Abstract
Macrolide resistance in Streptococcus pneumoniae due to efflux has emerged as an important worldwide clinical problem over the past decade. Efflux is mediated by the genes of the genetic element mega (macrolide efflux genetic assembly) and related elements, such as Tn1207.1. These elements contain two adjacent genes, mef (mefE or mefA) and the closely related mel gene (msrA homolog), encoding a proton motive force pump and a putative ATP-binding cassette transporter homolog, and are transcribed as an operon (M. Del Grosso et al., J. Clin. Microbiol. 40:774-778, 2004; K. Gay and D. S. Stephens, J. Infect. Dis. 184:56-65, 2001; and M. Santagati et al., Antimicrob. Agents Chemother. 44:2585-2587, 2000). Previous studies have shown that Mef is required for macrolide resistance in S. pneumoniae; however, the contribution of Mel has not been fully determined. Independent deletions were constructed in mefE and mel in the serotype 14 macrolide-resistant strains GA16638 (erythromycin [Em] MIC, 8 to 16 μg/ml) and GA17719 (Em MIC, 2 to 4 μg/ml), which contain allelic variations in the mega element. The MICs to erythromycin were significantly reduced for the independent deletion mutants of both mefE and mel compared to those of the parent strains and further reduced threefold to fourfold to Em MICs of <0.15 μg/ml with mefE mel double mutants. Using quantitative reverse transcription-PCR, the expression of mefE in the mel deletion mutants was increased more than 10-fold. However, in the mefE deletion mutants, the expression of mel did not differ significantly from the parent strains. The expression of both mefE and mel was inducible by erythromycin. These data indicate a requirement for both Mef and Mel in the novel efflux-mediated macrolide resistance system in S. pneumoniae and other gram-positive bacteria and that the system is inducible by macrolides.
Streptococcus pneumoniae is a leading cause of respiratory infections, which include otitis media, sinusitis, and pneumonia. Antibiotic treatment of these infections has become increasingly problematic due to an emergence of resistance to both penicillin and non-β-lactam antibiotics. During the last decade, a rapid increase in the resistance of S. pneumoniae to macrolides has been observed in the United States (3, 12, 13, 19, 43).
The major mechanisms of macrolide resistance in S. pneumoniae are target modification and drug efflux. Genetic determinants conferring macrolide resistance by target modification include erm and mutations in the 23S rRNA and ribosomal proteins. The erm(B) gene product methylates the peptidyl transferase center of newly synthesized 23S rRNA, thereby conferring cross-resistance to lincosamides and streptogramin B (MLS phenotype) (30, 42). Mutations in the 23S rRNA and ribosomal proteins L4 and L22 have also been reported and can confer macrolide-lincosamide (ML) and macrolide-streptogramin B (MS) resistance phenotypes when different mutations are combined (5, 21, 26, 28).
Throughout the world, rapidly increasing rates of macrolide resistance have been due primarily to the second major mechanism of macrolide resistance in S. pneumoniae, efflux linked to the gene product of mef (14, 20, 37, 39). Mef belongs to the major facilitator superfamily of efflux pumps and carries a proton motive force pump that is specific for 14- and 15-membered macrolides (M phenotype) (7, 40). Two variants, mefE and mefA, with >90% protein sequence homology, are found in isolates of S. pneumoniae (9, 10, 14, 15). Macrolide resistance due to the presence of MefE accounts for the majority of macrolide-resistant pneumococcal strains isolated in the United States (12, 13).
The genetic elements harboring both mefA and mefE in S. pneumoniae are localized on conjugative transposon-related elements (15, 36). The mefE gene is present on the 5.4- or 5.5-kb mega (macrolide efflux genetic assembly) element that confers macrolide resistance to susceptible S. pneumoniae (15), and mefA is found on the closely related elements Tn1207.1 (36) and Tn1207.3 (35). Other genetic elements have subsequently been identified to contain mega-like regions, including Tn2009, the chimeric element in S. pyogenes composed of a transposon inserted into a prophage (1, 11), and elements found in viridans streptococci (6). Both mefE and mefA are part of an operon in mega that includes a downstream gene, mel, a homolog of msrA (15, 36). In staphylococci, msrA encodes a 488-amino-acid ATP-binding cassette (ABC) transporter protein which results in an energy-dependent efflux of erythromycin (34). ABC transporter proteins typically contain two ATP-binding domains located cytoplasmically that interact with two hydrophobic domains (22). Both MsrA and Mel contain ATP-binding domains characteristic of ABC transporters; however, they lack hydrophobic segments carrying the transmembrane domains. Although MsrA is predicted to interact with chromosomally encoded transmembrane complexes, MsrA was sufficient in conferring resistance to macrolides and streptogramin B (MS phenotype) (32). In S. pneumoniae, mefE and mel are cotranscribed as an operon and are predicted to be a dual efflux pump in S. pneumoniae (15). The two allelic forms of mega, 5.4 or 5.5 kb, differ in the presence or absence of a 99-bp insertion between mefE and mel. Here we describe the requirement of both MefE, the proton motive force pump homolog, and Mel, the homolog of an ATP-binding cassette transporter, in macrolide efflux in S. pneumoniae.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1. Serotype 14 pneumococcal isolates GA16638 and GA17719 and other erythromycin-resistant S. pneumoniae isolates were obtained as part of an active, population-based surveillance program of invasive pneumococcal disease in metropolitan Atlanta. Surveillance and isolate collection methods have been described previously (14, 15, 18). The initial antimicrobial susceptibility of isolates was assessed according to guidelines established by the Clinical and Laboratory Standards Institute (formerly NCCLS) (8). Isolates not susceptible to erythromycin (MIC, ≥0.5 μg/ml) were further classified by antibiogram and molecular studies. GA16638 and GA17719 are M phenotype macrolide-resistant isolates (erythromycin [Em] MICs, 8 to 16 μg/ml and 2 to 4 μg/ml, respectively) originally obtained from blood. These strains contained single copies of mefE and mel and were negative for other known macrolide resistance mechanisms (38). MICs to clindamycin for both strains were ≤0.12 μg/ml. MICs to other agents for GA16638 and GA17719, respectively, were as follows: penicillin, 4 μg/ml and 2 to 4 μg/ml; chloramphenicol, 4 μg/ml and 4 μg/ml; vancomycin, 0.25 μg/ml and 0.25 μg/ml; cefotaxime, 1 to 2 μg/ml and 1 μg/ml; quinupristin-dalfopristin, <1 μg/ml and <1 μg/ml; ciprofloxacin, <2 μg/ml and <2 μg/ml; and telithromycin, 0.5 μg/ml and 0.06 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Derivationa | Reference or source |
|---|---|---|
| S. pneumoniae strains | ||
| GA16638 | Parent type 14, type 2 mega, Emr | This study |
| GA17719 | Parent type 14, type 1 mega, Emr | This study |
| KA3000/01 | pKA309 × GA16638 ΔmefE Ems | This study |
| KA3003/04 | pKA312 × GA16638 Δmel Ems | This study |
| KA3005/06 | pKA321 × GA16638 ΔmefE Δmel Ems | This study |
| ATCC 49619 | 26 | |
| E. coli DH5α plasmids | ||
| pCR2.1-TOPO | Vector for cloning of PCR products, Apr Knr | Invitrogen |
| pSF151 | 41 | |
| pWA101 | TOP10 pCR2.1::0.34-kb KG7/KG11 PCR product from GA16638 | This study |
| pWA103 | TOP10 pCR2.1::0.56-kb KG8/KG10 PCR product from GA16638 | This study |
| pKA309 | DH5α pSF151::double-fragment ligation of KpnI/XbaI insert of pWA101 and KpnI/PstI insert of pWA103 ligated to XbaI/PstI of vector | This study |
| pKA310 | TOP10 pCR2.1::0.66-kb KG20/KG41R PCR fragment from GA16638 | This study |
| pKA312 | DH5α pSF151::double-fragment ligation of SacI/EcoRV fragment from pWA103 ligated to EcoRV/SpeI fragment of pKA310 | This study |
| pKA321 | DH5α pSF151::0.34-kb EcoRI/SpeI fragment of pWA101 and 0.6-kb XbaI/BamHI fragment of pKA310 ligated to EcoRI/BamHI of vector, Knr | This study |
Emr, erythromycin resistant; Ems, erythromycin sensitive; Apr, ampicillin resistant; Knr, kanamycin resistant. × indicates transformation (or crossing) of the plasmid into the strain. Primers are listed in Table 2.
S. pneumoniae strains were grown in Todd-Hewitt broth supplemented with 0.5% yeast extract (Difco) or on blood agar base no. 2 (Difco) or Trypticase soy agar, each supplemented with 5% sheep erythrocytes (BBL, Fisher Scientific), at 37°C in 5% CO2. Escherichia coli strains were grown in L broth or on L agar at 37°C. For E. coli, ampicillin and kanamycin were used at concentrations of 100 μg/ml and 50 μg/ml, respectively. For S. pneumoniae, Em was used at concentrations between 0.5 and 4 μg/ml.
Construction of mefE and mel deletion mutants.
Efflux due to the presence of mefE was determined in previous studies by PCR (14). For the deletion of mefE, Taq polymerase (Sigma) was used to obtain pneumococcal PCR fragments of 0.34 kb with primers KG7 and KG11 and 0.56 kb with primers KG8 and KG10 from GA16638. The resulting PCR products were cloned into pCR2.1-TOPO (Invitrogen) to generate pWA101 and pWA103, respectively. Ligation of the KpnI/XbaI insert of pWA101 and the KpnI/PstI insert of pWA103 into the vector pSF151 yielded pKA309. For the deletion of mel, a 0.66-kb PCR fragment was generated using primers KG20 and KG41R and cloned into the pCR2.1-TOPO vector to yield pKA310. The SacI/EcoRV fragment from pWA103 was ligated to the EcoRV/SpeI fragment of pKA310 and subcloned into pSF151 to generate pKA312. To generate a mefE mel double knockout mutant, the 0.34-kb EcoRI/SpeI fragment of pWA101 and 0.6-kb XbaI/BamHI fragment of pKA310 were ligated and subcloned into pSF151. Plasmid DNA was isolated using the method described by Birnboim and Doly (2) or QIAGEN columns (QIAGEN, Inc.). Primers used in this study are listed in Table 2.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′→3′) |
|---|---|
| mefE-1 | 5′-GCT AGT GGA TCG TCA TGA TAG G-3′ |
| mefE-2 | 5′-TTC CCG AAA CGG CTA AAC TGG T-3′ |
| mefE-3F | 5′-ATA TGG GCA GGG CAA GCA G-3′ |
| mefE-4R | 5′-CAT TTG CAG GAT GGC ACT AGT G-3′ |
| mel-2F | 5′-GAA CGT AAG AGC CAA GCT GCA-3′ |
| mel-3R | 5′-GGC ACG TTC CGC AAT AAA TT-3′ |
| KG7 | See reference 15 |
| KG8 | See reference 15 |
| KG10 | 5′-ACA CCT AGC TTG CCT ACA AGT G-3′ |
| KG11 | 5′-GCA GAA TCT ATA CCC GAT GAT AGG-3′ |
| KG20 | 5′-CTG TTC TGG TTG GCG GAC TC-3′ |
| KG41R | 5′-CAT GTC TGA CTT ATC ACT AGA G-3′ |
| rpsE-1F | 5′-ACG TCG TCT TCG TTT CGC A-3′ |
| rpsE-2R | 5′-CGA CCA TTG TCA CCA AC-3′ |
| fabK-1F | 5′-TGA TGT GGA TGG TGG CTC TG-3′ |
| fabK-2R | 5′-GAA ACA AGC CCT GCG ATT TG-3′ |
For construction of the deletions in S. pneumoniae, pKA309 and pKA312 were used to delete mefE and mel, respectively. Plasmids were transformed into GA16638 by previously described methods (16, 17). Transformation mixtures were diluted and plated onto blood agar plates. Colonies were then replica patched onto blood agar plates and blood agar plates containing 4 μg/ml of Em and screened for susceptibility to Em. Colonies that were susceptible to Em were further confirmed by PCR. Deletions of 0.92 and 1.2 kb were constructed in mefE and mel, respectively (Fig. 1). Amplification with the primer-specific pairs KG11/KG10 for mefE and KG8/KG41R for mel resulted in a 1.8-kb fragment from the parent strain and a 0.87-kb fragment in the mefE mutants and a 2.3-kb fragment from the parent strain and 0.8-kb fragment in the mel deletion mutants. Two independent mutants were generated for mefE, KA3000 and KA3001, and independent mutants KA3003 and KA3004 were generated for mel. mefE mel double mutants were also constructed using pKA321. Independent deletion mutants were also constructed in strain GA17719 using the plasmids described above. Deletions were further confirmed by Southern hybridization using internal probes of the deleted regions for both mefE and mel. Probes flanking the deleted regions were also used to show retention of the restriction enzyme pattern in the mega locus. For Southern analyses, chemiluminescent detection from the Genius system (Roche Molecular Biochemicals) was used. Chromosomal DNA was isolated using QIAGEN genomic tips.
FIG. 1.
Schematic of mega element for strain GA16638 illustrating the locations of single mutations for mefE, mel, and a mefE mel double mutant. ORF, open reading frame.
MIC studies.
To determine Em MICs, subcultures of the parent and mutant strains were grown overnight on Trypticase soy agar blood agar plates. Several colonies were removed using a sterile swab, resuspended in Mueller-Hinton broth, and vortexed, and turbidity was adjusted to a 0.5 McFarland standard (optical density at 600 nm) using a spectrophotometer. Within 15 min after preparation of the suspension, a sterile swab was dipped into the suspension and rotated against the sides of the tube to remove excess fluid. A Mueller-Hinton agar plate (with 5% sheep blood) was then inoculated with the wet swab in three directions to completely cover the plate, which was allowed to dry for 10 to 15 min. An Em-containing Etest strip (Remel, Inc.) with an MIC range of 0.016 to 256 μg/ml was placed carefully on each plate. Plates were incubated for 20 h at 37°C in a 5% CO2 incubator. Quadruplicate cultures were used for each strain. Broth dilution MICs of the parent strains and mutants were also reconfirmed at the CDC using Clinical and Laboratory Standards Institute guidelines (8).
RNA extraction and real-time quantitative reverse transcription-PCR (RT-PCR).
RNA was isolated using QIAGEN RNA minicolumns according to the manufacturer's protocol. Briefly, cultures were grown to mid-exponential phase at 37°C in Todd-Hewitt broth supplemented with 0.5% yeast extract, with or without Em, at concentrations of 0.024 and 1.2 μg/ml. The RNA samples were further treated with DNase for 1 h at 37°C to remove contaminating chromosomal DNA. The digestion mixture was cleaned by following the QIAGEN mini-column protocol. To ensure that contaminating DNA was not present, the final RNA preparation was tested by standard PCR amplification using Taq polymerase with primers KG8 and KG10.
Expression of mefE and mel was determined using real-time quantitative RT-PCR. Reverse transcription was done according to the Gene Amp kit (Applied Biosystems) using 1 μg total RNA for the parent and mutant strains. Reactions were also performed without reverse transcriptase for each strain for use as negative controls. To quantify mRNA, cDNA templates were diluted twofold in 1× PCR buffer and used in subsequent experiments. Quantitative PCR was performed using the 2× SYBR green supermix (Bio-Rad) according to the manufacturer's protocol with 0.2 μM each of forward and reverse primers in a 25-μl total reaction mixture volume. Reactions were performed in 96-well microtiter plates using the iCycler (Bio-Rad). Amplification of the target gene was done for 40 cycles (94°C for 30 s, 55°C for 30 s, and 72°C for 30 s). Calculation of the results was performed by a modified method of Robertson et al. (31). The amount of target was normalized to a control target gene, fabK, which varied less than twofold for each strain. A calibration curve was generated by twofold serial dilutions of 1 μg total RNA for GA16638 containing a fragment encoding ribosomal protein RpsE. For each strain, three replicates were performed on duplicate and independent RNA samples. The change (n-fold) in expression was relative to the expression of the parent GA16638 strain grown without Em.
Accumulation and efflux assays.
For efflux determination, cultures of the parent and mutant strains were inoculated with 0.025 μg/ml Em to induce expression of the pump. When cultures reached mid-exponential phase (∼3 × 108 CFU/ml), 0.025 μg/ml of [14C]erythromycin (Perkin Elmer Life Sciences) was added to each culture. Samples of 2.5 ml were collected from each culture at 0, 10, 20, and 30 min and filtered using a Millipore 1225 sampling vacuum manifold (Fisher Scientific) onto glass microfiber membrane filters (Fisher Scientific). Filters were washed two times with 1% NaCl-1 mg/ml Em and air dried, and cell-associated [14C]Em was measured using liquid scintillation. A susceptible strain, GA16328, was also used in the assay.
Statistical analyses.
Statistical analyses were performed using an unpaired Student t test. Significant differences (P < 0.05) were determined between parent and mutant strains.
RESULTS
Both MefE and Mel are required for erythromycin resistance in mega-containing strains.
To ascertain the independent importance of mefE and mel in pneumococcal efflux macrolide resistance, deletions were constructed in both mefE and mel by allelic replacement in the type 14 (mega type 2) parent strain GA16638 (Etest erythromycin MIC, 15 [±1.0] μg/ml) (Table 3). For MefE, an internal deletion of 305 amino acids was constructed, which resulted in the loss of 75% of the predicted protein. For Mel, a 395-amino-acid truncation, resulting in an ∼80% loss of the predicted protein, was constructed (Fig. 1). Deletion mutations were confirmed by PCR and Southern hybridizations (data not shown). The Etest erythromycin MICs were reduced for both independent mefE (13-fold) and mel (22-fold) deletion mutants compared to that of the parent strain (Table 3). Reductions in MICs were also obtained when the mefE (twofold) and the mel (ninefold) mutations were constructed in the clinical isolate GA17719 (serotype 14) (Table 3) (MIC, 4.13 [±0.3] μg/ml), which has a type 1 mega insert (15). In the mefE mel double mutants of both strains, erythromycin MICs were further reduced threefold to fourfold (Table 3) to MICs of <0.15 μg/ml. Similar changes were seen when MICs were determined by microdilution methods (Table 3). Also, the telithromycin MICs of both strains with deletions of mefE or mel or both were reduced from 0.5 μg/ml (GA16638) and 0.06 μg/ml (GA17719) to ≤0.03 μg/ml in mel, mef, and dual mutants. The antibiograms of the mutants otherwise remained unchanged compared to those of the parent strains.
TABLE 3.
MICs of erythromycin for mega mutants
| Strain | Em MIC (μg/ml) (±SEM)a
|
|
|---|---|---|
| Etest | Microdilution | |
| GA16638 (type 2 mega, parent) | 15 (±1.00) | 8 |
| GA16638 ΔmefE | 1.13 (±0.16)b | 0.5 |
| GA16638 Δmel | 0.68 (±0.04)b | 0.25 |
| GA16638 ΔmefE Δmel | 0.15 (±0.01)b | 0.06 |
| GA17719 (type 1 mega, parent) | 4.13 (±0.13) | 2 |
| GA17719 ΔmefE | 2.13 (±0.13)b | 0.5 |
| GA17719 Δmel | 0.46 (±0.03)b | 0.25 |
| GA17719 ΔmefE Δmel | 0.13 (±0.00)b | ≤0.03 |
| ATCC 49619 | 0.09 (±0.01)b | ≤0.03 |
Standard errors of the means are based on quadruplicate cultures.
P < 0.0001 compared to results for the parent strains.
Expression of mefE and mel.
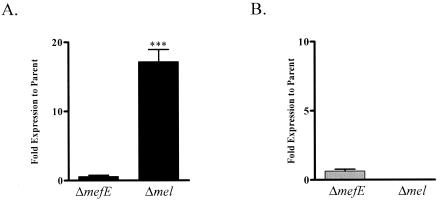
The mutations in mefE or mel may influence expression of the gene not mutated. The two allelic forms of mega, 5.4 and 5.5 kb, differ in their intergenic regions separating mefE and mel, which may also influence the expression of these genes. Using real-time quantitative RT-PCR, expression of mefE and mel was determined in the parent and mutant strains. In the wild-type strains, the genes were expressed as an operon and levels of expression of mefE and mel in the two allelic forms were similar. Mutations did not decrease the expression of the adjacent gene, and thus the construction of the mutations did not have a polar effect on expression. Expression of mefE in the mel mutants was increased more than 10-fold (Fig. 2A). The expression of mel in the mefE deletion mutants did not differ significantly from that in the parent strain (Fig. 2B). Expression profiles in GA16638 and GA17719 were similar with mefE and mel deletions.
FIG. 2.
Expression of mefE (A) and mel (B). RNA was isolated from mid-exponential cultures using the QIAGEN RNeasy minipreps. Three replicates were performed for each strain on duplicate and independent RNA samples. The amount of target is normalized to a control target gene, fabK, relative to an internal ribosomal calibrator. Data are expressed as percentages of the amount in the parent GA16638. Statistical analyses were done using the unpaired Student t test (***, P < 0.005).
Expression of mefE and mel is erythromycin inducible.
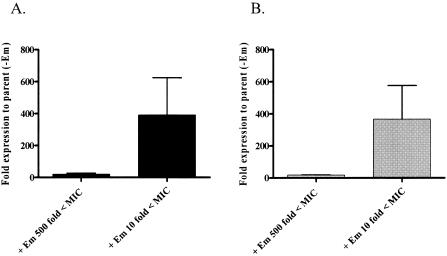
Parent strain GA16638 grown in the presence of subinhibitory concentrations of Em induced the expression of both mefE and mel (Fig. 3). Levels of induction of gene expression by Em varied when different concentrations of Em were used. A concentration of 0.024 μg/ml of Em (500-fold less than the MIC) resulted in a 20-fold increase in the expression of mefE and a 15-fold increase in the expression of mel. However, when GA16638 was grown with a concentration of 1.2 μg/ml Em (10-fold less than the MIC), expression of both mefE and mel was increased more than 300-fold. These results suggest a regulatory mechanism of mefE and mel in S. pneumoniae that is inducible by Em. No significant effect on mef or mel expression was observed when cultures were grown with the nonmacrolide antibiotic kanamycin (data not shown).
FIG. 3.
Inducible expression of mefE (A) and mel (B) in GA16638. RNA was isolated from mid-exponential cultures grown with Em that was either 10-fold (1.2 μg/ml) or 500-fold (0.024 μg/ml) less than the MIC. Expression is relative to the expression of mefE and mel in the parent strain grown without Em.
Accumulation and efflux of [14C]erythromycin.
Cell-associated [14C]erythromycin was increased in the mefE and mel mutant strains compared to the level in the parent strain (Fig. 4). The mefE and mel mutants and the double mutants consistently accumulated more erythromycin than the erythromycin-resistant parent strains. Differences between the mutants in accumulation were not demonstrated. A wild-type-susceptible strain showed increased accumulation of erythromycin (data not shown). The accumulation in the mutants indicated decreased efflux of [14C]erythromycin at all time points.
FIG. 4.
Cell-associated [14C]Em. Mid-exponential-phase cultures were incubated with 0.025 μg/ml of [14C]erythromycin. A culture volume of 2.5 ml was collected at 0, 10, 20, 30 min and filtered. Cell-associated [14C]Em was measured on air-dried filters using liquid scintillation. The data shown are from one experiment but were representative of at least three experiments.
DISCUSSION
Macrolide resistance mediated by efflux emerged as a major global problem in the 1990s (14, 20, 39, 44) in S. pneumoniae and is now one of the major mechanisms of macrolide resistance worldwide. Efflux-mediated macrolide resistance is mediated by the macrolide efflux genetic element, mega (15), and larger genetic elements, such as Tn1207.1, that contain mega or closely related homologs (1, 11, 35, 36). Two adjacent genes, mef and mel (msr), have been identified in these mega-containing elements. To determine the molecular basis of macrolide efflux in S. pneumoniae, independent deletion mutations in mefE and mel were constructed in S. pneumoniae. Mutations in either mefE or mel in GA16638, a serotype 14 type 2 mega insertion strain, resulted in significant (P < 0.0001) decreases in resistance to erythromycin. Levels of resistance to erythromycin were reduced up to 22-fold by independent mefE and mel deletion mutants. The mef and mel double mutant further decreased erythromycin resistance an additional threefold to fourfold. Similar results were obtained with the serotype 14 strain containing an allelic variant of mega that differs by 99 bp in the intergenic region between mefE and mel (15).
Interestingly, the expression of mefE in the Δmel mutants was increased more than 10-fold, but the increased expression of mefE in the Δmel mutants did not restore resistance to erythromycin. The increase observed in transcription due to the mutation in mel located downstream and in the same transcriptional unit suggests a regulatory role of Mel on mef and mel expression. Alternatively, mRNA stability of mefE is increased in the mel mutant. Either would be predicted to increase levels of MefE in a mel deletion background, but the predicted increase in MefE does not influence levels of resistance to erythromycin.
Previous studies have shown the requirement of mefE in S. pyogenes and pneumococcal resistance to erythromycin (7, 40). However, the genetic elements harboring mefE- or mefA-resistant determinants in S. pneumoniae and more recently, S. pyogenes, all reveal a similar genetic organization, with mel located downstream of mefE (1, 10, 11, 15, 35, 36). Because sequence analyses of these elements have become available only recently, it is predicted that mel was present in the original mefA isolates of S. pyogenes and a mefE isolate of S. pneumoniae (7). In view of the structure of the genetic elements, the erythromycin-susceptible isolates that were transformed to erythromycin-resistant isolates with genomic DNA of clinical isolates harboring mefE would have been likely to also contain mel. Thus, mel along with mefE is predicted to be present in mefE-containing, gram-positive clinical isolates that are erythromycin resistant. This hypothesis is supported by sequence data of the efflux erythromycin resistance elements (1, 11, 15).
The requirement of both mefE and mel in resistance to erythromycin supports a dual efflux pump model; however, the exact mechanism by which the two gene products function in mediating efflux remains unclear. Because the levels of resistance to erythromycin in ΔmefE and Δmel mutants are similar, and the expression of each gene in the mutant strains is either unaffected or increased, both MefE and Mel appear to be necessary for erythromycin resistance and are predicted to interact to drive the efflux of macrolides. The lower MICs in the Δmel mutant and ΔmefE Δmel double mutant may suggest that mel has some residual pump activity independent of mef, but this is not likely of clinical importance. The data also suggest that macrolide resistance (2, 4, or 16 μg or higher) requires mef and mel but is not sufficient to explain the range of MICs seen for MefE/Mel-containing isolates. These differences may depend on factors (e.g., expression of the operon, chromosomal location, posttranslational modification, or other phenotypes in the strain) other than the presence of the genes.
In staphylococci, the mel homolog msrA encodes an ABC transporter protein which results in an energy-dependent efflux of erythromycin (34). Previous studies have suggested that msrA interacts with another protein since it lacks the membrane-spanning domains characteristic of ABC transporter pumps; however, this putative protein has not been identified in Staphylococcus aureus (29, 32, 33). While both MsrA and Mel lack hydrophobic membrane-spanning domains of classical ABC transporters and have considerable homology at the predicted amino acid level, the question of whether they are functional homologs remains unclear. Mel confers an M phenotype in S. pneumoniae, while MsrA confers an MS phenotype in staphylococci (29), suggesting differences in the mechanisms of these proteins. More recently, mel [designated msr(D)] alone was found to be capable of conferring macrolide resistance in a susceptible pneumococcal strain by transformation (9) but did not fully restore the MIC resistance of the donor strain. The Mel transformants also had slightly increased resistance to ketolides. The strain used in that study, CP1250, is a derivative of the highly competent unencapsulated Rx that was chemically mutagenized using 1-methyl-3-nitro-1-nitrosoguanidine (25, 27). Our data also indicate a role for MefE/Mel in ketolide export. Efflux of telithromycin was recently demonstrated for S. pyogenes (4). In our studies, both Mef and Mel are required for maximal mef-mediated efflux of erythromycin. In support of this model, deletions of mefE and/or mel resulted in increased accumulation of radiolabeled [14C]erythromycin, suggesting a decrease in efflux. In additional studies of the parent strains, the accumulation of erythromycin was increased when inhibitors of both proton motive force pumps and ABC transporters, such as carbonyl cyanide m-chlorophenylhydrazone, sodium arsenate, and sodium orthovanadate, were added (K. D. Ambrose et al., unpublished). Further, the increased expression of mefE in the mel mutants did not restore erythromycin resistance. Thus, Mel is required for macrolide resistance in S. pneumoniae and functions with MefE as part of the efflux pump. In S. pneumoniae, Mef could be the membrane-spanning protein necessary for ABC transporters like Mel that lack hydrophobic membrane-spanning domains. This would represent a novel model of efflux in bacteria.
The Mef/Mel efflux pump is inducible by erythromycin. Complicating the emergence of pneumococcal macrolide resistance, the MICs of erythromycin for mefE mel-containing strains having drastically increased in invasive S. pneumoniae, with 88% of strains now having MICs of ≥8 μg/ml and 63.5% having MICs of ≥16 μg/ml (38). While several factors (e.g., encapsulated [serotype] background) may have contributed to this trend, levels of Mef and Mel expressed in isolates by erythromycin induction may contribute to this phenomenon or may have led to higher levels of constitutive expression in some isolates. Inducible efflux pumps have been described to occur in pathogenic bacteria, such as the MexXY efflux pump in Pseudomonas aeruginosa, which is inducible by erythromycin, tetracycline, or gentamicin (23). MsrA is inducible in staphylococci (24), and Daly et al. (9) recently showed that Mel (Msr) is inducible by erythromycin.
In conclusion, efflux mechanisms of macrolide resistance associated with the mega element have emerged as a major resistance mechanism in S. pneumoniae and other gram-positive pathogens. Macrolide-resistant S. pneumoniae harboring the 5.5- or 5.4-kb mega genetic element requires the presence of both of the mefE and mel gene products to confer high-level macrolide resistance.
Acknowledgments
We thank Lane Pucko and surveillance personnel of Georgia Emerging Infections Program, Active Bacterial Core Surveillance (ABCs), for technical and helpful assistance and William M. Shafer for helpful comments. We also thank Bernard Beall and Delois Jackson at the CDC and Larry Martin and Susu Zughaier for help with the antimicrobial susceptibility testing.
This work was supported by a Fellowships in Research Science and Teaching (FIRST) Award (to K.D.A.), Emory University, and a VA Merit Award (to D.S.S.).
REFERENCES
- 1.Banks, D. J., S. F. Porcella, K. D. Barbian, J. M. Martin, and J. M. Musser. 2003. Structure and distribution of an unusual chimeric genetic element encoding macrolide resistance in phylogenetically diverse clones of group A streptococcus. J. Infect. Dis. 188:1898-1908. [DOI] [PubMed] [Google Scholar]
- 2.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, S. D., D. J. Farrell, and I. Morrissey. 2004. Prevalence and molecular analysis of macrolide and fluoroquinolone resistance among isolates of Streptococcus pneumoniae collected during the 2000-2001 PROTEKT US study. J. Clin. Microbiol. 42:4980-4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canton, R., A. Mazzariol, M. I. Morosini, F. Baquero, and G. Cornaglia. 2005. Telithromycin activity is reduced by efflux in Streptococcus pyogenes. J. Antimicrob. Chemother. 55:489-495. [DOI] [PubMed] [Google Scholar]
- 5.Canu, A., B. Malbruny, M. Coquemont, T. A. Davies, P. C. Appelbaum, and R. Leclercq. 2002. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerdá Zolezzi, P., L. M. Laplana, C. R. Calvo, P. G. Cepero, M. C. Erazo, and R. Gómez-Lus. 2004. Molecular basis of resistance to macrolides and other antibiotics in commensal viridans group streptococci and Gemella spp. and transfer of resistance genes to Streptococcus pneumoniae. Antimicrob. Agents Chemother. 48:3462-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clancy, J., J. Petitpas, F. Dib-Hajj, W. Yuan, M. Cronan, A. V. Kamath, J. Bergeron, and J. A. Retsema. 1996. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol. Microbiol. 22:867-879. [DOI] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing. Fifteenth informational supplement, vol. 25, M100-S15. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 9.Daly, M. M., S. Doktor, R. Flamm, and D. Shortridge. 2004. Characterization and prevalence of MefA, MefE, and the associated msr(D) gene in Streptococcus pneumoniae clinical isolates. J. Clin. Microbiol. 42:3570-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Grosso, M., F. Iannelli, C. Messina, M. Santagati, N. Petrosillo, S. Stefani, G. Pozzi, and A. Pantosti. 2002. Macrolide efflux genes mef(A) and mef(E) are carried by different genetic elements in Streptococcus pneumoniae. J. Clin. Microbiol. 40:774-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Del Grosso, M., A. S. d'Abusco, F. Iannelli, G. Pozzi, and A. Pantosti. 2004. Tn2009, a Tn916-like element containing mef(E) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 48:2037-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doern, G. V., K. P. Heilmann, H. K. Huynh, P. R. Rhomberg, S. L. Coffman, and A. B. Brueggemann. 2001. Antimicrobial resistance among clinical isolates of Streptococcus pneumoniae in the United States during 1999-2000, including a comparison of resistance rates since 1994-1995. Antimicrob. Agents Chemother. 45:1721-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farrell, D. J., and S. G. Jenkins. 2004. Distribution across the USA of macrolide resistance and macrolide resistance mechanisms among Streptococcus pneumoniae isolates collected from patients with respiratory tract infections: PROTEKT US 2001-2002. J. Antimicrob. Chemother. 54(Suppl. 1):i17-i22. [DOI] [PubMed] [Google Scholar]
- 14.Gay, K., W. Baughman, Y. Miller, D. Jackson, C. G. Whitney, A. Schuchat, M. M. Farley, F. Tenover, and D. S. Stephens. 2000. The emergence of Streptococcus pneumoniae resistant to macrolide antimicrobial agents: a 6-year population-based assessment. J. Infect. Dis. 182:1417-1424. [DOI] [PubMed] [Google Scholar]
- 15.Gay, K., and D. S. Stephens. 2001. Structure and dissemination of a chromosomal insertion element encoding macrolide efflux in Streptococcus pneumoniae. J. Infect. Dis. 184:56-65. [DOI] [PubMed] [Google Scholar]
- 16.Hardy, G. G., M. J. Caimano, and J. Yother. 2000. Capsule biosynthesis and basic metabolism in Streptococcus pneumoniae are linked through the cellular phosphoglucomutase. J. Bacteriol. 182:1854-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Havarstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann, J., M. S. Cetron, M. M. Farley, W. S. Baughman, R. R. Facklam, J. A. Elliott, K. A. Deaver, and R. F. Breiman. 1995. The prevalence of drug-resistant Streptococcus pneumoniae in Atlanta. N. Engl. J. Med. 333:481-486. [DOI] [PubMed] [Google Scholar]
- 19.Hyde, T. B., K. Gay, D. S. Stephens, D. J. Vugia, M. Pass, S. Johnson, N. L. Barrett, W. Schaffner, P. R. Cieslak, P. S. Maupin, E. R. Zell, J. H. Jorgensen, R. R. Facklam, and C. G. Whitney. 2001. Macrolide resistance among invasive Streptococcus pneumoniae isolates. JAMA 286:1857-1862. [DOI] [PubMed] [Google Scholar]
- 20.Johnston, N. J., J. C. de Azavedo, J. D. Kellner, and D. E. Low. 1998. Prevalence and characterization of the mechanisms of macrolide, lincosamide, and streptogramin resistance in isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2425-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leclercq, R., and P. Courvalin. 2002. Resistance to macrolides and related antibiotics in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:2727-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linton, K. J., and C. F. Higgins. 1998. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol. Microbiol. 28:5-13. [DOI] [PubMed] [Google Scholar]
- 23.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:3322-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuoka, M., L. Janosi, K. Endou, and Y. Nakajima. 1999. Cloning and sequences of inducible and constitutive macrolide resistance genes in Staphylococcus aureus that correspond to an ABC transporter. FEMS Microbiol. Lett. 181:91-100. [DOI] [PubMed] [Google Scholar]
- 25.Morrison, D. A., S. A. Lacks, W. R. Guild, and J. M. Hageman. 1983. Isolation and characterization of three new classes of transformation-deficient mutants of Streptococcus pneumoniae that are defective in DNA transport and genetic recombination. J. Bacteriol. 156:281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagai, K., P. C. Appelbaum, T. A. Davies, L. M. Kelly, D. B. Hoellman, A. T. Andrasevic, L. Drukalska, W. Hryniewicz, M. R. Jacobs, J. Kolman, J. Miciuleviciene, M. Pana, L. Setchanova, M. K. Thege, H. Hupkova, J. Trupl, and P. Urbaskova. 2002. Susceptibilities to telithromycin and six other agents and prevalence of macrolide resistance due to L4 ribosomal protein mutation among 992 pneumococci from 10 central and Eastern European countries. Antimicrob. Agents Chemother. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pestova, E. V., L. S. Havarstein, and D. A. Morrison. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21:853-862. [DOI] [PubMed] [Google Scholar]
- 28.Reinert, R. R., A. Wild, P. Appelbaum, R. Lütticken, M. Y. Cil, and A. Al-Lahham. 2003. Ribosomal mutations conferring resistance to macrolides in Streptococcus pneumoniae clinical strains isolated in Germany. Antimicrob. Agents Chemother. 47:2319-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reynolds, E., J. I. Ross, and J. H. Cove. 2003. Msr(A) and related macrolide/streptogramin resistance determinants: incomplete transporters? Int. J. Antimicrob. Agents. 22:228-236. [DOI] [PubMed] [Google Scholar]
- 30.Roberts, M. C., J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, and H. Seppala. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robertson, G. T., W.-L. Ng, J. Foley, R. Gilmour, and M. E. Winkler. 2002. Global transcriptional analysis of clpP mutations of type 2 Streptococcus pneumoniae and their effects on physiology and virulence. J. Bacteriol. 184:3508-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross, J. I., E. A. Eady, J. H. Cove, and S. Baumberg. 1995. Identification of a chromosomally encoded ABC-transport system with which the staphylococcal erythromycin exporter MsrA may interact. Gene 153:93-98. [DOI] [PubMed] [Google Scholar]
- 33.Ross, J. I., E. A. Eady, J. H. Cove, and S. Baumberg. 1996. Minimal functional system required for expression of erythromycin resistance by msrA in Staphylococcus aureus RN4220. Gene 183:143-148. [DOI] [PubMed] [Google Scholar]
- 34.Ross, J. I., E. A. Eady, J. H. Cove, W. J. Cunliffe, S. Baumberg, and J. C. Wootton. 1990. Inducible erythromycin resistance in staphylococci is encoded by a member of the ATP-binding transport super-gene family. Mol. Microbiol. 4:1207-1214. [DOI] [PubMed] [Google Scholar]
- 35.Santagati, M., F. Iannelli, C. Cascone, F. Campanile, M. R. Oggioni, S. Stefani, and G. Pozzi. 2003. The novel conjugative transposon Tn1207.3 carries the macrolide efflux gene mef(A) in Streptococcus pyogenes. Microb. Drug Resist. 9:243-247. [DOI] [PubMed] [Google Scholar]
- 36.Santagati, M., F. Iannelli, M. R. Oggioni, S. Stefani, and G. Pozzi. 2000. Characterization of a genetic element carrying the macrolide efflux gene mef(A) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2585-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shortridge, V. D., G. V. Doern, A. B. Brueggemann, J. M. Beyer, and R. K. Flamm. 1999. Prevalence of macrolide resistance mechanisms in Streptococcus pneumoniae isolates from a multicenter antibiotic resistance surveillance study conducted in the United States in 1994-1995. Clin. Infect. Dis. 29:1186-1188. [DOI] [PubMed] [Google Scholar]
- 38.Stephens, D. S., S. M. Zughaier, C. G. Whitney, W. S. Baughman, L. Barker, K. Gay, D. Jackson, W. A. Orenstein, K. Arnold, A. Schuchat, and M. M. Farley. 2005. Incidence of macrolide resistance in Streptococcus pneumoniae after introduction of the pneumococcal conjugate vaccine: population-based assessment. Lancet 365:855-863. [DOI] [PubMed] [Google Scholar]
- 39.Sutcliffe, J., A. Tait-Kamradt, and L. Wondrack. 1996. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob. Agents Chemother. 40:1817-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tait-Kamradt, A., J. Clancy, M. Cronan, F. Dib-Hajj, L. Wondrack, W. Yuan, and J. Sutcliffe. 1997. mefE is necessary for the erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:2251-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tao, L., D. J. LeBlanc, and J. J. Ferretti. 1992. Novel streptococcal-integration shuttle vectors for gene cloning and inactivation. Gene 120:105-110. [DOI] [PubMed] [Google Scholar]
- 42.Weisblum, B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitney, C. G., M. M. Farley, J. Hadler, L. H. Harrison, C. Lexau, A. Reingold, L. Lefkowitz, P. R. Cieslak, M. Cetron, E. R. Zell, J. H. Jorgensen, and A. Schuchat. 2000. Increasing prevalence of multidrug-resistant Streptococcus pneumoniae in the United States. N. Engl. J. Med. 343:1917-1924. [DOI] [PubMed] [Google Scholar]
- 44.Widdowson, C. A., and K. P. Klugman. 1999. Molecular mechanisms of resistance to commonly used non-betalactam drugs in Streptococcus pneumoniae. Semin. Respir. Infect. 14:255-268. [PubMed] [Google Scholar]