Abstract
Neisseria gonorrhoeae becomes resistant to tetracycline by two major mechanisms: expression of a plasmid-encoded TetM protein and mutations in endogenous genes (chromosomally mediated resistance). Early studies by Sparling and colleagues (P. F. Sparling F. A. J. Sarubbi, and E. Blackman, J. Bacteriol. 124:740-749, 1975) demonstrated that three genes were involved in high-level chromosomally mediated tetracycline resistance (MIC of tetracycline ≥ 2 μg/ml): ery-2 (now referred to as mtrR), penB, and tet-2. While the identities of the first two genes are known, the tet-2 gene has not been identified. We cloned the tet-2 gene, which confers tetracycline resistance, from tetracycline-resistant clinical isolate N. gonorrhoeae FA6140 and show that resistance is due to a single point mutation (Val-57 to Met) in the rpsJ gene (rpsJ1) encoding ribosomal protein S10. Moreover, the identical mutation was found in six distinct tetracycline-resistant clinical isolates in which the MIC of tetracycline was ≥2 μg/ml. Site-saturation mutagenesis of the codon for Val-57 identified two other amino acids (Leu and Gln) that conferred identical levels of resistance as the Met-57 mutation. The mutation maps to the vertex of a loop in S10 that is near the aminoacyl-tRNA site in the structure of the 30S ribosomal subunit from Thermus thermophilus, and the residue equivalent to Val-57 in T. thermophilus S10, Lys-55, is within 8 to 9 Å of bound tetracycline. These data suggest that large noncharged amino acids alter the rRNA structure near the tetracycline-binding site, leading to a lower affinity of the antibiotic.
Neisseria gonorrhoeae, the etiological agent for the sexually transmitted infection gonorrhea, is responsible for nearly 350,000 infections each year (7). One of the hallmarks of N. gonorrhoeae is its ability to become resistant to many of the drugs used to treat infections. For example, both penicillin and tetracycline used to be prescribed routinely to treat gonococcal infections, but increasing resistance to these antimicrobials eventually led to their discontinuation. The antibiotics currently recommended for the treatment of gonococcal infections are the β-lactam antibiotics ceftriaxone and cefixime and the fluoroquinolones ciprofloxacin, ofloxacin, and levofloxacin (8). Recently, however, the increasing prevalence of fluoroquinolone resistance in gonococcal infections on the West Coast and in certain patient populations (i.e., men who have sex with men) has necessitated the Centers for Disease Control and Prevention to recommend that fluoroquinolones not be used to treat gonococcal infections in these areas or in men who have sex with men (5, 6).
Two types of resistance to penicillin and tetracycline are found in N. gonorrhoeae: plasmid-mediated resistance and chromosomally mediated acquisition of mutated genes or loci (30). Plasmid-mediated resistance to penicillin and tetracycline is due to the production of a β-lactamase (27) and the TetM protein (23), respectively. For chromosomally mediated resistance, resistance genes are mutated versions of normal, endogenous genes such that each mutant gene incrementally increases resistance to the antibiotic until treatment failure occurs. These resistance determinants can be transferred, often in a specific order, from a resistant strain to a recipient susceptible strain by DNA transformation and homologous recombination.
The mechanisms of chromosomally mediated penicillin resistance are complicated and not completely understood. For example, at least four resistance determinants are involved in chromosomally mediated high-level penicillin resistance (defined as a penicillin MIC ≥ 2 mg/ml): (i) the penA determinant, which encodes mutations in PBP 2 that decrease its rate of acylation by penicillin (31); (ii) the mtrR locus, which is a single-base-pair deletion in the promoter for the mtrR repressor that results in the overexpression of the MtrC-MtrD-MtrE efflux pump (14, 26); (iii) the penB determinant, which encodes mutations in the PIB outer membrane porin (13, 24); and (iv) the ponA determinant, which encodes a mutation in PBP 1 that, like penA, decreases its rate of acylation by penicillin (28). In addition, at least one other gene is involved in high-level penicillin resistance, but its identity has remained elusive.
In contrast to penicillin resistance, the mechanisms of chromosomally mediated tetracycline resistance are more straightforward. Sparling and colleagues (30) showed that tetracycline resistance of an antibiotic-susceptible strain such as FA19 could be increased to intermediate levels (tetracycline MIC [MICtet] ≤ 1 μg/ml) following transformation with the mtrR (previously known as ery-2) and penB resistance determinants from a chromosomally mediated penicillin- and tetracycline-resistant N. gonorrhoeae (CMRNG) clinical isolate. High-level resistance to tetracycline at the same MIC as that in the donor strain could be obtained by transformation of an additional resistance determinant, termed tet-2, but its identity was not determined.
We report here on the identification of the tet-2 gene (renamed rpsJ1) as a Val-57-to-Met point mutation in the rpsJ gene encoding ribosomal protein S10. Our results indicate that this mutation is widespread in chromosomally mediated tetracycline-resistant strains and that other large, noncharged amino acids afford identical levels of resistance as the Met-57 mutation. Examination of the 3-Å structure of the 30S ribosomal subunit from Thermus thermophilus bound with tetracycline (3) indicated that the mutation maps to the vertex of a loop in the S10 protein that projects toward the aminoacyl-tRNA site of the small subunit and is within 8 to 9 Å of the antibiotic. Although the amino acid appears to be too far from the antibiotic to have a direct effect on binding, these data suggest that the mutation modulates the rRNA-binding site for tetracycline, thus lowering the affinity of the antibiotic for the ribosome.
MATERIALS AND METHODS
Bacterial strains and plasmids.
FA19 (a tetracycline-susceptible strain with an MICtet = 0.25 μg/ml) and FA6140 (with an MICtet = 4 μg/ml) have been described previously and were obtained from Fred Sparling, University of North Carolina at Chapel Hill (10, 21). Construction of PR100 (FA19 penA mtrR penB) was described previously (28). Clinical isolates of N. gonorrhoeae in which the MIC of tetracycline was ≥2 μg/ml (but ≤8 μg/ml) were obtained from several sources. Strains 10026, 10030, L2313, L4617, and L9715 were kindly provided by Marcia Hobbs, University of North Carolina at Chapel Hill, and were from a Wilson County, North Carolina, surveillance program (12, 16). Sequence analysis of the mtr and por gene regions indicated that these strains had frameshift mutations in the mtrR transcriptional regulator and penB mutations at residues 120 and 121 of the por gene. Strains 111, 114, and 131, each of which had the mtrR promoter mutation and penB mutations at residues 120 and 121 of the por gene, were obtained from Joan Knapp at the Centers for Disease Control and Prevention. MS11 was kindly provided by Janne Cannon, University of North Carolina at Chapel Hill. pMH-S10*, which was pUC18 containing the entire coding sequence of S10 with the Val-57-to-Met mutation and a 155-bp downstream sequence that included a tandem repeat of the Neisseria 10-bp uptake sequence (11), was used to transfer the rpsJ1 gene to susceptible strains (Table 1). Transformation of N. gonorrhoeae was carried out by standard methods (24, 28).
TABLE 1.
Strains and plasmids used in this study
| Strain | Description | Selection agent (concn [μg/ml]) |
|---|---|---|
| pMH-S10* | pUC18 containing the rpsJ1 allele from FA6140 | |
| pUC18-pilQ2 | pUC18 containing bp 1333 to 2196 of pilQ2 and an additional 300 bp of downstream sequence (39) | |
| FA19 | Tetracycline-susceptible laboratory strain | |
| FA6140 | CMRNG clinical isolate | |
| FA19 penA4 | FA19 transformed with FA6140 DNA | Penicillin G (0.016) |
| FA19 penA4 rpsJ1 | FA19 penA4 transformed with pMH-S10* | Tetracycline (0.37) |
| FA19 penA4 mtrR | FA19 penA4 transformed with FA6140 DNA | Erythromycin (0.5) |
| FA19 penA mtrR rpsJ1 | FA19 penA4 mtrR transformed with pMH-S10* | Tetracycline (0.37) |
| PR100 | FA19 penA4 mtrR transformed with FA6140 DNA; same as FA19 penA4 mtrR penB2 | Tetracycline (0.37) |
| PR100 rpsJ1 | PR100 transformed with pMH-S10* | Tetracycline (2.0) |
| PR100 pilQ2 | PR100 transformed with pUC18-pilQ2 | Penicillin G (1.2) |
| PR100 pilQ2 rpsJ1 | PR100 pilQ2 transformed with pMH-S10* | Tetracycline (3.0) |
Growth conditions and MIC determinations.
N. gonorrhoeae was grown on GC medium base (GCB) plates (Difco/BD, Franklin Lakes, NJ) with supplements I and II (19) at 37°C in a humidified 5% CO2-95% air atmosphere. Escherichia coli was cultured on Luria broth plates or in 2x YT medium. MICs were determined by the spot method, as described previously (28). Briefly, cells were grown on GCB plates and resuspended in GCB broth with supplements I and II and 10 mM MgCl2 at an optical density at 600 nm of 1 × 107 cells/ml, and 5-μl aliquots were spotted onto GCB plates containing increasing concentrations of tetracycline. The MIC was defined as the lowest concentration at which no more than five colonies grew following incubation for 24 h. At least two to four individual colonies from each transformation were tested. The MICs were identical in individual colonies from each transformation and in separate experiments over several days.
Cloning of the tet-2 (rpsJ1) gene from FA6140.
To isolate the tet-2 gene, chromosomal DNA from CMRNG strain FA6140 was digested with ApoI, which produces a 5′-AATT overhang that can be ligated into an EcoRI site, and size fractionated on a 0.7% agarose gel. Ten fractions of ApoI fragments ranging in size from 0.5 to 6 kb were purified from the gel and tested for their ability to transform PR100 to tetracycline resistance on GCB plates containing 2 μg/ml tetracycline. DNA fragments from a size fraction between 1.4 and 1.8 kb gave the highest transformation frequencies, and these were used to create a minilibrary in the EcoRI-digested pMH2 vector. pMH2 contained the origin of replication from pACYC177 (9) and the multiple-cloning site from pUC18. Nine hundred sixty clones were transferred into 10 96-well plates, and plasmids isolated from 10 cultures, each of which contained all of the clones from 1 of the 10 plates, were used in the transformation assay. Of the original 10 cultures examined, 5 gave rise to high numbers of resistant colonies following transformation. Similar experiments with cultures grown from a single plate eventually resulted in the identification of a plasmid capable of transforming PR100 to tetracycline resistance at a high frequency.
Identification of rpsJ alleles in tetracycline-resistant clinical isolates.
The rpsJ genes from a set of tetracycline-resistant clinical strains were amplified by PCR with primers flanking the gene. Single colonies of each strain were diluted into 25 μl water, boiled for 5 min, and centrifuged at top speed for 5 min. A 1-μl aliquot was used as the template in the PCR with rpsJ gene-specific primers. PCR products were purified with a PCR purification kit (Marligen, Gaithersburg, MD) and submitted directly for sequencing by the University of North Carolina sequencing facility.
Random mutagenesis of codon 57 in rpsJ.
Random mutagenesis of codon 57 was accomplished by overlap-extension PCR as described previously (15, 24). The sense mutagenic primer (5′-AACATTTTGCGTTCTCCGCACNNNAACAAAACTTCCCGTGAACAA-3′) comprised 21 bp preceding codon 57, 3 random bases, and 20 bp following codon 57, while the antisense primer (5′-GTGCGGAGAACGCAAAATGTT-3′) was complementary to the first 21 bp of the sense primer. pMH-S10 was used as a template. In the first set of reactions, a sense vector primer (relative to the direction of the rpsJ gene) and the antisense mutagenic primer or the sense mutagenic primer and an antisense vector primer were used to amplify the same template in separate amplification reactions. The resulting fragments were isolated and combined, and the full-length rpsJ gene with a randomized codon 57 was amplified in a second PCR to which only the vector primers were added. The resulting mutant fragments were purified and either used directly in a transformation reaction or subcloned into pUC18 to generate a minilibrary of mutant fragments. Sequencing of the PCR fragment or the minilibrary revealed the presence of 4 bases of roughly equal intensity at each of the three positions of the codon. Resistant transformants were passaged, and the rpsJ gene was amplified from genomic DNA and submitted to DNA sequencing.
RESULTS
Cloning of the tet-2 gene.
Previous data from Sparling and coworkers demonstrated that chromosomally mediated tetracycline resistance could be transferred from a CMRNG strain to a susceptible strain by transformation and selection (30). One of the genes identified in the prior study, termed tet-2, was specific for tetracycline resistance, but its identity was not determined. To clone the tet-2 gene, we created a minilibrary of size-fractionated ApoI fragments (between 1.4 and 1.8 kb) from genomic DNA isolated from the CMRNG strain FA6140 (MICtet = 4 μg/ml) (10) and isolated clones capable of transforming PR100 (FA19 penA4 mtrR penB2) to high-level tetracycline resistance (see Materials and Methods for more details). Five clones were identified, each of which contained an identical 1,554-bp fragment encompassing a portion of the tufA gene (encoding Ef-Tu) and the entire rpsJ gene encoding ribosomal protein S10. PCR amplification and transformation confirmed that the rpsJ gene conferred tetracycline resistance, and sequencing of the rpsJ gene from each of the five clones revealed an identical G-to-A point mutation at codon 57 that changed Val to Met (Fig. 1).
FIG. 1.
Identification of the tet-2 gene as a mutation in the rpsJ gene encoding ribosomal protein S10. Clones capable of conferring tetracycline resistance to PR100 (FA19 penA4 mtrR penB2; see Table 1) were isolated as described in Materials and Methods. Five separate clones each contained a portion of the tufA gene and the entire rpsJ gene encoding S10. Transformation experiments with the two genes amplified by PCR demonstrated that only the rpsJ gene was capable of conferring tetracycline resistance. Sequencing identified a single G→A mutation that changes Val-57 to Met in the rpsJ gene from FA6140 (a CMRNG strain) compared to the sequence of FA19. The same mutation was found in the rpsJ gene of MS11, a laboratory gonococcal strain (33), from a BLAST search of GenBank (accession number L36380).
Unexpectedly, the rpsJ sequence of MS11 (GenBank accession number L36380), a commonly used gonococcal laboratory strain that was isolated from a patient with uncomplicated gonorrhea (33), also contained the Val-57-to-Met mutation, along with an AN→QT double mutation at residues 2 and 3 (Fig. 1). To verify that the Val-57-to-Met mutation was responsible for tetracycline resistance, we transformed PR100 with MS11 DNA. As expected, MS11 DNA transformed PR100 to tetracycline resistance at a high frequency (∼1 × 10−4), and the MICs of these transformants were identical to those constructed with the isolated mutant rpsJ gene (data not shown). Moreover, PCR and transformation experiments showed that the Val-57-to-Met mutation and not the double mutation at the second and third residues conferred resistance to tetracycline. These data demonstrate that the mutant allele of rpsJ is identical to tet-2, that it is present in MS11, and that resistance is due to a single amino acid mutation at Val-57. This mutant allele is referred to hereafter as rpsJ1.
Identification of rpsJ1 in tetracycline-resistant clinical isolates.
To determine the extent of the rpsJ1 allele in chromosomally mediated tetracycline-resistant strains, we amplified and sequenced the rpsJ genes from a set of clinical isolates in which the MIC of tetracycline was ≥2 μg/ml but ≤8 μg/ml. Five of these strains, each with an MICtet of 2 μg/ml, were from a Wilson County, North Carolina, surveillance program carried out from 1992 to 1994 (12), while three additional strains (strains 111, 114, and 131), each with an MICtet of 4 μg/ml, were from an outbreak in Cincinnati, Ohio, in 1994. Each of the strains examined contained an ATG codon at position 57, identical to that found in FA6140 and MS11. No other differences were noted.
Spontaneous mutations in the rpsJ gene.
To determine if we could select for spontaneous rpsJ mutations in the laboratory, we plated FA19 (1 × 109 CFU) on GCB plates containing tetracycline at concentrations 1.2- to 1.8-fold higher than its MIC, but no resistant colonies were observed. To increase the frequency of mutation, we repeated the experiment with FA19 in which the mutS gene was deleted (28). Several hundred small colonies growing on the plates were pooled, the rpsJ gene was amplified from the genomic DNA of the pooled clones, and the resulting fragments were used to transform FA19 to tetracycline resistance. Whereas the rpsJ1 gene PCR product gave rise to thousands of resistant colonies, no resistant transformants were observed with the rpsJ gene amplified from the pool of spontaneous mutants. These data suggest that the frequency of spontaneous mutation in rpsJ is low and that the rpsJ1 allele arose in the 1970s as a rare event and was then disseminated throughout the species by homologous recombination.
High-level tetracycline resistance requires three genes.
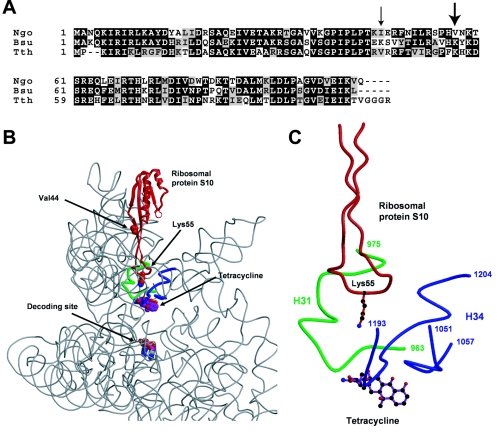
The level of tetracycline resistance in N. gonorrhoeae depends on the genetic background (30). To determine the effects of different resistance determinants in tetracycline resistance, we transformed a series of strains with pMH-S10*, which harbors the rpsJ1 allele (Table 1). The MICs of tetracycline for the recipient strains FA19, FA19 penA4, and FA19 penA4 mtrR were all identical (0.25 μg/ml), while the MIC for strain PR100 (FA19 penA4 mtrR penB2) was 1.0 μg/ml (Fig. 2). When the rpsJ1 allele was introduced into the first three strains, the MIC of tetracycline increased fourfold to 1 μg/ml (Fig. 2). When the rpsJ1 allele was introduced into PR100, which contains both mtrR and penB2, resistance again increased fourfold (to 4 μg/ml). The MIC of tetracycline for PR100 rpsJ1 was identical to that for FA6140, suggesting that mtrR, penB, and rpsJ1 are both necessary and sufficient for strains to reach high-level chromosomally mediated tetracycline resistance.
FIG. 2.
MICs of tetracycline for gonococcal strains with different genetic backgrounds. The open bars indicate the MIC of tetracycline for each of the indicated parental strains (see Table 1 for more information). The black bars indicate the MIC of tetracycline following transformation of each strain with the rpsJ1 allele encoding S10 with a Val-57-to-Met mutation. In each case, transfer of the rpsJ1 allele increased tetracycline resistance by three- to fourfold. The MICs were determined in three separate experiments with at least four individual transformants for each strain; no variation in the MICs was observed in the separate experiments.
We previously identified a spontaneous mutation in PR100, termed penC, which increased the MIC of tetracycline twofold (28), and have shown that resistance is due to a point mutation in the pilQ gene (pilQ2) that interferes with the stability of the PilQ secretin and decreases antibiotic influx (39). To examine the influence of the pilQ2 mutation on tetracycline resistance mediated by rpsJ1, we transformed PR100 pilQ2 with pMH-S10* and determined the MIC of tetracycline of the resulting strain. As shown in Fig. 2, the MIC of tetracycline for strain PR100 pilQ2 increased threefold (from 2 to 6 μg/ml) upon acquisition of the rpsJ1 allele. Thus, irrespective of the genetic background, the rpsJ1 determinant in N. gonorrhoeae increases tetracycline resistance between three- and fourfold.
Role of residue 57 in mediating tetracycline resistance in S10.
To understand at a molecular level how mutations at position 57 confer tetracycline resistance to N. gonorrhoeae, we randomized the codon for Val-57 and used the resulting rpsJ mutant PCR fragments to transform PR100 to increased tetracycline resistance. Twenty-two clones were isolated, and the rpsJ genes from these isolates were amplified and sequenced. From these 22 clones, three amino acids at position 57 were identified: Met, Leu, and Gln. Each of these mutations conferred identical levels of tetracycline resistance to PR100 (4 μg/ml) or FA19 (1 μg/ml), suggesting that large, uncharged amino acids at this position increase tetracycline resistance and that each of these amino acids affects resistance in an identical manner.
Each of the amino acids identified in our randomized codon screen were both large and hydrophobic, but because it was possible that not all codons were represented in our random screen, the necessity of having an amino acid that was both large and hydrophobic was unclear. Thus, we employed site-directed mutagenesis to change Val-57 to the large but charged amino acid Lys, which is the residue found in this position in the Thermus thermophilus S10 protein (Fig. 3A). Transformation of either FA19 or PR100 with the rpsJ gene containing the Val-57-to-Lys mutation and selection on 1.1 to 2 times the MIC of tetracycline did not yield resistant colonies at a rate greater than that for the buffer controls, even though the rpsJ1 allele generated hundreds of resistant colonies in the same experiment. These data demonstrate that the amino acid at position 57 must be both large and hydrophobic to increase the MIC of tetracycline in N. gonorrhoeae.
FIG. 3.
Proposed structural mechanism of the rpsJ1 mutation for increasing tetracycline resistance. (A) Sequence alignment of S10 from N. gonorrhoeae (Ngo), B. subtilis (Bsu), and T. thermophilus (Tth) using ClustalX (18). The small arrow identifies the position of the Lys-46-to-Glu mutation in B. subtilis that confers tetracycline resistance, while the larger arrow indicates the position of the Val-57 to Met mutation in N. gonorrhoeae characterized in this study. (B) A low-magnification view of the 30S ribosomal subunit structure from T. thermophilus (3) showing the rRNA backbone (gray) and the S10 protein (red). Also shown is the mRNA decoding site (marked by nucleotides A1492 and A1493) (4) and the major binding site of tetracycline. The two residues indicated in S10, Val-44 and Lys-55 (shown in Corey-Pauling-Koltum [CPK] format), are equivalent to Lys-46 of B. subtilis and Val-57 of N. gonorrhoeae, respectively. (C) A closer view of the small subunit showing rRNA helices H31 (green) and H34 (blue) that form the binding site for tetracycline along with the S10 ribosomal protein. Lys-55 forms contacts with H31 and H34. Panels B and C were made with molscript (20) and Raster3d (22).
Proposed mechanism of rpsJ1-mediated tetracycline resistance.
Tetracycline is known to bind to the 30S ribosome near the aminoacyl-tRNA binding site, thereby interfering with the binding of aminoacyl-tRNAs and blocking peptide chain elongation (29). Recently, high-resolution structures of the individual 30S and 50S ribosomal subunits and both subunits together have provided significant insight into ribosomal function and the mechanisms of inhibitors of protein synthesis (1, 37, 38). Of particular relevance to this study is the 3.0-Å structure of the Thermus thermophilus 30S ribosomal subunit bound with tetracycline (3, 4), which allows an examination at the structural level of the potential effect of a mutation at Val-57 of ribosomal protein S10.
S10 is an elongated protein (∼63 Å in its longest dimension) with two distinct regions: a mixed α/β globular region at one end of the protein and a long antiparallel β ribbon connected by a large loop at the other end (Fig. 3B). In the 30S structure, the loop at the end of the antiparallel β strands is situated near the top of the groove that contains the aminoacyl- and peptidyl-tRNA functional sites (A and P sites, respectively). In T. thermophilus S10, Lys-55 is equivalent to Val-57 of S10 from N. gonorrhoeae (Fig. 3A), and this residue is situated at the vertex of the loop extending toward the A site of the 30S subunit. The bound tetracycline is within 8.5 Å of the ɛ-NH2 group of Lys-55 (Fig. 3C), consistent with our results demonstrating that changes in this amino acid influence the binding of the antibiotic. Although the side chain is probably too far removed to cause a direct blocking of tetracycline binding, it is likely that large, hydrophobic side chains at this position alter the RNA structure in the vicinity of the bound tetracycline, hence decreasing the binding affinity of tetracycline.
DISCUSSION
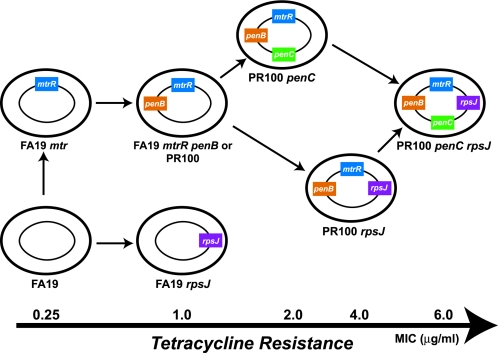
Tetracyclines were one of the first “broad-spectrum” antibiotics developed, and although they remain useful for the treatment of certain bacterial infections, their use has been seriously compromised by the widespread emergence of resistant strains. Most strains become resistant to tetracycline not by mutations in RNA or ribosomal proteins but by the expression of tetracycline-specific efflux pumps or by the production of proteins (TetM and TetO) that mimic the elongation factors and allow the release of the antibiotic from the ribosome (29). N. gonorrhoeae, however, is an exception; in this organism high-level chromosomally mediated resistance to tetracycline is mediated by a combination of three gene mutations: (i) the mtrR mutation, which results in overexpression of an nonspecific efflux pump (MtrC-MtrD-MtrE) that promotes the efflux of a range of hydrophobic agents and detergents; (ii) the penB determinant, which is a mutated porin IB that decreases the influx of tetracycline into the cell, and (iii) the rpsJ1 allele, which we have characterized in this study (Fig. 4). In addition, the penC (pilQ2) resistance determinant also increases tetracycline resistance, but the clinical significance of this mutation remains unproven (28). Of these, only the rpsJ1 determinant is specific for tetracycline resistance, making it the first resistance gene specific for this antibiotic in chromosomally mediated tetracycline-resistant gonococcal strains. Although the combination of these mutations does not confer a level of tetracycline resistance as high as that observed with tetracycline-specific efflux pumps or the TetM determinant, the mtrR-penB-rpsJ1 gene triad is highly effective and provides levels of resistance above those clinically achievable at the site of infection.
FIG. 4.
Schematic showing the various ways in which chromosomally mediated genes can increase tetracycline resistance. The colored boxes represent the various resistance genes discussed in the text. Chromosomally mediated clinical isolates that are classified as tetracycline resistant (MICtet ≥ 2 μg/ml) contain the penB, mtrR, and rpsJ1 resistance determinants. The MICs of resistant strains shown at the bottom are approximate; the actual MICs depend on which mtr mutation (14) and penB allele (24) are present. See the text for details.
Our results indicate that the rpsJ1 allele acts independently of other resistance factors and increases the MIC of tetracycline between three- and fourfold, no matter what the genetic background. In contrast, the other determinants involved in tetracycline resistance, mtrR, penB, and penC, increase tetracycline resistance only in certain backgrounds. For example, the MIC of tetracycline does not increase in strains upon acquisition of either the mtrR or penB resistance determinants alone but increases only when both resistance determinants are present (25, 30, 34). Likewise, the penC mutation does not increase tetracycline resistance unless both mtrR and penB are already present (39). The differences between rpsJ1 and the other determinants likely reflect the fact that rpsJ1 acts directly at the site of action of tetracycline, thereby increasing resistance by a similar amount beyond the existing level in the parental strain. In contrast, the other determinants all act by limiting the concentration of tetracycline in the cell, and there appears to be a synergism between the MtrC-MtrD-MtrE efflux pump and the porin IB protein such that both genes are necessary to decrease cellular levels of tetracycline (25).
The mechanism by which the rpsJ1 mutation arose and propagated throughout the species appears to be different from that for the penA resistance determinant. In penA-mediated resistance, interspecies recombination and horizontal transfer have resulted in a large array of different mutant penA alleles (2, 17, 32), such that the coding sequences of resistant penA alleles differ from those of wild-type strains even in positions that do not change the protein sequence. In contrast, the same Val-57-to-Met mutation was observed in multiple strains with distinct geographical locations that were isolated from the late 1970s through 1994. Interestingly, both Gln and Leu confer similar levels of resistance, but these changes were not observed in any of the clinical isolates. Because we were unable to readily select spontaneous mutations of the rpsJ gene in the laboratory, it seems reasonable to assume that the Val-57-to-Met mutation was a rare spontaneous mutation in an early isolate from the 1970s (when chromosomally mediated tetracycline resistance emerged) that then disseminated throughout the species by horizontal transfer. A similar mechanism is likely to have occurred with the ponA1 allele encoding PBP 1, which has a single-base-pair mutation (and subsequent amino acid change) within 2,400 bp of coding sequence (28).
In the absence of a high-resolution structure of the 30S ribosomal subunit from N. gonorrhoeae, the exact molecular mechanism by which the Val-57-to-Met mutation increases resistance to tetracycline cannot be conclusively proven. However, given the reasonably high level of homology between the T. thermophilus and N. gonorrhoeae S10 proteins (55% identity, 75% similarity), crystal structures of the 30S subunit from T. thermophilus provide a useful framework for understanding the effect of this mutation. In the structure of the 30S subunit in complex with tetracycline, the primary binding site of the antibiotic is observed in the A site, which comprises the RNA helices H31, H34, and H44 (Fig. 3C) (3). Although this site is formed exclusively by RNA, the surrounding ribosomal proteins, including S4, S6, and S14 as well as S10, presumably help maintain its precise architecture. From sequence alignments (Fig. 3A), Lys-55 in S10 from T. thermophilus is equivalent to Met-57 of gonococcal S10. Lys-55 is located at the apex of a large β ribbon, which points toward the tetracycline-binding site and contacts both H31 and H34. This residue is too distant, however, for any mutation to clash directly with tetracycline. Instead, such a mutation would likely perturb the local rRNA structure within this region by altering the contacts between S10 and the RNA helices H31 and H34. Importantly, such a modulation of RNA structure must lower the affinity of rRNA for tetracycline without any deleterious effects on protein synthesis. A similar scenario applies to other chromosomally mediated resistance genes, where a narrow window exists in which the gene mutation increases resistance without significantly disrupting the normal function of the protein. For example, mutations in the penA gene decrease the ability of PBP 2 to be inhibited by penicillin but do not prevent its normal function of peptide cross-linking.
Only one previous study has identified a mutation in a ribosomal protein that gives rise to tetracycline resistance. Williams and Smith (36) identified a tetracycline-resistant strain of Bacillus subtilis following ethyl methanesulfonate mutagenesis that showed an altered mobility of the ribosomal protein S10 on two-dimensional gel electrophoresis. This mutation was subsequently mapped to a Lys-46-to-Glu alteration in S10 (35). These data are consistent with a role of S10 in tetracycline binding, although the mutations are in distinct regions of the protein. The mutation in the B. subtilis S10 protein maps onto the antiparallel β ribbon of the T. thermophilus S10 structure toward the globular domain (Fig. 3A and B), but the mutation site is not in a conserved residue (its equivalent in T. thermophilus S10 is Val-44) and its role in ribosomal structure is difficult to predict. Surprisingly, the Lys-46-to-Glu mutation in B. subtilis S10 confers high levels of resistance to tetracycline (>50 μg/ml) relative to that conferred by the Val-57-to-Met mutation in gonococcal S10 (36). This suggests that the B. subtilis mutation causes a much more significant perturbation to the tetracycline-binding site on the ribosome than the rpsJ1 mutation. Consistent with this hypothesis, colonies with the Lys-46-to-Glu mutation grew considerably slower than wild-type cells (36), suggesting that the mutation causes a disruption in the positioning of the S10 protein in the ribosome and a subsequent decrease in the rate of protein synthesis.
In summary, our results show that tet-2, a determinant associated with chromosomally mediated tetracycline resistance, is a mutation in the rpsJ gene (rpsJ1) that encodes ribosomal protein S10. Thus, in chromosomally mediated tetracycline-resistant isolates, three resistance determinants, mtrR, penB, and rpsJ1, are both necessary and sufficient to achieve high-level tetracycline resistance. The MIC of tetracycline for specific isolates differs according to which of these determinants are present (Fig. 4). In addition, the MIC can also differ depending on the type of mtr mutation (strains with deletions or mutations in the mtrR transcriptional regulator are less resistant to a variety of hydrophobic agents than strains with a single-base-pair deletion in the mtrR promoter region) and on which penB allele is present (14, 24). In contrast to mtrR and penB, which also contribute to penicillin resistance, the rpsJ1 allele is specific for tetracycline resistance and likely acts directly by lowering the affinity of the ribosome for the antibiotic through alteration of its binding site. These results highlight the complex mechanisms by which gonococcal strains become resistant to tetracycline.
Acknowledgments
This work was supported by Public Health Service grants AI-36901 from the National Institute of Allergy and Infectious Diseases (to R.A.N.) and GM-66861 from the National Institute of General Medical Sciences (to C.D.).
REFERENCES
- 1.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289:905-920. [DOI] [PubMed] [Google Scholar]
- 2.Bowler, L. D., Q. Y. Zhang, J. Y. Riou, and B. G. Spratt. 1994. Interspecies recombination between the penA genes of Neisseria meningitidis and commensal Neisseria species during the emergence of penicillin resistance in N. meningitidis: natural events and laboratory simulation. J. Bacteriol. 176:333-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brodersen, D. E., W. M. Clemons, Jr., A. P. Carter, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103:1143-1154. [DOI] [PubMed] [Google Scholar]
- 4.Carter, A. P., W. M. Clemons, D. E. Brodersen, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407:340-348. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2004. Increases in fluoroquinolone-resistant Neisseria gonorrhoeae among men who have sex with men—United States, 2003, and revised recommendations for gonorrhea treatment, 2004. Morb. Mortal. Wkly. Rep. 53:335-338. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2002. Increases in fluoroquinolone-resistant Neisseria gonorrhoeae—Hawaii and California, 2001. Morb. Mortal. Wkly. Rep. 51:1041-1044. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2004. Sexually transmitted disease surveillance 2003 supplement: Gonococcal Isolate Surveillance Project (GISP) annual report—2003. Centers for Disease Control and Prevention, Atlanta, Ga.
- 8.Centers for Disease Control and Prevention. 2002. Sexually transmitted disease treatment guidelines 2002. Morb. Mortal. Wkly. Rep. 51:36-41. [Google Scholar]
- 9.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danielsson, D., H. Faruki, D. Dyer, and P. F. Sparling. 1986. Recombination near the antibiotic resistance locus penB results in antigenic variation of gonococcal outer membrane protein I. Infect. Immun. 52:529-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elkins, C., C. E. Thomas, H. S. Seifert, and P. F. Sparling. 1991. Species-specific uptake of DNA by gonococci is mediated by a 10-base-pair sequence. J. Bacteriol. 173:3911-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox, K. K., J. C. Thomas, D. H. Weiner, R. H. Davis, P. F. Sparling, and M. S. Cohen. 1999. Longitudinal evaluation of serovar-specific immunity to Neisseria gonorrhoeae. Am. J. Epidemiol. 149:353-358. [DOI] [PubMed] [Google Scholar]
- 13.Gill, M. J., S. Simjee, K. Al-Hattawi, B. D. Robertson, C. S. Easmon, and C. A. Ison. 1998. Gonococcal resistance to beta-lactams and tetracycline involves mutation in loop 3 of the porin encoded at the penB locus. Antimicrob. Agents Chemother. 42:2799-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagman, K. E., W. Pan, B. G. Spratt, J. T. Balthazar, R. C. Judd, and W. M. Shafer. 1995. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology 141:611-622. [DOI] [PubMed] [Google Scholar]
- 15.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 16.Hobbs, M. M., T. M. Alcorn, R. H. Davis, W. Fischer, J. C. Thomas, I. Martin, C. Ison, P. F. Sparling, and M. S. Cohen. 1999. Molecular typing of Neisseria gonorrhoeae causing repeated infections: evolution of porin during passage within a community. J. Infect. Dis. 179:371-381. [DOI] [PubMed] [Google Scholar]
- 17.Ito, M., T. Deguchi, K. S. Mizutani, M. Yasuda, S. Yokoi, S. Ito, Y. Takahashi, S. Ishihara, Y. Kawamura, and T. Ezaki. 2005. Emergence and spread of Neisseria gonorrhoeae clinical isolates harboring mosaic-like structure of penicillin-binding protein 2 in central Japan. Antimicrob. Agents Chemother. 49:137-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 19.Kellogg, D. S., W. L. Peacock, W. E. Deacon, L. Browh, and C. I. Perkle. 1963. Neisseria gonorrhoeae. I. Virulence genetically linked to colonial variation. J. Bacteriol. 85:1274-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 21.Maness, M. J., and P. F. Sparling. 1973. Multiple antibiotic resistance due to a single mutation in Neisseria gonorrhoeae. J. Infect. Dis. 128:321-330. [DOI] [PubMed] [Google Scholar]
- 22.Merritt, E. A., and M. E. P. Murphy. 1994. Raster3D Version 2.0: a program for photorealistic molecular graphics. Acta Crystallogr. D 50:869-873. [DOI] [PubMed] [Google Scholar]
- 23.Morse, S. A., S. R. Johnson, J. W. Biddle, and M. C. Roberts. 1986. High-level tetracycline resistance in Neisseria gonorrhoeae is result of acquisition of streptococcal tetM determinant. Antimicrob. Agents Chemother. 30:664-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olesky, M., M. Hobbs, and R. A. Nicholas. 2002. Identification and analysis of amino acid mutations in porin IB that mediate intermediate-level resistance to penicillin and tetracycline in Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 46:2811-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olesky, M., R. L. Rosenberg, and R. A. Nicholas. Submitted for publication.
- 26.Pan, W., and B. G. Spratt. 1994. Regulation of the permeability of the gonococcal cell envelope by the mtr system. Mol. Microbiol. 11:769-775. [DOI] [PubMed] [Google Scholar]
- 27.Phillips, I. 1976. Beta-lactamase producing penicillin-resistant gonococcus. Lancet ii:656-657. [DOI] [PubMed] [Google Scholar]
- 28.Ropp, P. A., M. Hu, M. Olesky, and R. A. Nicholas. 2002. Mutations in ponA, the gene encoding penicillin-binding protein 1, and a novel locus, penC, are required for high-level chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 46:769-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scholar, E., and W. B. Pratt. 2000. The antimicrobial drugs, 2nd ed. Oxford University Press, Inc., New York, N.Y.
- 30.Sparling, P. F., F. A. J. Sarubbi, and E. Blackman. 1975. Inheritance of low-level resistance to penicillin, tetracycline, and chloramphenicol in Neisseria gonorrhoeae. J. Bacteriol. 124:740-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spratt, B. G. 1988. Hybrid penicillin-binding proteins in penicillin-resistant strains of Neisseria gonorrhoeae. Nature 332:173-176. [DOI] [PubMed] [Google Scholar]
- 32.Spratt, B. G., Q. Y. Zhang, D. M. Jones, A. Hutchison, J. A. Brannigan, and C. G. Dowson. 1989. Recruitment of a penicillin-binding protein gene from Neisseria flavescens during the emergence of penicillin resistance in Neisseria meningitidis. Proc. Natl. Acad. Sci. USA 86:8988-8992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swanson, J. 1973. Studies on gonococcus infection. IV. Pili: their role in attachment of gonococci to tissue culture cells. J. Exp. Med. 137:571-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veal, W. L., R. A. Nicholas, and W. M. Shafer. 2002. Overexpression of the MtrC-MtrD-MtrE efflux pump due to an mtrR mutation is required for chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. J. Bacteriol. 184:5619-5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei, Y., and D. H. Bechhofer. 2002. Tetracycline induces stabilization of mRNA in Bacillus subtilis. J. Bacteriol. 184:889-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams, G., and I. Smith. 1979. Chromosomal mutations causing resistance to tetracycline in Bacillus subtilis. Mol. Gen. Genet. 177:23-29. [DOI] [PubMed] [Google Scholar]
- 37.Wimberly, B. T., D. E. Brodersen, W. M. J. Clemons, R. J. Morgan-Warren, A. P. Carter, C. Vonrhein, T. Hartsch, and V. Ramakrishnan. 2000. Structure of the 30S ribosomal subunit. Nature 407:327-339. [DOI] [PubMed] [Google Scholar]
- 38.Yusupov, M. M., G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. Cate, and H. F. Noller. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292:883-896. [DOI] [PubMed] [Google Scholar]
- 39.Zhao, S., D. M. Tobiason, M. Hu, H. S. Seifert, and R. A. Nicholas. 2005. The penC mutation conferring antibiotic resistance in Neisseria gonorrhoeae arises from a mutation in the PilQ secretin that interferes with multimer stability. Mol. Microbiol. 57:1238-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]