Abstract
A T578I mutation in MexB compromised the protein's contribution to antimicrobial resistance and negatively impacted its interaction with MexA. Mutations causing single amino acid changes in the C-terminal domain of MexA (R221H, L245F, E254K, and V259I) suppressed the antimicrobial susceptibility of a MexBT578I-expressing Pseudomonas aeruginosa strain and restored a MexA interaction with MexBT578I. These data confirm the importance of the MexA C-terminal region in MexB binding and the likely significance of the region surrounding T587I of MexB in MexA interaction.
Pseudomonas aeruginosa is an opportunistic human pathogen characterized by an innate resistance to multiple antimicrobials (6), resistance increasingly attributable, at least in part, to the operation of broadly specific, multidrug efflux systems of the resistance-nodulation-division (RND) family (16). Several RND family multidrug efflux systems have been described in Pseudomonas aeruginosa, although the major system contributing to intrinsic multidrug resistance is encoded by the mexAB-oprM operon (16, 17). The MexAB-OprM efflux system consists of an inner membrane drug-proton antiporter (the RND component) (MexB), an outer membrane channel-forming component (OprM), and a periplasmic membrane fusion protein (MFP) (MexA) (16, 17). Crystal structures have been reported for MexA (2, 7) and OprM (1), and a MexB structure (11) has been derived from modeling on the available structure of the homologous AcrB protein (14), although details of pump assembly, including the identities of interacting domains of individual pump constituents, remain largely unknown.
In vivo interactions between MexA and MexB (12, 15) and MexA and OprM (12) have been confirmed, and the MexAB-OprM tripartite complex has been recovered from P. aeruginosa in the absence of cross-linking (12; D. Nehme and K. Poole, unpublished data). Interestingly, MexA association with MexB is dependent upon the presence of OprM (12, 15), although MexA-OprM association may be independent of MexB (12). Similarly, genetic (5) and biochemical (9, 23, 24) studies have confirmed in vivo interactions between AcrA, AcrB, and TolC in Escherichia coli, and an AcrAB-TolC complex is also recoverable from E. coli without prior cross-linking (23). A C-terminal domain of AcrA is implicated in the binding of this MFP to its cognate RND component, AcrB (4, 24), and while mutations in the corresponding region of MexA have been isolated and shown to abrogate MexA function (15), the importance of this region vis-à-vis MexB binding has not been established. The three-dimensional model of MexB identifies a region of the protein likely to be involved in MexA binding, and indeed, a mutation here (T578I) compromised MexB activity (11). To assess, then, the involvement of the MexA C-terminal domain in MexB binding, MexA suppressors of the T578I mutation in MexB were recovered and mapped. We report here the recovery of several C-terminal MexA suppressor mutations that restore binding to the MexBT578I protein in vivo.
The strains and plasmids used in this study are listed in Table 1. All bacterial strains were grown as indicated previously (15). Plasmids derived from pRK415 were maintained with tetracycline (10 μg/ml, E. coli; 30 μg/ml, P. aeruginosa K2275), while plasmids derived from pMMB206 were maintained with chloramphenicol (10 μg/ml, E. coli; 10 μg/ml, P. aeruginosa K2275). Plasmid pDN34 encoding MexBT578I was constructed by cloning a 4.5-kb EcoRI fragment from pJKM15 (11) carrying the mexB(T578I) gene into EcoRI-restricted pMMB206. Plasmid pDN39 encoding MexBE864K was similarly constructed by cloning the mexB(E864K) gene from pJKM16 (11) into EcoRI-restricted pMMB206. PCR was performed according to published protocols (15) except for the addition of 5% (vol/vol) dimethyl sulfoxide to the reaction mixture and the use of an annealing temperature of 60°C. DNA sequencing was performed by ACGT Corporation (Toronto, Ontario, Canada). The antimicrobial susceptibilities of P. aeruginosa K2275 and its plasmid-containing derivatives were assessed as described previously (15) using an isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM)-supplemented growth medium. The expression of the MexA and MexB proteins in P. aeruginosa and E. coli strains was assessed following sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western immunoblotting of whole-cell extracts prepared from overnight LB cultures (19) with anti-MexA (15) and anti-MexB (22) antisera.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Properties or genotypea | Source or reference |
|---|---|---|
| P. aeruginosa | ||
| K870 | Spontaneous Smr derivative of wild-type strain PAO1 | 18 |
| K2275 | K870 ΔmexR ΔmexAB | 15 |
| K1589 | K870 ΔmexR ΔmexB | 8 |
| E. coli | ||
| DH5α | φ80dlacZΔM15 Δ(lacZYA-argF) endA1 recA1 | 3 |
| S17-1 | thi pro hsdR recA Tra+ | 21 |
| Plasmids | ||
| pRK415 | P. aeruginosa-E. coli shuttle cloning vector; Tcr | 10 |
| pDN3 | pRK415::mexA | 15 |
| pDN30 | pRK415::mexA(R221H) | This study |
| pDN31 | pRK415::mexA(L245F) | This study |
| pDN32 | pRK415::mexA(E254K) | This study |
| pDN33 | pRK415::mexA(V259I) | This study |
| pJKM15 | pRK415::mexB(T578I) | 11 |
| pJKM16 | pRK415::mexB(E864K) | 11 |
| pMMB206 | P. aeruginosa-E. coli shuttle cloning vector; Cmr | 13 |
| pDN25 | pMMB206::mexB-His | 15 |
| pDN38 | pMMB206::mexA-mexB-His | This study |
| pDN34 | pMMB206::mexB(T578I) | This study |
| pDN35 | pMMB206::mexB(T578I)-His | This study |
| pDN36 | pMMB206::mexA-mexB(T578I)-His | This study |
| pDN37 | pMMB206::mexA(L245F)-mexB(T578I)-His | This study |
| pDN39 | pMMB206::mexB(E864K) | This study |
Tcr, tetracycline resistance; Cmr, chloramphenicol resistance; mexB-His, mexB gene was engineered to produce a MexB protein with a C-terminal hexahistidine tag; mexB(T578I)-His, mexB gene encoding MexB with a T578I substitution was engineered to produce a MexB protein with a C-terminal hexahistidine tag. Amino acid changes in the MexA or MexB proteins encoded by the indicated plasmids are shown in parentheses.
A T578I mutation in MexB compromises interaction with MexA.
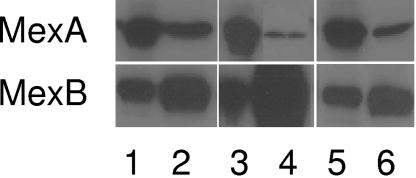
A previous study conducted in this lab identified a T578I mutation in MexB that severely compromised its ability to provide antibiotic resistance in a P. aeruginosa strain lacking a chromosomally encoded MexB protein; this mutation dramatically reduced the resistance of the strain to a wide range of MexA-MexB-OprM antimicrobial substrates (11) (Table 2). Interestingly, T578 occurs in a region of MexB that corresponds to a region in the homologous AcrB protein of the E. coli AcrAB-TolC pump implicated in interaction with its MFP component, AcrA (14). Accordingly, we predicted that MexBT578I was compromised in its ability to provide antibiotic resistance because it was unable to interact with MexA. To assess this directly, P. aeruginosa K2275 expressing plasmid-encoded MexA and wild-type MexB-His (histidine-tagged MexB) (from pDN38) or MexA and MexBT578I-His (from pDN36) was extracted with detergent, and the extracts were incubated with Ni-nitrilotriacetic acid (Ni-NTA) agarose beads (QIAGEN, Mississauga, Ontario, Canada) to recover the histidine-tagged MexB proteins as described previously (15). Corecovery of MexA (assessed using immunoblotting) was then used as a measure of MexA binding to the corresponding MexB protein in vivo (15). The mexB(T578I) gene was first histidine tagged by excising the 3′ end of wild-type mexB-His from plasmid pDN25 as a KpnI-HindIII fragment and cloning it into KpnI-HindIII-restricted pDN34 to yield pDN35. This effectively swapped the untagged 3′ end of mexB(T578I) with a His-tagged 3′ end. To introduce the wild-type mexA gene into plasmids pDN25 and pDN35, the mexA gene was amplified from plasmid pDN3 using primers JT-28-EcoRI (5′-GAATTCGAATTCGAATGTAAGTATTTTGCCTGC-3′; tandem EcoRI sites underlined) and JT-27 (5′-GAGCTCGAGCTCGATCACCCACGCGAAAATGG-3′). The PCR product was digested with EcoRI, freeing a mexA-containing 730-bp fragment from the PCR product, and cloned into EcoRI-restricted pDN25. The resulting vector, pDN38, carried the wild-type mexA gene upstream of mexB-His, with both genes under the control of the resident plac promoter of pDN38. The same mexA-containing fragment was cloned into pDN35 upstream of the mexB(T578I)-His gene of this vector, yielding pDN36 in which mexA and mexB(T578I)-His were similarly controlled by plac. Plasmids pDN38 and pDN36 were subsequently mobilized into P. aeruginosa K2275 from E. coli DH5α using a triparental mating procedure (25) and plasmid-containing isolates selected on chloramphenicol (10 μg/ml) and imipenem (0.5 μg/ml; to counterselect donor and helper E. coli strains). As seen previously, MexA was readily recovered together with wild-type MexB-His on Ni-NTA agarose beads (Fig. 1, lane 2, top panel), confirming the ability of these proteins to interact in vivo. In contrast, very little MexA was recovered together with MexBT578I-His (Fig. 1, lane 4, top panel), despite the even higher level of MexBT578I-His recovered from the Ni-NTA agarose beads in this experiment compared with that of wild-type MexB-His (Fig. 1, bottom panel, compare lanes 2 and 4). Clearly, MexBT578I-His was less able to bind MexA than its wild-type counterpart was.
TABLE 2.
Antibiotic susceptibility of P. aeruginosa K2275 expressing mutant MexA and MexB proteinsa
| Plasmids | MexAB proteins expressedb | MIC (μg/ml) forc:
|
|||
|---|---|---|---|---|---|
| CAR | NOV | NAL | CEF | ||
| pDN3, pDN25 | MexAWT, MexBWT | 128 | 512 | 128 | 8 |
| pDN3, pDN34 | MexAWT, MexBT578I | 4 | 32 | 16 | 1 |
| pDN30, pDN34 | MexAR221H, MexBT578I | 128 | 256 | 128 | 8 |
| pDN31, pDN34 | MexAL245F, MexBT578I | 128 | 256 | 128 | 8 |
| pDN32, pDN34 | MexAE254K, MexBT578I | 128 | 256 | 128 | 8 |
| pDN33, pDN34 | MexAV259I, MexBT578I | 128 | 256 | 128 | 8 |
| pDN30, pDN39 | MexAR221H, MexBE864K | 2 | 16 | 8 | 1 |
| pDN31, pDN39 | MexAL245F, MexBE864K | 2 | 16 | 8 | 1 |
| pDN32, pDN39 | MexAE254K, MexBE864K | 2 | 16 | 8 | 1 |
| pDN33, pDN39 | MexAV259I, MexBE864K | 2 | 16 | 8 | 1 |
| pDN3, pDN39 | MexAWT, MexBE864K | 2 | 16 | 8 | 1 |
P. aeruginosa K2275 harboring the indicated plasmids was used to perform antibiotic susceptibility testing as described in the text. IPTG was included in the growth medium to induce MexB expression from pMMB206-derived plasmids.
The MexA and MexB proteins expressed from the indicated plasmids are shown, with mutations indicated in subscript. MexAWT, wild-type MexA.
CAR, carbenicillin; NOV, novobiocin; NAL, nalidixic acid; CEF, cefoperazone.
FIG. 1.
Western immunoblot assessing in vivo binding of wild-type MexA and MexAL245F to MexBT578I-His. Cell envelopes from P. aeruginosa strain K2275 carrying plasmid pDN38 (pMMB206::mexA-mexB-His) (lanes 1 and 2), pDN36 [pMMB206::mexA-mexB(T578I)-His] (lanes 3 and 4), and pDN37 [pMMB206::mexA(L245F)-mexB(T578I)-His] (lanes 5 and 6) were extracted as described in the text. Triton X-100-soluble extracts of cell envelope preparations were incubated with Ni-NTA agarose and Triton X-100-soluble cell envelope extracts (odd-numbered lanes), and elution fractions off Ni-NTA agarose (even-numbered lanes) were immunoblotted and developed with antibodies to MexA (top panel) and MexB (bottom panel).
Isolation of MexA suppressors of MexBT578I.
To assess further the significance of T578 vis-à-vis MexB interaction with MexA and, possibly, to identify regions or residues of MexA important for this interaction, attempts were made to recover MexA suppressors of MexBT578I. Thus, mexA-carrying plasmid pDN3 was submitted to hydroxylamine chemical mutagenesis as described previously (15). The pool of mutagenized plasmids was introduced into E. coli S17-1 via electroporation (20) and mobilized into P. aeruginosa K2275 (ΔmexR ΔmexAB) harboring the MexBT578I-encoding plasmid pDN34 via conjugation (15). Selection of suppressor mutations was performed by spreading the conjugation mixture on LB agar containing tetracycline (10 μg/ml), carbenicillin (20 μg/ml), imipenem (0.5 μg/ml; as counterselection against the donor E. coli), and 1 mM IPTG [to induce transcription of mexB(T578I)]. P. aeruginosa K2275 expressing the plasmid-encoded MexBT578I (i.e., a MexA-MexBT578I-OprM pump) is unable to grow in the presence of 20 μg/ml carbenicillin, while P. aeruginosa expressing a wild-type functional MexAB-OprM system can. Therefore, potential MexA suppressors would restore growth of K2275(pDN34) on carbenicillin. Of several transconjugants carrying possible MexA suppressors, four showed increased resistance to several antimicrobials known to be substrates for MexAB-OprM (Table 2), consistent with these transconjugants harboring a MexAB-OprM pump with restored activity. Isolation of the mutagenized pDN3 from each of these transconjugants and their subsequent reintroduction into P. aeruginosa K2275 confirmed that restored multidrug resistance in K2275 was indeed dependent upon the mutagenized mexA gene in each instance. Nucleotide sequencing of these mexA genes confirmed single mutations in each of the genes producing single amino acid changes in MexA (R221H, L245F, E254K and V259I; Table 2). The MexA suppressor mutations were, however, specific to MexBT578I and did not rescue the hypersusceptibility phenotype attributable to a MexB mutation (E864K; Table 2) situated in the predicted vestibule region of the MexB trimer (11). A representative MexA suppressor, mexA(L245F), was introduced into plasmid pDN35 as described above for wild-type mexA, using plasmid pDN31 as a template for PCR amplification of the gene, and the resultant vector, pDN37, encoding both MexAL245F and MexBT578I-His, was mobilized into P. aeruginosa K2275. As expected, MexAL245F showed markedly improved binding to MexBT578I-His (Fig. 1, lane 6) relative to wild-type MexA.
MexA suppressor mutations map near the putative MexB-binding domain.
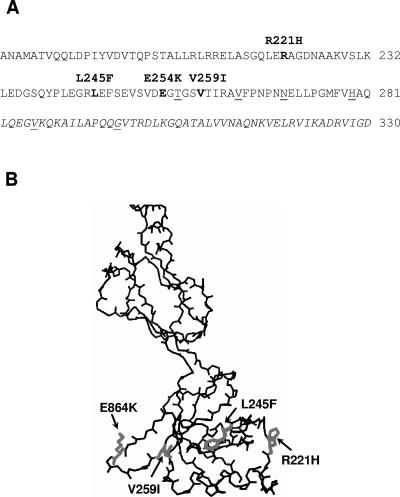
A C-terminal domain of AcrA was previously shown to be instrumental in the interaction of this MFP component of the AcrAB-TolC efflux system with AcrB, its cognate RND partner (4). Interestingly, the four suppressor mutations isolated in the course of this study map very close to the corresponding region of MexA, with three of the changes (L245F, E254K, and V259I) within 30 amino acids of this region (Fig. 2A). This region was previously implicated in MexB binding and was the site of several amino acid changes that negatively impacted MexA function (15) (Fig. 2A). Interestingly, too, the mutant residues all map to a common face of the MexA structure (Fig. 2B), a face that in light of the above data may well be involved in MexB interaction. Still, as these occur outside the proposed interaction domain and upstream of mutations previously shown to impact MexA function, it may also be that these mutations have instead a common influence on the disposition of the “downstream” MexB-binding region (not resolved in the available MexA crystal structure), indirectly facilitating improved interaction with the MexBT578I protein. In possible agreement with this, the suppressor MexAs were still functional with wild-type MexB (i.e., the cloned mexA suppressor genes complemented the multidrug-susceptible phenotype of a ΔmexA P. aeruginosa strain) (data not shown).
FIG. 2.
Mutations in MexA that suppress the antibiotic susceptibility phenotype of MexBT578I-expressing P. aeruginosa. (A) Linear sequence of the MexA C-terminal region (residues 184 to 330). Suppressor mutations are highlighted above the corresponding residue, which is shown in bold type in the MexA sequence shown. Previously identified (15) residues in MexA whose mutation compromised MexA function are underlined. The region of MexB that aligns with a proposed AcrB-binding domain of AcrA (4) is italicized. (B) Three-dimensional structure of monomeric MexA29-259 (PDB identification number 1VF7; http://www.ncbi.nlm.nih.gov) with the mutated residues indicated in gray.
Acknowledgments
This work was supported by an operating grant from the Canadian Cystic Fibrosis Foundation (CCFF). D.N. was the recipient of a CCFF studentship.
REFERENCES
- 1.Akama, H., M. Kanemaki, M. Yoshimura, T. Tsukihara, T. Kashiwagi, H. Yoneyama, S. I. Narita, A. Nakagawa, and T. Nakae. 2004. Crystal structure of the drug-discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: dual modes of membrane anchoring and occluded cavity end. J. Biol. Chem. 17:52816-52819. [DOI] [PubMed] [Google Scholar]
- 2.Akama, H., T. Matsuura, S. Kashiwagi, H. Yoneyama, S. Narita, T. Tsukihara, A. Nakagawa, and T. Nakae. 2004. Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J. Biol. Chem. 279:25939-25942. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Short protocols in molecular biology, 2nd ed. John Wiley & Sons, Inc., New York, N.Y.
- 4.Elkins, C. A., and H. Nikaido. 2003. Chimeric analysis of AcrA function reveals the importance of its C-terminal domain in its interaction with the AcrB multidrug efflux pump. J. Bacteriol. 185:5349-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerken, H., and R. Misra. 2004. Genetic evidence for functional interactions between TolC and AcrA proteins of a major antibiotic efflux pump of Escherichia coli. Mol. Microbiol. 54:620-631. [DOI] [PubMed] [Google Scholar]
- 6.Hancock, R. E. W., and D. P. Speert. 2000. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Res. Updates 3:247-255. [DOI] [PubMed] [Google Scholar]
- 7.Higgins, M. K., E. Bokma, E. Koronakis, C. Hughes, and V. Koronakis. 2004. Structure of the periplasmic component of a bacterial drug efflux pump. Proc. Natl. Acad. Sci. USA 101:9994-9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirakata, Y., R. Srikumar, K. Poole, N. Gotoh, T. Suematsu, S. Kohno, S. Kamihira, R. E. Hancock, and D. P. Speert. 2002. Multidrug efflux systems play an important role in the invasiveness of Pseudomonas aeruginosa. J. Exp. Med. 196:109-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Husain, F., M. Humbard, and R. Misra. 2004. Interaction between the TolC and AcrA proteins of a multidrug efflux system of Escherichia coli. J. Bacteriol. 186:8533-8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 11.Middlemiss, J. K., and K. Poole. 2004. Differential impact of MexB mutations on substrate selectivity of the MexAB-OprM multidrug efflux pump of Pseudomonas aeruginosa. J. Bacteriol. 186:1258-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mokhonov, V. V., E. I. Mokhonova, H. Akama, and T. Nakae. 2004. Role of the membrane fusion protein in the assembly of resistance-nodulation-cell division multidrug efflux pump in Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 322:483-489. [DOI] [PubMed] [Google Scholar]
- 13.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 14.Murakami, S., R. Nakashima, E. Yamashita, and A. Yamaguchi. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587-593. [DOI] [PubMed] [Google Scholar]
- 15.Nehme, D., X. Z. Li, R. Elliot, and K. Poole. 2004. Assembly of the MexAB-OprM multidrug efflux system of Pseudomonas aeruginosa: identification and characterization of mutations in mexA compromising MexA multimerization and interaction with MexB. J. Bacteriol. 186:2973-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poole, K. 2004. Efflux-mediated multiresistance in Gram-negative bacteria. Clin. Microbiol. Infect. 10:12-26. [DOI] [PubMed] [Google Scholar]
- 17.Poole, K. 2004. Efflux pumps, p. 635-674. In J.-L. Ramos (ed.), Pseudomonas, vol. I. Genomics, life style and molecular architecture. Kluwer Academic, Plenum Publishers, New York, N.Y. [Google Scholar]
- 18.Poole, K., K. Tetro, Q. Zhao, S. Neshat, D. Heinrichs, and N. Bianco. 1996. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob. Agents Chemother. 40:2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redly, A., and K. Poole. 2003. Pyoverdine-mediated regulation of FpvA synthesis in Pseudomonas aeruginosa: involvement of a probable ECF sigma factor, FpvI. J. Bacteriol. 185:1261-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Simon, R., U. Priefer, and A. Puehler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 22.Srikumar, R., T. Kon, N. Gotoh, and K. Poole. 1998. Expression of Pseudomonas aeruginosa multidrug efflux pumps MexA-MexB-OprM and MexC-MexD-OprJ in a multidrug-sensitive Escherichia coli strain. Antimicrob. Agents Chemother. 42:65-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tikhonova, E. B., and H. I. Zgurskaya. 2004. AcrA, AcrB, and TolC of Escherichia coli form a stable intermembrane multidrug efflux complex. J. Biol. Chem. 279:32116-32124. [DOI] [PubMed] [Google Scholar]
- 24.Touze, T., J. Eswaran, E. Bokma, E. Koronakis, C. Hughes, and V. Koronakis. 2004. Interactions underlying assembly of the Escherichia coli AcrAB-TolC multidrug efflux system. Mol. Microbiol. 53:697-706. [DOI] [PubMed] [Google Scholar]
- 25.Zhao, Q., X.-Z. Li, R. Srikumar, and K. Poole. 1998. Contribution of outer membrane efflux protein OprM to antibiotic resistance in Pseudomonas aeruginosa independent of MexAB. Antimicrob. Agents Chemother. 42:1682-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]