Abstract
Fluoroquinolones are an important class of antibiotics for the treatment of infections arising from the gram-positive respiratory pathogen Streptococcus pneumoniae. Although there is evidence supporting interspecific lateral DNA transfer of fluoroquinolone target loci, no studies have specifically been designed to assess the role of intraspecific lateral transfer of these genes in the spread of fluoroquinolone resistance. This study involves a comparative evolutionary perspective, in which the evolutionary history of a diverse set of S. pneumoniae clinical isolates is reconstructed from an expanded multilocus sequence typing data set, with putative recombinants excluded. This control history is then assessed against networks of each of the four fluoroquinolone target loci from the same isolates. The results indicate that although the majority of fluoroquinolone target loci from this set of 60 isolates are consistent with a clonal dissemination hypothesis, 3 to 10% of the sequences are consistent with an intraspecific lateral transfer hypothesis. Also evident were examples of interspecific transfer, with two isolates possessing a parE-parC gene region arising from viridans group streptococci. The Spain 23F-1 clone is the most dominant fluoroquinolone-nonsusceptible clone in this set of isolates, and the analysis suggests that its members act as frequent donors of fluoroquinolone-nonsusceptible loci. Although the majority of fluoroquinolone target gene sequences in this set of isolates can be explained on the basis of clonal dissemination, a significant number are more parsimoniously explained by intraspecific lateral DNA transfer, and in situations of high S. pneumoniae population density, such events could be an important means of resistance spread.
Fluoroquinolones were originally introduced in the mid-1980s principally for the treatment of gram-negative bacterial infections. Subsequent development of fluoroquinolones was targeted to also be effective against gram-positive cocci. Much of the reason for this development interest was related to the increasing resistance of the important respiratory tract pathogen Streptococcus pneumoniae to other antibiotics, particularly beta-lactams. Fluoroquinolone resistance in S. pneumoniae remains relatively low, but it is nonetheless increasing, particularly in some geographic areas, such as Hong Kong (16). Resistance to fluoroquinolones in gram-positive cocci, such as S. pneumoniae, is thought to be mainly due to spontaneous mutations that occur in target loci due to errors of polymerase during replication (17). The acquisition of new genetic material via lateral gene transfer (LGT; also referred to as recombination) is generally assumed to be of limited importance in fluoroquinolone resistance in gram-positive bacteria (17).
Several recent studies have specifically set out to examine the role of interspecific lateral DNA transfer of fluoroquinolone target loci from viridans group streptococci to S. pneumoniae. Ferrandiz et al. (12) and Balsalobre et al. (1) present evidence for interspecific lateral DNA transfer involving the parE-parC gene region from viridans group streptococci to S. pneumoniae. Balsalobre et al. (1) concluded that 11% of the 46 isolates examined were putative interspecific recombinants and suggest that viridans group streptococci act as donors of fluoroquinolone resistance genes to S. pneumoniae via horizontal transfer. Recently, de la Campa et al. (5) reported on the quinolone resistance-determining regions (QRDRs) of parE, parC, and gyrA from a set of 75 ciprofloxacin-resistant isolates from Spain, 5 of which are of viridans group streptococcus origin. In contrast, another study concluded that the interspecific lateral transfer of fluoroquinolone target loci was not occurring at all (30). Thus, although there is evidence for the lateral transfer of fluoroquinolone target loci from viridans group streptococci to S. pneumoniae, the frequency and importance of the phenomenon in the spread of resistance remain somewhat uncertain.
If interspecific lateral transfer of fluoroquinolone target loci is known to occur, then it is likely that intraspecific lateral transfer among S. pneumoniae isolates is also occurring. Indeed, it has been shown that the frequency of homologous recombination in S. pneumoniae decreases with the sequence divergence between donor and recipient (20, 25). Furthermore, it is clear that penicillin binding proteins (PBPs) are transferred both intraspecifically (4, 22) and interspecifically (7, 14). Nonetheless, we are not aware of any demonstration of intraspecific DNA exchange involving the fluoroquinolone target loci of S. pneumoniae clinical isolates. Undoubtedly, part of the reason for this is the fact that intraspecific events are much more difficult to detect than interspecific events, since differences in the donor and recipient sequences can be much more subtle and require an approach that concomitantly indexes both the evolutionary history of the isolates and that of the resistance genes. Several recent publications have commented on the importance of clonal dissemination in the spread of fluoroquinolone resistance in S. pneumoniae (15, 28, 31), while other studies from North America report a wider genetic diversity, with fewer isolates clearly associated with the international clones (21, 40). A complete understanding of the role of clonal spread is not possible without knowledge of the role of intraspecific lateral DNA transfer of fluoroquinolone target loci. At present, we are not aware of any published studies that attempt to address intraspecific lateral DNA transfer of S. pneumoniae fluoroquinolone target loci.
A classic means of detecting lateral DNA transfer is to assess branching congruence or incongruence in comparisons of a control evolutionary history (the best estimate of the true evolutionary history for the organisms in question) to that derived from a particular locus (gene history) obtained from the same biological entities (for a review, see reference 23). Incongruent histories support lateral DNA transfer, and congruent histories do not. This approach is analogous to that used in previous studies of intraspecific lateral transfer of PBPs in S. pneumoniae, except that the earlier studies did not compare reconstructed evolutionary histories but instead genotyped the isolates and assessed whether different clones had identical PBP gene sequences (horizontal transfer) or were in fact indistinguishable based on overall genetic relatedness and their PBP sequence (clonal spread) (4, 22). Evolutionary histories provide an additional level of information regarding relationships and, consequently, the prospect of a more thorough picture of clonal spread versus horizontal transfer. However, in the case of S. pneumoniae the reconstruction of a control evolutionary history is complicated by recombination and by the relatively low level of sequence variation present in housekeeping genes between different isolates (10). Although both these variables hamper phylogenetic reconstruction, they are much less of a problem with evolutionary networks, which are well suited to the representation of relationships between sequences of relatively low divergence, typical of population data, and allow the incorporation of the nonbifurcating genealogical information that can be associated with population-level divergences (3, 10). It is also possible to at least partially ameliorate the problems of recombination. A large number of methods for the detection of recombinant sequences are now available, and some of these have been applied to bacterial population sequence data (13). Once recombinant sequence regions are detected, they can be excised before the reconstruction of evolutionary histories. Thus, we believe that it is possible to reconstruct control evolutionary histories for S. pneumoniae isolates by employing network reconstruction of relatively large amounts of housekeeping sequence (with the multilocus sequence typing [MLST] data as a subset) from each isolate after the sequence has been carefully screened for putative recombinants. Such a control history can then serve as a meaningful framework for comparison against evolutionary networks of other genes of interest, in this case, fluoroquinolone target loci.
It is important to try and assess the existence and ultimately estimate the frequency of intraspecific lateral transfer of fluoroquinolone target loci because it could be a very important means of resistance spread in situations in which the population density of S. pneumoniae is high. Children represent such a situation; studies of day care and pediatric chronic care centers have found that children are often colonized with high densities of pneumococci in the nasopharynx (26, 27, 39). At present, the potential pediatric indications for fluoroquinolones include chronic ear infection, invasive gastrointestinal infection, complicated urinary tract infections, and pseudomonal bronchopulmonary complications in cystic fibrosis (26). There is, however, increasing pressure on regulatory authorities to approve fluoroquinolones for more general use in the pediatric population (26). Therefore, frequent intraspecific lateral transfer of fluoroquinolone target loci between S. pneumoniae isolates, combined with the approval of fluoroquinolones for more general use in the pediatric population, could represent an important set of circumstances for the development of fluoroquinolone resistance.
The purpose of this study is to use molecular evolution principles and techniques in an attempt to gain insight into the frequency with which S. pneumoniae fluoroquinolone target loci may be spread via intraspecific lateral DNA transfer. The evolutionary history of a diverse set of S. pneumoniae clinical isolates from the Alexander Project collection (11, 19) is derived from network reconstructions of an expanded MLST data set that includes a concatenated 8-kb sequence alignment of housekeeping genes, with putative recombinants excluded. This control history is then assessed against evolutionary networks of each of the four fluoroquinolone target loci from the same set of isolates. Congruent histories support a clonal dissemination hypothesis for a given fluoroquinolone target locus, and discordant histories suggest lateral DNA transfer.
MATERIALS AND METHODS
Sequence data collection.
The susceptibility data and descriptions of the tests for the Alexander Project have been published elsewhere (11, 19). Our choice of isolates from the Alexander Project collection involved a random subsample of 42 fluoroquinolone-resistant isolates (MIC ≥ 8 μg/ml for either ciprofloxacin or ofloxacin from the total of 170 fluoroquinolone-resistant isolates), 4 fluoroquinolone-intermediate isolates (ciprofloxacin and ofloxacin MICs = 4 μg/ml), and 14 susceptible isolates (ciprofloxacin and ofloxacin MICs < 4 μg/ml) and spanned the years 1992 to 2000. Two of the fluoroquinolone-resistant isolates were subsequently determined to have a parE-parC gene region that arose from an interspecific DNA exchange event involving viridans group streptococci and were not included in the evolutionary reconstructions. Table 1 includes the details for the 60 isolates used in this study. The majority of isolates (n = 44) were nasopharyngeal in origin, the origins of 11 were unknown, and 5 were from blood. Colony PCR was performed with 45 μl of Invitrogen High Fidelity PCR Supermix per colony. The control histories were reconstructed from a concatenated set of housekeeping gene sequences; the resulting alignment of nine loci for the 58 S. pneumoniae clinical isolates, prior to any exclusions, was 9,473 bp long. This included all or nearly all of the sequences of the recP (transketolase), gki (glucose kinase), gdh (glucose-6-phosphate dehydrogenase), xpt (xanthine phosphoribosyltransferase), spi (signal peptidase I), and aroE (shikimate dehydrogenase) genes used for MLST (8); we did not sample ddl (the usual seventh MLST locus) because previous studies have shown that it is frequently recombinant (9). In addition to the sequencing of larger fragments of the customary MLST loci, we also included the genes neighboring aroE, spi, and xpt, which are aroB (3-dehydroquinate synthase), rnhb (RNase HII), and pbux (xanthine permease), respectively. The complete aroE and aroB genes were amplified with primers aroE+1f (5′-CACTGCGGATGTGACTGGTTCGA-3′) and aroE+2r (5′-CCCTCAATAATAGCTGTTAGACGGGG-3′). The complete spi and rnhb genes were amplified with primers spi+1f (5′-GATAGAAGAAGAGGCTGAGATTGGT-3′) and spi+1r (5′-CAATCTCACGGCTGAGCTGAGTT-3′). A fragment encompassing all of the xpt gene and most of the pbux gene was amplified with primers xpt+1f (5′-GTCTATGATACCACTACAACGGGA-3′) and xpt+1r (5′-CGGCATTGAGGACAATAGCGAGT-3′). We amplified gki (all but 33 bp of the 3′ end), gdh (all but 27 bp of the 3′ end and 40 bp of the 5′ end), and recP (all but 76 bp of the 3′ end) using the following respective primer pairs: gki1f (5′-CACGCAAACCTTTGCATAAGTGA-3′) and gki2r (5′-ACAAGTGATGCTGCTCCGATAAC-3′), gdh2f (5′-TGTTACAATTTTCGGTGCGAGTG-3′) and gdh2r (5′-TCTAAGCGACCATCTTGACGATA-3′), and recP1f (5′-CTTTGGAGGATTTCCGTATGTTG-3′) and recP2r (5′-CTGCCAATACTTTTGGTGCTGGG-3′).
TABLE 1.
Details for the isolates included in this study, including MICs, laboratory identification, and Alexander Project identification
| Laboratory identification no.a | Alexander Project identification no. | NS profileb | MIC (μg/ml) of fluoroquinolone antibioticsc
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CIP | GAT | GEM | GRE | LEV | MOX | OFL | TRV | |||
| 0.14GE00 | 2000321143 | 1 | 0.25 | 0.02 | 1 | 0.12 | 2 | |||
| 0.15GR00 | 2000381105 | 1 | 0.25 | 0.03 | 1 | 0.25 | 2 | |||
| 0.16GR98 | 1998381349 | 1 | 0.02 | 2 | ||||||
| 0.19IS99d | 1999441130 | 1 | 0.02 | 0.25 | 1 | 2 | 0.3 | |||
| 0.29JA98 | 1998641094 | 0.5 | 0.02 | 2 | ||||||
| 0.31JA99 | 1999643069 | 1 | 0.02 | 0.12 | 1 | 1 | 0.1 | |||
| 0.34PL96e | 623116 | 1 | 2 | |||||||
| 0.45SL99 | 1999432187 | 1 | 0.02 | 0.12 | 1 | 2 | 0.1 | |||
| 0.47SP00 | 2000451124 | 2 | 0.5 | 0.03 | 1 | 0.12 | 2 | |||
| 0.49SP94e | 405196 | 1 | 2 | |||||||
| 0.57SW99d | 1999347003 | 2 | 0.03 | 0.25 | 1 | 2 | 0.3 | |||
| 0.68Oh92e | 214012 | 1 | 2 | |||||||
| 0.74Oh94e | 414009 | 1 | 2 | |||||||
| 0.7FR93e | 304081 | 1 | 2 | |||||||
| 128.2Ca00 | 2000145215 | Co | >8 | >1 | 0.12 | >2 | >1 | >8 | ||
| 132.1Ny98 | 1998111761 | F | >16 | 0.5 | >16 | |||||
| 132.1US00 | 2000167017 | F | 8 | >1 | 0.06 | 2 | 0.25 | 8 | ||
| 136.1PL98 | 1998401181 | Ch | >16 | 0.25 | 16 | |||||
| 215.1Oh00 | 2000154003 | E | >8 | >1 | 0.12 | >2 | >1 | >8 | ||
| 292.1AT96e | 622286 | E, D, Co | 8 | 8 | ||||||
| 294.1JA99 | 1999643075 | E, Ch | 8 | 0.12 | 1 | 2 | 8 | 0.5 | ||
| 319.1JA00 | 2000643031 | E, Ch, Co | 8 | >1 | 0.06 | 2 | 0.5 | 8 | ||
| 319.1JA99 | 1999643052 | E, Ch, Co | 8 | 0.12 | 2 | 4 | 8 | 0.5 | ||
| 319.2JA99 | 1999643061 | E, Ch, Co | 8 | 0.12 | 1 | 2 | 8 | 0.5 | ||
| 322.1JA98 | 1998641212 | E, F, D | 4 | 0.06 | 4 | |||||
| 432.1JA00 | 2000642018 | E, F, D | >8 | >1 | 0.25 | >2 | >1 | >8 | ||
| 458.1JA00 | 2000643086 | P, E, F, D | 4 | 0.5 | 0.03 | 2 | 0.25 | 4 | ||
| 524.1Ne00 | 2000143007 | P, E, F, Co | >8 | >1 | 0.12 | >2 | 1 | >8 | ||
| 526.1Tx99 | 1999107002 | P, E, F, D, Co | 8 | 0.06 | 0.5 | 1 | 4 | 0.3 | ||
| 543.1Ca99 | 1999127017 | P, E, F, D, Co | 8 | 0.06 | 1 | 2 | 4 | 0.5 | ||
| 544.1Tx99 | 1999130044 | P, E, F, Ch, D | 8 | 0.03 | 0.25 | 2 | 4 | 0.3 | ||
| 581.1Ny99 | 1999111025 | P, E, F, Ch, D, Co | 16 | 0.25 | 4 | 4 | 16 | 2 | ||
| 582.1CH00e | 2000631055 | P, E, F, Ch, D, Co | >8 | >1 | 0.12 | >2 | >1 | >8 | ||
| 584.1Ca99 | 1999101066 | P, E, F, Ch, D, Co | 8 | 0.06 | 1 | 2 | 4 | 0.5 | ||
| 586.1CH99 | 1999631055 | P, E, F, Ch, D, Co | 16 | 0.25 | 4 | 4 | 16 | 2 | ||
| 588.1JA99 | 1999641020 | P, E, F, D, Co | 16 | 0.12 | 4 | 4 | 16 | 2 | ||
| 636.1Ca99 | 1999131027 | P, E, F, Ch, D, Co | 16 | 0.12 | 4 | 4 | 16 | 2 | ||
| 638.2CH99 | 1999631091 | P, E, F, Ch, D, Co | 16 | 0.25 | 4 | 4 | 16 | 2 | ||
| 638.3CH99 | 1999631096 | P, E, F, Ch, D, Co | 16 | 0.25 | 4 | 4 | 16 | 2 | ||
| 67.1FR92e | 204027 | 4 | 8 | |||||||
| 67.1JA00 | 2000641008 | 4 | >1 | 0.12 | >2 | 1 | >8 | |||
| 672.1US00 | 2000167005 | P, E, F, Co | >8 | >1 | 0.25 | >2 | >1 | >8 | ||
| 68.1AT98 | 1998331178 | 8 | 0.06 | 4 | ||||||
| 68.1PR00 | 2000371002 | 8 | 0.5 | 0.06 | 2 | 0.25 | 4 | |||
| 68.1SW99d | 1999347026 | 8 | 0.06 | 1 | 2 | 4 | 0.5 | |||
| 68.1Wa00 | 2000148301 | 8 | >1 | 0.06 | 2 | 0.25 | 4 | |||
| 687.1CH99d | 1999631063 | P, E, F, Ch, D, Co | 16 | 0.25 | 4 | 4 | 16 | 2 | ||
| 700.1CH00e | 2000631075 | P, E, F, Ch, D, Co | 8 | >1 | 0.06 | >2 | >1 | >8 | ||
| 71.1SW99 | 1999342029 | Co | 4 | 0.06 | 2 | 2 | 4 | 1 | ||
| 72.1In00 | 2000153215 | F | 1 | 0.5 | 0.03 | 1 | 0.25 | 8 | ||
| 720.1CH00 | 2000631003 | P, E, F, Ch, D, Co | >8 | >1 | 0.25 | >2 | >1 | >8 | ||
| 720.3CH00e | 2000631046 | P, E, F, Ch, D, Co | 8 | >1 | 0.06 | >2 | >1 | >8 | ||
| 720.4CH00e | 2000631079 | P, E, F, Ch, D, Co | 8 | >1 | 0.06 | >2 | >1 | >8 | ||
| 73.1Tx98 | 1998107007 | F | 4 | 0.03 | 4 | |||||
| 94.1Nd00 | 2000141213 | F | 4 | >1 | 0.25 | >2 | >1 | >8 | ||
| 95.1BR98d | 1998211720 | >16 | 0.25 | >16 | ||||||
| 95.1In00 | 2000153209 | >8 | >1 | 0.25 | >2 | >1 | >8 | |||
| 95.1Ma00 | 2000133002 | >8 | >1 | 0.25 | >2 | >1 | >8 | |||
| 434.1FR99 | 1999361022 | P, E, F, Co | 8 | 0.12 | 0.5 | 2 | 4 | 0.25 | ||
| 591.1Ca99 | 1999131017 | P, E, F, Co | 16 | 1 | 4 | 4 | 16 | 2 | ||
The laboratory identification number is that used in the present study and is described as follows: an isolate with a zero to the left of the decimal place (e.g., 0.47SP00) is susceptible to all antibiotics used for testing in the Alexander Project (minimum of 15, depending on the year). As the numbers to the left of the decimal place increase, so do the number of antibiotics to which that isolate is resistant. The two-letter code following the number(s), immediately to the right of the decimal place, is associated with the country code or the state code, if it is an isolate from the United States (GE, Germany; GR, Greece; IS, Israel; JA, Japan; PL, Poland; SL, Slovakia; SP, Spain; SW, Switzerland; FR, France; US, United States (state unknown); AT, Austria; CH, China (Hong Kong); PR, Portugal; BR, Brazil; Oh, Ohio; Ca, California; Ny, New York; Ne, Nebraska; Tx, Texas; Wa, Washington; Nd, North Dakota; Ma, Massachusetts). The final two numbers in the code correspond to the year collected. Unless indicated otherwise, all isolates are nasopharyngeal.
NS profile, nonsusceptibility information for various antibiotics representing several different classes: P, penicillin; E, erythromycin; F, cefaclor; Ch, chloramphenicol; D, doxycycline; Co, co-trimoxazole. The MICs for nonsusceptibility for each of these antibiotics were defined as follows: penicillin, ≥0.12 μg/ml; erythromycin, ≥0.5 μg/ml; cefaclor, ≥2.0 μg/ml; chloramphenicol, ≥8.0 μg/ml; doxycycline, ≥4.0 μg/ml; co-trimoxazole, ≥1 μg/ml.
CIP, ciprofloxacin; GAT, gatifloxacin; GEM, gemifloxacin; GRE, grepafloxacin; LEV, levofloxacin; MOX, moxifloxacin; OFL, ofloxacin; TRV, trovafloxacin).
Isolate from blood.
Sample source unknown.
In addition to the housekeeping genes, we sampled the four fluoroquinolone target genes from the same set of 58 isolates: parE (topoisomerase IV subunit B), parC (topoisomerase IV subunit A), gyrA (DNA gyrase subunit A), and gyrB (DNA gyrase subunit B). The entire gyrB, gyrA, and parE genes were amplified with the following respective primer pairs: gyrAp1 (5′-ACGTTTTAGTGGTTTAGAGG-3′) and gyrAp4 (5′-TGAGCATCCCATTGAGAAGTTAGA-3′), gyrBp1 (5′-ATTGGCACTGTATGGTATCAC-3′) and gyrBp4 (5′-AGCGCCTCTAATCTCCCCTCGTT-3′), and parEp1 (5′-GCCGATAACAGCCAAAAGTCC-3′) and parEp4 (5′-AATTCGTCCCGTGTTCAGTTAC-3′). A fragment encompassing all of parC except 6 bp of the 3′ end was amplified with primers parCp1 (5′-TTTATGGGCTTTGTATCTTATGTC-3′) and parCp4 (5′-GTTTTCCCAATCAATCGTTACTGT-3′).
The PCR products were purified with a QIAGEN QIAquick PCR purification kit, according to the manufacturer's instructions. The products were sequenced on both strands by using Applied Biosystems machines 3730, 3700, and 3100 and Applied Biosystems Big Dye.
Reconstruction of evolutionary histories.
Homologous DNA sequence alignments were generated by using ClustalX, version 1.83 (38), with any necessary additional manual editing in Genedoc, version 2.6.0.2. The nine housekeeping genes were concatenated into a single alignment, and then the program PLATO (13) was used to determine if any portion of the alignment was evolving in an anomalous fashion due to either recombination or selection. Recombination can be difficult to detect with the low levels of sequence divergence typical of S. pneumoniae housekeeping genes, and PLATO is likely to miss some of the more subtle instances of recombination (33). However, we have found that it performs very well in detecting sequences known to be frequently recombinant in S. pneumoniae (ddl, for example [9]), and it has a low propensity to detect recombination when it is not present (33). Thus, PLATO may miss some subtle instances of recombinant sequences, but at the same time, it has a high likelihood of detecting the majority of recombinant sequences in our alignments. Since PLATO does not tolerate polytomies, identical sequences were removed from the housekeeping data set prior to PLATO runs and were then added back to the alignment for further phylogenetic analyses. Multiple PLATO analyses were performed to ensure convergence by using the following parameters: 10 steps for the sliding window, 1,000 replications of Monte Carlo simulation, and an HKY model of sequence evolution. Any regions of the housekeeping alignment that PLATO found to be statistically anomalous were removed. The individual genes within this concatenated alignment encompassed the following regions: aroE = positions 1 to 855, aroB = positions 856 to 1923, gdh = positions 1924 to 3337, gki = positions 3338 to 4272, pbux = positions 4273 to 5489, recP = positions 5490 to 7390, rnhb = positions 7391 to 8276, spi = positions 8277 to 8891, and xpt = positions 8892 to 9473.
The program TCS (3) was used to implement the method of Templeton et al. (37) for the reconstruction of statistical parsimony networks for both the housekeeping alignment and each of the fluoroquinolone target gene alignments. To avoid ambiguities in haplotype assignment, all missing positions were removed from each alignment prior to TCS analysis (37). In order to provide a further means of verifying our clonal group designations, phylogenies were also reconstructed. For the final concatenated housekeeping alignment, as well as the four resistance gene alignments, the best-fitting model of sequence evolution and the corresponding values for the rate matrix, shape of the gamma distribution, and proportion of invariant sites were estimated by the program MODELTEST (32). Phylogenies were reconstructed by the maximum-likelihood (ML) and neighbor-joining (NJ) methods implemented in PAUP*4.0b (36). For the ML method, starting trees were obtained by neighbor joining, with nearest-neighbor interchange used as the branch-swapping algorithm. Bootstrap support values were obtained with 1,000 replicates for NJ analyses and 100 replicates for ML analyses. In addition, Bayesian phylogenetic reconstruction was performed using Mr. Bayes (18) with 3,000,000 generations, a sampling frequency of every 100 generations, GTR + gamma + invariants, four Markov chains, random starting trees, and a burn-in of 2,000,000 generations. Multiple Bayesian analyses were performed to ensure convergence. Phylogenetic statistical comparisons were made by the conservative Shimodaira-Hasegawa (SH) test (34, 35).
Nucleotide sequence accession numbers.
The sequences obtained in this study have been deposited in GenBank under the following accession numbers: DQ174780 to DQ175475 and DQ176454 to DQ176511.
RESULTS AND DISCUSSION
Control history.
Within the concatenated alignment of nine housekeeping genes encompassing 9,473 bp, PLATO identified several anomalous evolving regions, which were excluded before phylogenetic analysis. These regions, with their accompanying PLATO Z values (Z values greater than 4.2526 were judged to be significant; the value indicated is the lowest from the various independent runs) included the following: positions 238 to 244, Z = 11.7949; positions 1874 to 1881, Z = 7.0216; positions 1971 to 1977, Z = 13.71133; positions 2350 to 2354, Z = 4.4019; positions 2792 to 2796, Z = 4.5986; positions 3083 to 3092, Z = 6.4793; positions 3124 to 3128, Z = 5.7274; positions 3185 to 3189, Z = 19.5443; positions 3212 to 3233, Z = 11.5082; positions 3313 to 3317, Z = 4.6513; positions 3790 to 3796, Z = 5.8212; positions 3844 to 3935, Z = 19.8386; positions 4617 to 4638, Z = 4.8284; positions 4922 to 4927, Z = 5.3199; positions 5337 to 5343, Z = 6.4382; positions 5993 to 5999, Z = 9.1872; positions 6229 to 6233, Z = 5.8387; positions 6303 to 6307, Z = 13.2928; positions 6672 to 6676, Z = 8.4217; positions 7157 to 7220, Z = 4.5825; positions 7267 to 7271, Z = 6.8452; positions 7561 to 8555, Z = 18.5434; positions 9033 to 9038, Z = 17.4671; and positions 9206 to 9210, Z = 4.3690. After the exclusion of these 1,310 characters the resulting alignment was 8,163 bp in length. Nearly all of the rnhb locus, as well as the 5′ end of spi, were excluded (rnhb is immediately upstream of spi) due to their anomalous character. Individual gene trees involving rnhb provided clear evidence for several strongly supported conflicting nodes in comparison to trees constructed from the 8,163-bp housekeeping alignment. It seems likely, therefore, that this 993-bp stretch encompassing most of rnhb and the 5′ end of spi is frequently recombinant.
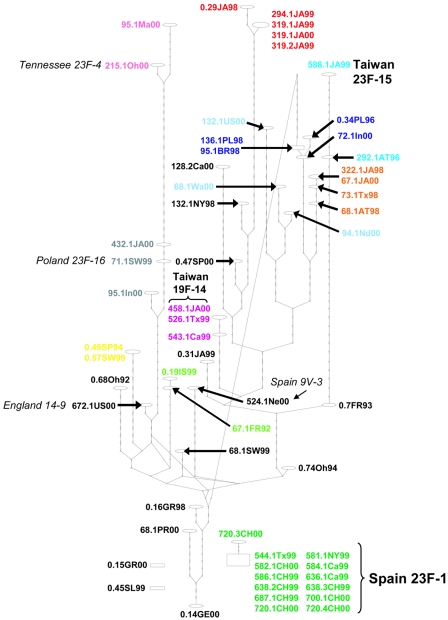
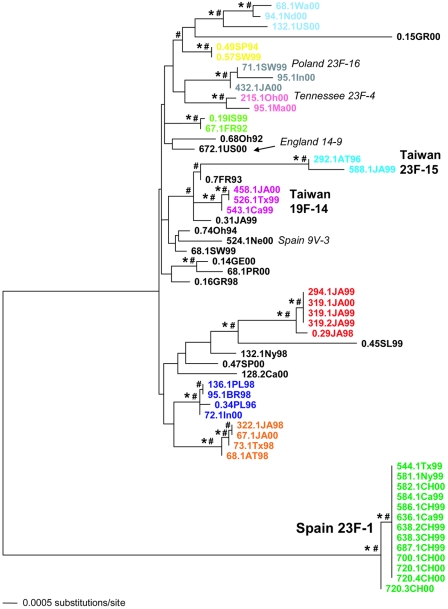
The control statistical parsimony network (3, 37) reconstructed from the 8,163 bp of concatenated housekeeping sequence data depicts two very distinct groups (Fig. 1). The main body of the network includes an assortment of isolates from around the world with various antibiotic resistance phenotypes, including isolates susceptible to all classes of antibiotics, as well as fluoroquinolone-nonsusceptible isolates (Table 1). A second group, disconnected from the rest of the network, includes only fluoroquinolone-nonsusceptible isolates, collected in 1999 and 2000 from China and the United States, that were so dissimilar from the other isolates that too many steps (>19) were required to accommodate their connection with the rest of the network. Based on our comparison of the necessary housekeeping sequence data to the MLST database (8), the latter group is quite clearly the globally distributed, multidrug-resistant clone known as Spain 23F-1 (MLST allelic profile 4-4-2-4-4-1-1; we amplified and sequenced the MLST portion of ddl from representative isolates of this clone to verify the ddl allele). The control phylogeny (Fig. 2) depicts a very similar arrangement, with two very distinct clades and a very long branch separating Spain 23F-1 from the rest of the isolates. To our knowledge this is the first indication that the Spain 23F-1 clone may be highly genetically distinct from much of the rest of S. pneumoniae. The reason that our data reveal this previously unrecognized difference appears to lie in the high level of sequence divergence of the aroE and aroB genes of Spain 23F-1 from the sequences of the aroE and aroB genes of the rest of the isolates. The customary MLST approach (8, 29) includes an approximately 400-bp piece of aroE, as well as similarly sized pieces of six other loci, whereas our approach involves sequence data derived from the complete aroE and aroB loci, concomitant with seven other complete (or very nearly complete) loci, of which the MLST sequence data are a subset. The fact that we have sequenced all of aroE, as well as aroB, results in the long branch leading to Spain 23F-1. The elimination of aroE and aroB from this data set eliminates the long branch, and reduction of the data set down to only the MLST sequence set does not reveal this long branch. Explanations for this difference in Spain 23F-1 aroE and aroB are not clear. Reconstruction of the phylogeny of various Streptococcus aroE and aroB sequences available in GenBank does not support an interspecific lateral transfer argument; the Spain 23F-1 aroE and aroB sequences tend to be monophyletic with other S. pneumoniae sequences and merely represent a divergent allele. However, it is possible that the putative donor sequence is simply not yet available in GenBank. Selection is a possibility, although many of the synapomorphies for Spain 23F-1 are synonymous substitutions. aroE codes for shikimate dehydrogenase and aroB codes for 3-dehydroquinate synthase, both of which are involved in the aromatic biosynthetic pathway; however, the selection cause-and-effect issues for such loci within a specific strain are far from clear.
FIG. 1.
Statistical parsimony network (TCS) of the concatenated 8,163-bp, nonrecombinant housekeeping gene sequence alignment. Ovals represent different haplotypes, with the laboratory identifications for the isolates comprising that haplotype indicated adjacently. The number of steps (synonymous with intermediate or unsampled haplotypes) separating different haplotypes is represented by the nodes appearing on the branches. The rectangle represents the TCS determination of the haplotype most likely to be ancestral for this set of sequences. Spain 23F-1 is disconnected from the rest of the network because it requires too many steps (>19) to connect this group with the main body of the network. Clone names have been added, whenever this is possible, through a comparison to the allelic profiles listed for the 26 international clones on the PMEN website (http://www.sph.emory.edu/PMEN/index.html). Isolates that have alleles for the six MLST genes that we sequenced (ddl unknown) identical to a PMEN profile are indicated in boldface; those matching an international clone for five alleles (ddl unknown) are indicated in italics. Isolates of the same putative clone, based on our evolutionary criteria, are similarly colored in order to track the evolution of each of their fluoroquinolone target genes in Fig. 3. Our colored clonal groups, based on a consensus of closely adjacent haplotypes apparent in this housekeeping network and strongly supported monophyletic groups with short (or no) internodes in the case of phylogenies (Fig. 2), correspond very closely with the clonal group designation based on MLST criteria (29).
FIG. 2.
Control phylogeny (ML tree) derived from concatenated alignment of nonrecombinant housekeeping gene sequence data 8,163 bp in length. *, NJ and ML bootstrap support >82%; #, Bayesian posterior probability >0.98. Isolates of the same putative clone are similarly colored in order to track the evolution of each of their fluoroquinolone target genes in Fig. 3. The color scheme is identical to that in Fig. 1.
We used the information from both the housekeeping network (Fig. 1) and phylogeny (Fig. 2) to demarcate several clonal groups (where a clonal group can be either a clone or isolates from a few closely related clones), the members of which are colored similarly in Fig. 3 in order to track the evolution of their fluoroquinolone target genes. These colored clonal groups are based on a consensus of closely adjacent haplotypes apparent in the housekeeping network (Fig. 1) and strongly supported monophyletic groups with short (or no) internodes in the case of the phylogeny (Fig. 2). In general, our clonal groups correspond very closely to the clonal designations based on MLST criteria, which suggests that two isolates are different clones if they differ at three or more alleles of the seven MLST loci (29). An exception to this would include our “light blue” clonal group, where isolate 132.1US00 differs from the other two isolates by at least four alleles and would therefore be a different clone, based on MLST criteria. However, our network (Fig. 1) and phylogeny (Fig. 2) very clearly associate this isolate with the other two, and therefore, we assign isolate 132.1US00 to the same clonal group as isolates 68.1Wa00 and 94.1Nd00. Our purpose here is to follow the history of the fluoroquinolone resistance-associated genes of isolates which share a recent common ancestry; all of the similarly colored isolates in the network (Fig. 1) and phylogeny (Fig. 2) fit that criterion. Adoption of the MLST criteria for clonal designation (with the exclusion of ddl) results in the conservative estimate that the housekeeping network (Fig. 1) includes a minimum of 24 clones, 17 of which are fluoroquinolone nonsusceptible. These clones are scattered throughout the network and occasionally include fully susceptible isolates as part of the clone, suggesting that fluoroquinolone nonsusceptibility has evolved on numerous different occasions in this set of isolates. However, approximately 50% of the fluoroquinolone nonsusceptible isolates can be accounted for by only a few clones (three or four). Nonsusceptible clones occasionally come from the same geographic location (e.g., the “red” clone from Japan with isolates 319.1JA99S, 319.2JA99S, 294.1JA99S, and 319.1JA00S) but more often include isolates from different geographic locations (e.g., the “purple” [Taiwan 19F-14] clone with isolates 458.1JA00S, 526.1Tx99S, and 543.1Ca99S). In addition to Spain 23F-1, it is possible to attach several of our isolates to some of the Pneumococcal Molecular Epidemiology Network (PMEN) international clones. For example, nonsusceptible isolates from the Taiwan 23F-15 and the Taiwan 19F-14 clones are represented (six identical alleles; ddl unknown), with others at least closely related (vary at one locus; ddl unknown) to Poland 23F-16, England 14-9, Spain 9V-3, and Tennessee 23F-4 (Fig. 1 and 2). Fluoroquinolone resistance has previously been reported for all of these clones (2, 15, 28). In our set, Spain 23F-1 is the clone with the largest number of fluoroquinolone nonsusceptible isolates (n = 13). Ten nonsusceptible isolates are members of clones currently uncharacterized by the PMEN.
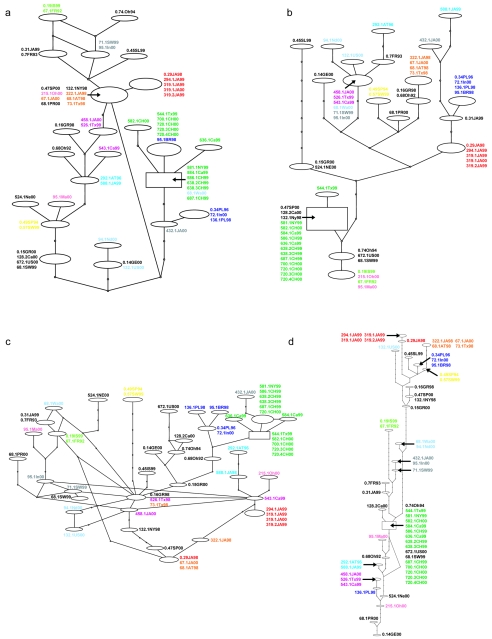
FIG. 3.
Statistical parsimony network (TCS) for each of the respective fluoroquinolone target loci; (a) parE; (b) gyrA (c) parC; (d) gyrB. Ovals represent different haplotypes, with the laboratory identifications for the isolates comprising that haplotype indicated adjacently. The number of steps (intermediate or unsampled haplotypes) separating different haplotypes are represented by the nodes appearing on the branches. The rectangle represents the TCS determination of the haplotype most likely to be ancestral for this set of sequences. The color scheme is identical to that in Fig. 1.
Lateral transfer of fluoroquinolone target loci.
In the process of collecting the comparative sequence data from the Alexander Project collection isolates, we identified several interspecific lateral DNA transfer examples involving the parE and parC gene regions of viridans group streptococci similar to those described previously (1, 12). Sequencing of the parE gene of isolates 434.1FR99S and 591.1Ca99S yielded a gene sequence similar to that of S. mitis, and thus, we sequenced the intergenic region between parE and parC of these two isolates in order to compare to earlier reports involving interspecific transfer of this region (1). Comparative analysis indicated that both our isolates, similar to those described by Balsalobre et al. (1), have a complete viridans group streptococcus ant gene immediately downstream of parE. Between ant and parC, two of the isolates of Balsalobre et al. (specifically, isolates 4589 and 3870) possess an approximately 875-bp insert (homologous to the sequence of S. mutans GTP pyrophosphokinase) which is not shared with our isolates, although one of ours (isolate 434.1FR99S) does have a 97-bp homologous piece of the 3′ end of this insert. Balsalobre et al. (1) reported 5 cases (out of 46) of S. pneumoniae strains with interspecific DNA exchange involving the parE, parC gene region, and we have found an additional 2 such cases (out of 60). Of these seven examples, there are five different genomic arrangements for this region, suggesting that interspecific recombination in this gene region may not be that rare of an occurrence. Recently, de la Campa et al. (5) reported on the QRDRs of parE, parC, and gyrA from a set of 75 isolates, 5 of which were of viridans group streptococcus origin. These various examples serve to highlight the fact that foreign DNA does appear to be playing a role in shaping the parE- parC gene region within S. pneumoniae, although the degree to which this is the case appears to be variable between studies and may be related to the environmental situation from which the isolates were sampled (30).
It seems highly likely that if interspecific lateral gene transfer of fluoroquinolone target loci is occurring between S. pneumoniae and viridans group streptococci, intraspecific recombination is also occurring. This view stems from the fact that the frequency of homologous recombination in S. pneumoniae decreases with the sequence divergence between donor and recipient (25) and that high population densities of S. pneumoniae are common in certain environments and situations (26, 27, 39). Homologous recombination involving fluoroquinolone loci could involve recombination points within or outside the target locus. Recombinant points either outside the gene or at least proximal to the 5′ and 3′ ends involving a donor and a recipient that differ in sequence, result in a different branching position for that isolate, relative to the control history, in networks reconstructed from sequence data for that particular fluoroquinolone target locus. Recombinant points within the gene may not result in a different branching position in the resulting evolutionary history because the recombinant piece may be relatively small and may not carry sufficient evolutionary signal to alter its position compared to the control history. Thus, a convincing alternative placement for a fluoroquinolone target locus relative to the control history results in a conservative estimate of the extent of intraspecific lateral gene transfer for this set of isolates. Our criterion for lateral transfer is the convincing disruption of a clonal group (same color in the control network) in networks derived from the fluoroquinolone target genes. This disruption can take two forms: (i) dispersion of one or more control clonal group members across the fluoroquinolone target gene network (category 1) or (ii) inclusion of a novel (different colored) isolate(s) in a control clonal group on the fluoroquinolone target gene network (category 2). For category 1, convincing disruption refers to the isolates of the same color in highly disparate positions (separated by a large number of substitutions and no longer nearest neighbors) on the fluoroquinolone gene network. For category 2 the most convincing evidence is from isolates that have identical or nearly identical haplotypes on a fluoroquinolone gene network but that are not closely related on the control housekeeping network. This conservative approach means that category 1 transfers are evaluated by using only the colored clonal groups and not singleton clones. A focus only on clonal groups for category 1 transfers will underrepresent the absolute number of LGT events for the entire data set. A further point of significance with regard to these definitions is that for category 1 the putative donor and recipient involved in the LGT are not determinable, whereas in category 2 they generally are.
A comparison of the control histories to each of the gene histories derived for the fluoroquinolone target loci indicates that there are regions of both branching similarity and differences (Fig. 1; Fig. 3a to d). For example, there is a group of four Japanese fluoroquinolone nonsusceptible isolates (indicated in red) from 1999 and 2000 which are of the same haplotype for all fluoroquinolone target loci as well as the housekeeping genes (Fig. 1; Fig. 3a to d). This sort of arrangement for the control history (Fig. 1) clearly supports these isolates as members of the same clone, and the same arrangement for each of the fluoroquinolone target loci (Fig. 3a to d) indicates that the fluoroquinolone target genes of these isolates were acquired through common ancestry and descent. A similar example involves the susceptible isolates 0.49SP94 and 0.57SW99, which have identical haplotypes for all statistical parsimony networks (Fig. 1; Fig. 3a to d). Indeed, the majority of fluoroquinolone gene sequences from the isolates of the colored clones group together and thus support the view of clonal dissemination of the fluoroquinolone target loci. Other cases support lateral DNA transfer involving one or more isolates of a clonal group and one or more fluoroquinolone target genes but never all four. For example, there is an international fluoroquinolone nonsusceptible clone apparent in the control history which includes two isolates from Japan, one isolate from Texas, and another isolate from Italy (indicated in orange in Fig. 1). These isolates have the identical haplotype for both gyrase genes, indicating no lateral transfer of these loci. However, the history underlying their parE genes is somewhat different (Fig. 3a). All four of these “orange” isolates have the same parE sequence; however, unlike the isolates with the control history, four additional isolates (isolates O.47SP00, 215.1Oh00, 132.1NY98, and 68.1PR00) also share this same haplotype and are very widely separated from the “orange” clone on the housekeeping network (Fig. 1). This is an example of our category 2 (see above) for lateral transfer and indicates that members of the “orange” clone have acted as donors in lateral DNA transfer events involving these other lineages. An example of our category 1 lateral transfer would involve the “blue” clonal group, composed of isolates 136.1PL98, 95.1BR98, 0.34PL96, and 72.1In00 (Fig. 1); the gyrB network clearly dissociates 136.1PL98 from the rest of the clones, with placement at virtually opposite ends of the network, indicating that 136.1PL98 has received a recombinant gyrB locus (Fig. 3d). Another example involves the “pink” clonal group (isolates 95.1Ma00 and 215.1Oh00) and the “light green” clonal group (isolates 0.19IS99 and 67.1FR92), which have identical gyrA sequences (Fig. 2b) but which are very dissimilar on the control network (Fig. 1). This suggests lateral transfer of gyrA involving these two clones, but the directionality cannot be assessed.
One of the advantages of using phylogenetic trees is that there are statistical tests available that allow one to make comparisons between topologies (branching arrangement) and thus assess whether the highest likelihood topology is significantly different from a constrained topology that reflects an alternative hypothesis of evolutionary history. A similarly agreed upon set of tests are presently not available for network comparisons. Using the conservative SH test (34, 35), we compared the highest-likelihood trees reconstructed from the fluoroquinolone target sequence data against trees reconstructed from the same data, but with the constraint that they had to contain the 11 clonal groups present in the control phylogeny. If the fluoroquinolone target loci in this set of isolates were vertically inherited, then they should conform to the clonal groups depicted on the control histories and the SH test run for each gene would not be significant. If the fluoroquinolone target genes are laterally transferred, then their evolutionary history will not conform to the clonal groups on the control phylogeny and the SH test will be significant. The result indicates that for all four fluoroquinolone target genes for this set of 58 isolates the null hypothesis of no difference between the constrained or clonal phylogeny and the highest-likelihood phylogeny was rejected, and thus, we accept the alternative hypothesis of lateral DNA transfer: gyrA, difference in -ln L = 14.8934 (P = 0.024); gyrB, difference in -ln L = 106.8542 (P = 0.000); parC, difference in -ln L = 56.8522 (P = 0.009); and parE, difference in -ln L = 82.7417 (P = 0.000). It is important to realize that this test does not incorporate any information regarding interclonal group relationships but, instead, merely assesses clonal group composition. Furthermore, the resulting gene phylogenies and statistical tests, employing principles very different from those involved in the network reconstruction, are in complete agreement with the conclusions derived from the networks. Thus, this phylogenetic statistical approach serves to corroborate our network-based conclusions regarding the lateral transfer of fluoroquinolone target loci.
Networks, on the other hand, have the conceptual advantage that they do not force the data into a tree-like (bifurcating) structure; however, it can be much more difficult to make comparisons between them. This is arguably somewhat exacerbated in our situation by the difference in the length of the data set used to reconstruct the control history and that used for each of the fluoroquinolone target loci. More specifically, in a comparison of networks, the confidence that one can place in a particular LGT event is related to the number of steps (nucleotide substitutions) that distinguish the isolates in question on the different networks. In a further effort to ensure that our network-based conclusions on transfer are as conservative as possible, we reconstructed additional networks specifically designed to assess whether the larger number of sequence positions in our housekeeping data (8,163 bp) compared to the number of sequence positions of individual fluoroquinolone target loci (1.6 to 2.3 kb, depending on the gene) were in any way biasing our conclusions on transfer. This was accomplished by creating multiple (n = 20), randomly reduced (jackknifing) housekeeping data sets (using the 8,163-bp data set as input) that match the number of sequence positions for each of the respective fluoroquinolone target loci, repeating the network reconstruction for each of those smaller data sets, and then evaluating whether each individual transfer hypothesis was still supported with these data sets of comparable size. All apparent instances of either category 1 or category 2 transfer were also supported by using these more reduced data sets and in some instances arguably provided more convincing evidence. For example, the adjacent isolates comprising the turquoise clone (Taiwan 23F-15) (isolates 292.1AT96 and 588.1JA99) are separated by 10 steps in the 8,163-bp housekeeping network (Fig. 1) and by 17 steps on the gyrA network (Fig. 3b); however, in housekeeping networks based on the same number of sequence positions as gyrA, these two isolates are separated by only two steps (n = 20; mean = 2.45; standard deviation = 1.39). Thus, random reduction of the data set simply reinforced the view that these two isolates have gyrA sequences that are very dissimilar (17 steps removed) from that expected from their evolutionary history (same clone; approximately 2 steps).
Another possible explanation for fluoroquinolone target loci from different clones with similar sequences is convergent molecular substitutions brought about by similar antibiotic selection pressure. If that were the case, then the majority of such substitutions should be amino acid changing in nature, and thus, evaluation of the lateral DNA transfer hypothesis by using only synonymous substitutions should yield a different result. Statistical parsimony networks reconstructed by using only synonymous substitutions tended to yield the same conclusions regarding lateral transfer events, indicating that convergent molecular selection due to antibiotic selective pressure is not the more parsimonious explanation. A conservative estimate of the number of lateral transfer events of fluoroquinolone target loci involving the lineages in this data set is approximately 17 (summarized in Table 2). The number of examples of lateral transfer for each of the loci seems roughly similar except for parC, for which the current data set supports fewer instances (Table 2).
TABLE 2.
Summary of LGT events involving fluoroquinolone loci from the set of 58 S. pneumoniae isolates examined in this study
| Gene | Transfer category | Description of transfer evidence (minimum number of necessary LGT events) |
|---|---|---|
| gyrA | 1 | Split of Taiwan 23F-15: 292.1AT98 from 588.1JA99 (one LGT) |
| gyrA | 2 | Taiwan 19F-14 (purple clone, isolates 458.1JA00, 526.1TX99, and 543.1CA99) and Poland 23F-16 (grey clone, isolates 71.1SW99 and 95.1ln00) with identical haplotypes (one LGT) |
| gyrA | 2 | Light green clone (isolates 0.19IS99 and 67.1FR92) and Tennessee 23F-4 (pink clone, isolates 95.1Ma00 and 215.1OH00) with identical haplotypes (one LGT) |
| gyrA | 2 | 0.47SP00, 132.1NY98, 128.2Ca000, and Spain 23F-1 with identical haplotypes (three LGTs) |
| gyrB | 1 | Split of blue clone (isolates 0.34PL96, 72.1ln00, and 95.1BR98) from 136.1PL98 (one LGT) |
| gyrB | 1 | Split of Tennessee 23F-4 (pink clone): 215.1Oh00 from 95.1Ma00 (one LGT) |
| gyrB | 2 | 0.74Oh94, 672.1US00, 68.1SW99, and Spain 23F-1 with identical haplotypes (three LGTs) |
| parE | 2 | 0.47SP00, 132.1NY98, 68.1PR00, 215.1Oh00, and orange clone (isolates 322.1JA98, 68.1AT98, 73.1TX98, and 67.1JA00) with identical haplotypes (three LGTs) |
| parE | 2 | 95.1BR98, 68.1Wa00, and Spain 23F-1 with identical haplotypes (two LGTs) |
| parC | 1 | Split of Poland 23F-16 (grey clone): 432.1JA00 from 71.1SW99 and 95.1ln0010 (one LGT) |
Many of the fluoroquinolone nonsusceptible isolates in this analysis carried substitutions in the QRDRs of the four loci, recognized as being important in conferring resistance to this class of antibiotics. This included the following substitutions and proportion of isolates, for each of the respective loci: parC, D78A/N (4.5%) and S79F/Y (54.5%), D83Y/G/N (11.4%); parE, D435N (13.6%), P454S (2.3%), and E474K (2.3%); gyrA, S81F/Y (52.3%) and E85K (2.3%); and gyrB, R379L (2.3%). Approximately 23% of the nonsusceptible isolates had wild-type parC; such proportions of fluoroquinolone nonsusceptible isolates without first-step parC mutations are in rough agreement with those from other studies (24). The vast majority of the isolates without parC mutations had mutations in gyrA and parE, with the most common parE mutations being L290F and A326V.
An important and interesting group to consider in the context of the lateral transfer of fluoroquinolone target genes is the Spain 23F-1 clone. We do not find any examples of these isolates “losing” their fluoroquinolone genes through recombination with another allele; note that “dark green” isolates are never dispersed in networks derived from their fluoroquinolone genes (Fig. 3). Instead, we find isolates from outside this clone with the same fluoroquinolone target genes as the Spain 23F-1 group (our category 2 indication of lateral transfer). This is true for all the fluoroquinolone target genes except parC (Fig. 3c). These lateral transfer events involve isolates throughout Europe and the United States covering a range of years. The fact that we see Spain 23F-1 as a donor of fluoroquinolone resistance genes and never as a recipient suggests the possibility that selection is somehow inhibiting recombination in Spain 23F-1. This might seem a bit counterintuitive, since Spain 23F-1 may have acquired its fluoroquinolone resistance character through a recombinational history. However, now that it has developed resistance to the current crop of fluoroquinolones, the selective pressure is different, and it might well be more beneficial to retain these particular alleles than lose them through recombination. It has been argued elsewhere that bacterial mismatch repair, which affects rates of recombination and mutation, might be in a state of evolutionary flux in bacteria (6). This could result from some environmental situations in which it is more beneficial to incorporate new DNA more efficiently, whereas in other situations it might be a better strategy to keep the genome more stable, thereby selecting for efficient mismatch repair. Antibiotics represent such a scenario; new antibiotics represent an enormous selective pressure, but for resistant clones the pressure becomes one of not losing resistance determinants through an undesirable recombination event. One would, however, need a much larger isolate sample size to properly evaluate this recipient hypothesis. Whatever the cause-effect explanations, Spain 23F-1, one of the most widely dispersed, highly successful, multidrug-resistant S. pneumoniae clones (29), is not only a health menace because of these characteristics, but also because its fluoroquinolone resistance determining genes are being laterally transferred to other lineages. Undoubtedly, this is also true for other major and minor clones; but the ubiquitous nature of the Spain 23F-1 clone, concomitant with the lateral transfer of the fluoroquinolone resistance associated genes arising from members of this clone, means that Spain 23F-1 is a doubly important source of fluoroquinolone resistance spread.
Our data and analysis indicate that fluoroquinolone resistance associated genes are being laterally transferred between S. pneumoniae isolates (minimum of 17 LGT events in a set of 58 isolates). This in turn suggests a potentially important means of resistance spread; however, the degree to which it is actually responsible for the spread of resistance remains a question. The histories of the fluoroquinolone target genes for the isolates evaluated in this data set are more often consistent with clonal dissemination than with lateral DNA transfer. For example, if we confine ourselves to a consideration of just our designated clonal groups, in the case of parE, of the 38 nonsusceptible isolates within these groups, 32 of them possess parE sequences which are consistent with a clonal dissemination hypothesis for this locus, 4 are consistent with lateral DNA transfer, and 2 are ambiguous. Roughly similar results are apparent for the other loci (clonal, LGT, and ambiguous): gyrA, 33, 3, and 2, respectively; gyrB, 34, 2, and 2, respectively; and parC: 33, 1, and 4, respectively.
Although the majority of fluoroquinolone target gene sequences in this S. pneumoniae data set can be explained on the basis of clonal dissemination, there are still a significant number which are more parsimoniously explained by intraspecific lateral DNA transfer; and in situations of high S. pneumoniae population density, such LGT events could be a very important means of resistance spread. The accurate assessment of this phenomenon is not trivial; but in the process of attempting to do so, we have made a concerted effort to be highly conservative in our identification of LGT loci. The degree to which LGT has played and is continuing to play a role in the spread of resistance to fluoroquinolones and other classes of antibiotics deserves further attention. Studies specifically assessing the relative frequency of intraspecific lateral DNA transfer of resistance conferring loci in different environmental situations would be of particular interest.
Acknowledgments
We thank the GSK US-based sequencing facility, under the management of Ganesh Sathe, for all their efforts on this project.
REFERENCES
- 1.Balsalobre, L., M. J. Ferrandiz, J. Linares, F. Tubau, and A. G. de la Campa. 2003. Viridans group streptococci are donors in horizontal transfer of topoisomerase IV genes to Streptococcus pneumoniae. Antimicrob. Agents Chemother. 47:2072-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canton, R., M. Morosini, M. C. Enright, and I. Morrissey. 2003. Worldwide incidence, molecular epidemiology and mutations implicated in fluoroquinolone-resistant Streptococcus pneumoniae: data from the global PROTEKT surveillance programme. J. Antimicrob. Chemother. 52:944-952. [DOI] [PubMed] [Google Scholar]
- 3.Clement, M., D. Posada, and K. A. Crandall. 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9:1657-1659. [DOI] [PubMed] [Google Scholar]
- 4.Coffey, T. J., C. G. Dowson, M. Daniels, J. Zhou, C. Martin, B. G. Spratt, and J. M. Musser. 1991. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol. Microbiol. 5:2255-2260. [DOI] [PubMed] [Google Scholar]
- 5.de la Campa, A. G., L. Balsalobre, C. Ardanuy, A. Fenoll, E. Perez-Trallero, J. Linares, and the Spanish Pneumococcal Infection Study Network G03/103. 2004. Fluoroquinolone resistance in penicillin-resistant Streptococcus pneumoniae clones, Spain. Emerg. Infect. Dis. 10:1751-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denamur, E., G. Lecointre, P. Darlu, O. Tenaillon, C. Acquaviva, C. Sayada, I. Sunjevaric, R. Rothstein, J. Elion, F. Taddei, M. Radman, and I. Matic. 2000. Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell 103:711-721. [DOI] [PubMed] [Google Scholar]
- 7.Dowson, C. G., T. J. Coffey, and B. G. Spratt. 1994. Origin and molecular epidemiology of penicillin-binding-protein-mediated resistance to beta-lactam antibiotics. Trends Microbiol. 2:361-366. [DOI] [PubMed] [Google Scholar]
- 8.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 9.Enright, M. C., and B. G. Spratt. 1999. Extensive variation in the ddl gene of penicillin-resistant Streptococcus pneumoniae results from a hitchhiking effect driven by the penicillin-binding protein 2b gene. Mol. Biol. Evol. 16:1687-1695. [DOI] [PubMed] [Google Scholar]
- 10.Feil, E. J., E. C. Holmes, D. E. Bessen, M. S. Chan, N. P. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felmingham, D., and J. Washington. 1999. Trends in the antimicrobial susceptibility of bacterial respiratory tract pathogens—findings of the Alexander Project 1992-1996. J. Chemother. 11:5-21. [DOI] [PubMed] [Google Scholar]
- 12.Ferrandiz, M. J., A. Fenoll, J. Linares, J., and A. G. de la Campa. 2000. Horizontal transfer of parC and gyrA in fluoroquinolone-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:840-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grassly, N. C., and E. C. Holmes. 1997. A likelihood method for the detection of selection and recombination using sequence data. Mol. Biol. Evol. 14:239-247. [DOI] [PubMed] [Google Scholar]
- 14.Hakenbeck, R., K. Kaminski, A. Konig, M. van der Linden, J. Paik, P. Reichmann, and D. Zahner. 1999. Penicillin-binding proteins in beta-lactam-resistant Streptococcus pneumoniae. Microb. Drug Resist. 5:91-99. [DOI] [PubMed] [Google Scholar]
- 15.Ho, P. L., W. C. Yam, T. K. Cheung, W. W. Ng, T. L. Que, D. N. Tsang, T. K. Ng, and W. H. Seto. 2001. Fluoroquinolone resistance among Streptococcus pneumoniae in Hong Kong linked to the Spanish 23F clone. Emerg. Infect. Dis. 7:906-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho, P. L., R. W. Yung, D. N. Tsang, T. L. Que, M. Ho, W. H. Seto, T. K. Ng, W. C. Yam, and W. W. Ng. 2001. Increasing resistance of Streptococcus pneumoniae to fluoroquinolones: results of a Hong Kong multicentre study in 2000. J. Antimicrob. Chemother. 48:659-665. [DOI] [PubMed] [Google Scholar]
- 17.Hooper, D. C. 2002. Fluoroquinolone resistance among gram-positive cocci. Lancet Infect. Dis. 2:530-538. [DOI] [PubMed] [Google Scholar]
- 18.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs, M. R., D. Felmingham, P. C. Appelbaum, and R. N. Gruneberg and The Alexander Project Group. 2003. The Alexander Project 1998-2000: susceptibility of pathogens isolated from community-acquired respiratory tract infection to commonly used antimicrobial agents. J. Antimicrob. Chemother. 52:229-246. [DOI] [PubMed] [Google Scholar]
- 20.Janoir, C., I. Podglajen, M. D. Kitzis, C. Poyart, and L. Gutmann. 1999. In vitro exchange of fluoroquinolone resistance determinants between Streptococcus pneumoniae and viridans streptococci and genomic organization of the parE-parC region in S. mitis. J. Infect. Dis. 180:555-558. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, C. N., W. H. Benjamin, Jr., S. A. Moser, S. K. Hollingshead, X. Zheng, M. J. Crain, M. H. Nahm, and K. B. Waites. 2003. Genetic relatedness of levofloxacin-nonsusceptible Streptococcus pneumoniae isolates from North America. J. Clin. Microbiol. 41:2458-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kell, C. M., J. Z. Jordens, M. Daniels, T. J. Coffey, J. Bates, J. Paul, C. Gilks, and B. G. Spratt. 1993. Molecular epidemiology of penicillin-resistant pneumococci isolated in Nairobi, Kenya. Infect. Immun. 61:4382-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, W. H. 1997. Molecular evolution. Sinauer Associates, Sunderland, Mass.
- 24.Lim, S., D. Bast, A. McGeer, J. de Azavedo, and D. E. Low. 2003. Antimicrobial susceptibility breakpoints and first-step parC mutations in Streptococcus pneumoniae: redefining fluoroquinolone resistance. Emerg. Infect. Dis. 9:833-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majewski, J., P. Zawadzki, P. Pickerill, F. M. Cohan, and C. G. Dowson. 2000. Barriers to genetic exchange between bacterial species: Streptococcus pneumoniae transformation. J. Bacteriol. 182:1016-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandell, L. A., L. R. Peterson, R. Wise, D. Hooper, D. E. Low, U. B. Schaad, K. P. Klugman, and P. Courvalin. 2002. The battle against emerging antibiotic resistance: should fluoroquinolones be used to treat children? Clin. Infect. Dis. 35:721-727. [DOI] [PubMed] [Google Scholar]
- 27.Mannheimer, S. B., L. W. Riley, and R. B. Roberts. 1996. Association of penicillin-resistant pneumococci with residence in a paediatric chronic care facility. J. Infect. Dis. 174:513-519. [DOI] [PubMed] [Google Scholar]
- 28.McGee, L., C. E. Goldsmith, and K. P. Klugman. 2002. Fluoroquinolone resistance among clinical isolates of Streptococcus pneumoniae belonging to international multiresistant clones. J. Antimicrob. Chemother. 49:173-176. [DOI] [PubMed] [Google Scholar]
- 29.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefevre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pletz, M. W., L. McGee, B. Beall, C. G. Whitney, and K. P. Klugman. 2005. Interspecies recombination in type II topoisomerase genes is not a major cause of fluoroquinolone resistance in invasive Streptococcus pneumoniae isolates in the United States. Antimicrob. Agents Chemother. 49:779-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pletz, M. W., L. McGee, J. Jorgensen, B. Beall, R. R. Facklam, C. G. Whitney, and K. P. Klugman. 2004. Levofloxacin-resistant invasive Streptococcus pneumoniae in the United States: evidence for clonal spread and the impact of conjugate pneumococcal vaccine. Antimicrob. Agents Chemother. 48:3491-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 33.Posada, D., and K. A. Crandall. 2001. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl. Acad. Sci. USA 98:13757-13762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimodaira, H. 2002. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51:492-508. [DOI] [PubMed] [Google Scholar]
- 35.Shimodaira, H., and M. Hasegawa. 1999. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17:1246-1247. [DOI] [PubMed] [Google Scholar]
- 36.Swofford, D. L. 2002. PAUP* Version 4.0b10. Sinauer Associates, Sunderland, Mass.
- 37.Templeton, A. R., K. A. Crandall, and C. F. Sing. 1992. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132:619-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yagupsky, P., N. Porat, D. Fraser, F. Prajgrod, M. Merires, L. McGee, K. P. Klugman, and R. Dagan. 1998. Acquisition, carriage, and transmission of pneumococci with decreased antibiotic susceptibility in young children attending a day care facility in southern Israel. J. Infect. Dis. 177:1003-1012. [DOI] [PubMed] [Google Scholar]
- 40.Zhanel, G. G., A. Walkty, K. Nichol, H. Smith, A. Noreddin, and D. J. Hoban. 2003. Molecular characterization of fluoroquinolone resistant Streptococcus pneumoniae clinical isolates obtained from across Canada. Diagn. Microbiol. Infect. Dis. 45:63-67. [DOI] [PubMed] [Google Scholar]