against all viruses in this panel with the exception of the Y188L mutant, for which the IC50 was 21 nM, and the V106I E138K P236L triple mutant (Table 2). NVP exhibited diminished activity against the majority of the NNRTI resistance-associated mutant HIV-1 strains, which is consistent with published literature (8). Also, consistent with published literature, EFV demonstrated significantly diminished activity against strains containing L100I, K103N, or Y188L (3, 4).
TABLE 2.
Antiviral activity of GW678248 against HIV-1 containing nonnucleoside reverse transcriptase inhibitor resistance mutations in HeLa-CD4 MAGI assay
| HIV-1 RT mutation(s) | IC50 (nM)a of:
|
||
|---|---|---|---|
| GW678248 | Nevirapine | Efavirenz | |
| None | 0.52 ± 0.4 (33) | 88 ± 49 (17) | 0.80 ± 0.4 (39) |
| L100I | 0.5 ± 0.4 (5) | 500 ± 130 (3) | 21 ± 11 (14) |
| K101E | 0.96 ± 0.07 (3) | 520 ± 130 (3) | 2.5 ± 1.1 (8) |
| K103N | 1.0 ± 0.9 (21) | 5,800 ± 3,600 (9) | 25 ± 13 (41) |
| V106A | 3.4 ± 1.7 (19) | 8,200 ± 3,700 (7) | 1.8 ± 1.1 (29) |
| V106I | 1.1 ± 0.8 (8) | 150 ± 130 (4) | 0.8 ± 0.6 (10) |
| V106M | 1.3 ± 1.9 (7) | 2,900 ± 3,600 (3) | 8.6 ± 4 (7) |
| V108I | 0.9 ± 0.6 (8) | 290 ± 170 (7) | 2.1 ± 1.3 (11) |
| E138K | 1.3 ± 0.7 (5) | 65 ± 42 (6) | 0.86 ± 0.3 (8) |
| Y181C | 0.7 ± 0.5 (21) | 10,300 ± 4,100 (10) | 1.6 ± 1 (23) |
| Y188C | 0.11 ± 0.07 (6) | 2,300 ± 1,300 (10) | 1.4 ± 0.7 (11) |
| Y188L | 21 ± 6 (9) | >10,000 (2) | 240 ± 160 (5) |
| G190A | 0.9 ± 0.3 (9) | 12,000 ± 7,200 (4) | 7.0 ± 3 (13) |
| G190E | 0.7 ± 0.5 (7) | 95 (1) | 1.2 ± 0.6 (4) |
| P225H | 0.3 ± 0.2 (5) | 1,200 (1) | 1.7 ± 0.8 (7) |
| P236L | 1.1 ± 0.4 (5) | 56 ± 13 (2) | 0.8 ± 0.9 (9) |
| K103N, L100I | 1.5 ± 0.8 (7) | 7,900 ± 1,700 (4) | 930 ± 500 (7) |
| K103N, V108I | 0.63 ± 0.2 (6) | 23,000 ± 14,000 (4) | 93 ± 75 (14) |
| K103N, Y181C | 1.4 ± 0.9 (10) | >8,000 (6) | 46 ± 19 (22) |
| K103N, G190A | 4.1 ± 3.0 (11) | >10,000 (3) | 490 ± 230 (11) |
| K103N, P225H | 0.7 ± 0.2 (6) | 9,600 ± 2,100 (3) | 150 ± 110 (14) |
| V106A, Y181C | 0.9 ± 0.5 (6) | >10,000 (5) | 3.1 ± 2.5 (8) |
| V106I, Y181C | 4.9 ± 2.2 (6) | >10,000 (3) | 3.6 ± 1.1 (8) |
| V106I, P236L | 7 ± 5 (5) | NDb | 0.7 ± 0.3 (2) |
| V106I, E138K, P236L | 86 ± 62 (2) | 79 (1) | 2.3 ± 1.2 (3) |
| V108I, Y181C | 0.9 ± 0.5 (7) | >10,000 (3) | 3.1 ± 1.5 (6) |
IC50s are means ± standard deviations. Values in parentheses = n.
ND, not determined.
Additional characterization of GW678248 demonstrated that this compound was active against a panel of nucleoside reverse transcriptase inhibitor-resistant mutants, including strains resistant to all marketed nucleosides (IC50, ≤2 nM) (Table 3), and against a panel of PI-resistant mutants, including multi-PI-resistant strains (IC50, ≤2.4 nM; strains described in Materials and Methods; data not shown), indicating a lack of cross-resistance, as expected.
TABLE 3.
Antiviral activity of GW678248 against HIV-1 containing nucleoside resistance mutations in HeLa-CD4 MAGI assaya
| Strain (RT mutations) | Nucleoside resistance phenotype | IC50 (nM)b of:
|
||
|---|---|---|---|---|
| GW678248c | Nevirapined | Efavirenze | ||
| RTMN (M41L T215Y K219Q) | AZT | 0.4 | 35 ± 26 | 0.4 ± 0.3 |
| RTMCY (D67N K70R T215Y) | AZT | 0.5 | 39 ± 44 | 0.4 ± 0.3 |
| MDR151 (A62V V75I F77L Q151M) | All | 2.0 | 160 ± 120 | 1.8 ± 1.3 |
| M184V (M184V) | 3TC | 0.7 | 64 ± 60 | 0.7 ± 0.5 |
| DRSM34 (M41L D67N K70R M184V H208Y L210W T215Y K219E) | AZT, 3TC | 0.7 | 68 ± 62 | 0.4 ± 0.5 |
| ABC4 (K65R L74V M184V Y115F) | ABC, 3TC, DDI | 0.5 | 74 ± 66 | 0.9 ± 0.5 |
| 74V/184V (L74V M184V) | DDI, DDC, 3TC, ABC | 0.7 | 76 ± 30 | 0.6 ± 0.3 |
Abbreviations: AZT, zidovudine; 3TC, lamivudine; ABC, abacavir; DDI, didanosine; DDC, zalcitabine.
IC50s are means ± standard deviations.
n = 1.
n = 4.
n = 7.
Cytotoxicity and selectivity index.
Cell growth inhibition (cytotoxicity) assessments of GW678248 were made with human PBMCs (PHA-activated or unactivated) and a variety of human cell lines of B-cell origin (B-lymphoblastoid cell line IM-9 and monocytic lymphomic B-cell line U-937), T-cell origin (lymphoblastic leukemia T-cell line Molt-4 and human T-cell leukemia virus type 1-immortalized leukemia cell line MT-4), lung fibroblast origin (MRC-5), and hepatocellular carcinoma origin (HepG2). A 50% cytotoxicity concentration for GW678248 was not achieved for any of the cell types tested due to the fact that the compound solubility limit was reached prior to the attainment of 50% inhibition of cell growth. GW678248 precipitation was observed at 5 μM; the 50% cytotoxicity concentration was therefore estimated to be >2.5 μM, which was the highest concentration tested that did not exceed solubility (data not shown). From these results, the selectivity index is calculated to be >2,500 for GW678248 against WT HIV-1 and mutant derivatives having Y181C or K103N residue substitutions in RT.
Effects of human serum albumin and α-1 acid glycoprotein on anti-HIV-1 activity.
To estimate the effects of serum binding proteins on the GW678248 IC50 value, the HeLa-CD4 MAGI assay was conducted in the presence or absence of 45 mg/ml HSA and 1 mg/ml AAG. These data, shown in Table 4, indicate a 6.8-fold ± 2.2-fold shift in the GW678248 IC50 in the presence of HSA plus AAG. This value for GW678248 compares favorably with the 4.4 ± 0.3-fold shift observed for NVP but is substantially less than that observed with EFV, which demonstrated a 16-fold ± 2-fold shift under the same assay conditions. HSA was found to affect the IC50 more than AAG when each agent was tested individually.
TABLE 4.
Fold shift in antiviral activity against HIV-1 HxB2 in the presence of human serum binding proteins
| Compound | Fold shifta
|
|||
|---|---|---|---|---|
| −HSA
|
+HSAb
|
|||
| −AAG | +AAGc | −AAG | +AAG | |
| GW678248Xd | 1 | 4.4 ± 1.2 | 7.5 ± 1.5 | 6.8 ± 2.2 |
| Efavirenze | 1 | 3.7 ± 0.9 | 12 ± 2 | 16 ± 2 |
| Nevirapinef | 1 | 0.8 ± 0.2 | 4.8 ± 1 | 4.4 ± 0.3 |
Values are the average fold shift (IC50 in the presence of human proteins/IC50 in the absence of human proteins) ± standard deviations.
45 mg/ml HSA added to standard assay conditions.
1 mg/ml human AAG added to standard assay conditions.
n ≥ 6.
n ≥ 6.
n = 3.
DISCUSSION
Central to the design of a screen for a new-generation NNRTI is determination of activity directly against the molecular target, including key drug-resistant mutant forms, in biochemical assays. The clinically observed RT mutations that confer resistance to marketed NNRTIs (7) alter the NNRTI binding site architecture (9, 10). Notable among these amino acid substitutions are K103N, Y181C (or I), Y188C (or L), V106A, V108I, G190A (or S or E), and P236L. The added selection for secondary mutations, such as L100I, K101E, and P225H, produces even higher levels of drug resistance. The biochemical studies of GW678248 were conducted with a panel of NNRTI-resistant mutants. This panel consisted of WT and RT containing the single substitutions L100I, K103N, V106A/I, V108I, E138K, Y181C, Y188C, and P236L and the double substitutions V106A Y181C and V108I Y181C. These studies indicated that GW678248 had potent activity against the molecular target and a favorable resistance profile against key NNRTI resistance-conferring mutations.
In vitro antiviral assays corroborated and extended the biochemical studies.
The isogenic virus panel included the same RT mutations as the biochemical panel and was extended to include K101E, V106M, Y188L, G190A, G190E, P225H, K103N L100I, K103N Y181C, K103N G190A, K103N P225H, V106I Y181C, V106I P236L, and V106I E138K P236L. GW678248 was a potent inhibitor of the majority of these NNRTI-resistant strains. Of the single-amino-acid substitutions, the most resistant to GW678248 was Y188L. Y188L, which arises as a consequence of a two-nucleotide change, constitutes an alternate pathway for EFV resistance. Although Y188L confers greater resistance than K103N, the K103N substitution is selected in the majority of patients receiving EFV (3, 4). The triple mutation V106I E138K P236L was included in the panel because initial resistance passage experiments selected for this mutation (9a). Indeed, this combination of mutations conferred the greatest level of resistance, 86 nM, or ∼170-fold compared with the WT.
Another NNRTI in development, TMC125, has in vitro potencies similar to those of GW678248 against WT HIV-1 and strains containing NNRTI resistance mutations (1). Although direct comparisons between IC50s determined under different assay conditions in different labs can be potentially misleading, the relative severalfold change of each compound against HIV-1 strains containing key NNRTI resistance mutations compared to the wild type in each assay system suggests that both compounds have similar though not identical resistance profiles. Based on a comparison of the published data on TMC125 (1) and that presented here, TMC125 has a sevenfold shift in IC50 against Y181C compared to a 1.3-fold shift for GW678248. Activities of both compounds are within two- or threefold of WT activity levels against K103N or L100I and within three- or fourfold of WT activity levels against the K103N Y181C double mutant. TMC125 has a 19-fold shift in IC50 against the L100I K103N mutant compared to a 3-fold shift for GW678248. In contrast, GW67828 has a 40-fold shift against the Y188L mutant compared to a 5-fold shift for TMC125. Although both compounds have high potency against WT HIV and certain strains resistant to the current generation of NNRTIs, it is likely that these drugs will not have identical resistance profiles. Cross-resistance between them remains to be determined.
The human protein binding experiments indicated that the in vitro IC50 of GW678248 was only moderately affected when additional proteins were added to standard assay conditions. Methods for determining the in vitro effects of protein binding are not standardized and are of undetermined clinical relevance (5). However, the shift in IC50 of GW678248 that was due to human serum proteins was less than that observed for EFV and thus not considered to be a risk for clinical development. GW678248 did not exhibit cytotoxicity in several cell lines tested or in PBMCs up to the limit of solubility. The selectivity index of WT antiviral activity over cytotoxicity was >2,500.
The biochemical and in vitro assay results described herein demonstrate that GW678248 is a potent and selective inhibitor of WT HIV-1 and the major NNRTI-resistant strains. This compound has potency superior to that of the benchmark compounds NVP and EFV against the vast majority of HIV-1 strains tested. Likewise, the high selectivity index based on lack of cytotoxicity and high antiviral potency is favorable for clinical development. Accordingly, these results are highly relevant for, and support, GW695634, the prodrug of GW678248, which is currently in phase II clinical studies and intended for use in combination with other antiretroviral drugs. This drug is expected to provide benefit to HIV-1-infected patients, including those who have virus resistant to the current NNRTIs.
Acknowledgments
We thank Jerry Jeffrey for constructing the V106M mutant and corresponding antiviral data and Laurence Robinson for providing the viruses used in the protease inhibitor-resistant virus panel. We thank Dave Stammers and Dave Stuart, Oxford University, for their contributions to the GSK NNRTI program. We also thank the GW695634 Project Team for input and guidance and both Paul Wilmott (Caudex) and Barbara Rutledge for editorial assistance.
REFERENCES
- 1.Andries, K., H. Azijn, T. Thielemans, D. Ludovici, M. Kukla, J. Heeres, P. Janssen, B. De Corte, J. Vingerhoets, R. Pauwles, and M.-P. de Bethune. 2004. TMC125, a novel next-generation nonnucleoside reverse transcriptase inhibitor active against nonnucleoside reverse transcriptase inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48:4680-4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Averett, D. R. 1989. Anti-HIV compound assessment by two novel high capacity assays. J. Virol. 23:263-276. [DOI] [PubMed] [Google Scholar]
- 3.Bacheler, L., S. Jeffrey, G. Hanna, R. D'Aquila, L. Wallace, K. Logue, B. Cordova, K. Hertogs, B. Larder, R. Buckery, D. Baker, K. Gallagher, H. Scarnati, R. Tritch, and C. Rizzo. 2001. Genotypic correlates of phenotypic resistance to efavirenz in virus isolates from patients failing nonnucleoside reverse transcriptase inhibitor therapy. J. Virol. 75:4999-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bacheler, L. T., E. D. Anton, P. Kudish, D. Baker, J. Bunville, K. Krakowski, L. Bolling, M. Aujay, X. V. Wang, D. Ellis, M. F. Becker, A. L. Lasut, H. J. George, D. R. Spalding, G. Hollis, and K. Abremski. 2000. Human immunodeficiency virus type 1 mutation selected inpatients failing efavirenz combination therapy. Antimicrob. Agents Chemother. 44:2475-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boffito, M., D. J. Black, T. F. Blaschke, M. Rowland, R. J. Bertz, J. G. Gerber, and V. Miller. 2003. Protein binding in antiretroviral therapies. AIDS Res. Hum. Retrovir. 19:825-835. [DOI] [PubMed] [Google Scholar]
- 6.Chan, J. H., G. A. Freeman, J. H. Tidwell, K. R. Romines, L. T. Schaller, J. R. Cowan, S. S. Gonzales, G. S. Lowell, C. W. Andrews III, D. J. Reynolds, M. StClair, R. J. Hazen, R. G. Ferris, K. L. Creech, G. B. Roberts, S. A. Short, K. Weaver, G. W. Koszalka, and L. R. Boone. 2004. Novel benzophenones as non-nucleoside reverse transcriptase inhibitors of HIV-1. J. Med. Chem. 47:1175-1182. [DOI] [PubMed] [Google Scholar]
- 7.D'Aquila, R. T., J. M. Schapiro, F. Brun-Vezinet, B. Clotet, B. Conway, L. M. Demeter, R. M. Grant, V. A. Johnson, D. R. Kuritzkes, C. Loveday, R. W. Shafer, and D. D. Richman. 2003. Drug resistance mutations in HIV-1. Top. HIV Med. 11:92-96. [PubMed] [Google Scholar]
- 8.De Clercq, E. 2004. Non-nucleoside reverse transcriptase inhibitors (NNRTIs): past, present and future. Chem. Biodivers. 1:44-64. [DOI] [PubMed] [Google Scholar]
- 9.Esnouf, R., J. Ren, C. Ross, Y. Jones, D. Stammers, and D. Stuart. 1995. Mechanism of inhibition of reverse transcriptase by non-nucleoside inhibitors. Nat. Struct. Biol. 2:303-308. [DOI] [PubMed] [Google Scholar]
- 9a.Hazen, R. J., R. J. Harvey, M. H. St. Clair, R. G. Ferris, G. A. Freeman, J. H. Tidwell, L. T. Schaller, J. R. Cowan, S. A. Short, K. R. Romines, J. H. Chan, and L. R. Boone. Anti-human immunodeficiency virus type I activity of the nonnucleoside reverse transcriptase inhibitor GW678248 in combination with other antiretrovirals, against clinical isolate viruses, and in in vitro selection for resistance. Antimicrob. Agents Chemother., in press. [DOI] [PMC free article] [PubMed]
- 10.Huang, H., R. Chopra, G. Verdine, and S. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 11.Kellam, P., and B. A. Larder. 1994. Recombinant virus assay: a rapid, phenotypic assay for assessment of drug susceptibility of human immunodeficiency virus type 1 isolates. Antimicrob. Agents Chemother. 38:23-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pauwels, R., E. De Clercq, J. Desmyter, J. Balzarini, P. Goubau, P. Herdewijn, H. Vanderhaeghe, and M. Vandeputte. 1987. Sensitive and rapid assay on MT-4 cells for detection of antiviral compounds against the AIDS virus. J. Virol. Methods 16:171-185. [DOI] [PubMed] [Google Scholar]
- 14.Robinson, L. H., R. E. Myers, B. W. Snowden, M. Tisdale, and E. D. Blair. 2000. HIV type 1 protease cleavage site mutations and viral fitness: implications for drug susceptibility phenotyping assays. AIDS Res. Hum. Retrovir. 16:1149-1156. [DOI] [PubMed] [Google Scholar]