Abstract
A VIM-5-producing Enterobacter cloacae isolate (EDV/1) was identified in a collection of clinical strains stored before 2002. The gene, blaVIM-5, was located on a 2,712-bp BamHI-HindIII fragment of a 23-kbp (approximately) nonconjugative plasmid (pEDV5) in a class 1 integron as a single gene cassette.
Transferable metallo-β-lactamases (MβLs) are important resistance determinants in Acinetobacter spp. and Pseudomonas aeruginosa in hospitals (15, 16). MβLs, not yet frequent, are also disseminating among the members of the family Enterobacteriaceae (4, 11). Transferable MβLs belong to two major groups, IMP and VIM alleles. Recently, a novel variant of the VIM family, VIM-5, was detected in Klebsiella pneumoniae and P. aeruginosa clinical strains in Turkey (1, 10). We detected a VIM-5-producing Enterobacter cloacae isolate in a collection of clinical strains isolated before 2002 in a university hospital located in the southeast part of Turkey.
E. cloacae EDV/1 was identified in a collection of clinical isolates obtained from the Hospital of Dicle University (Turkey). It was isolated before 2002. The Escherichia coli strain ER2267 obtained from New England Biolabs was used as a host for cloning procedures, and the rifampin-resistant E. coli strain J-53-2 was used for transconjugation experiments (18). Plasmids pACYCDuet-1 (Novagen, Darmstadt, Germany) and pUC19 were the vectors used for cloning and subcloning experiments.
The MICs of antibiotics were determined by the broth microdilution method according to the guidelines of the Clinical and Laboratory Standards Institute (formerly NCCLS). Plasmid isolation and transconjugation experiments were described elsewhere (18). Hybridization and detection steps were accomplished with the digoxigenin (DIG)-dUTP detection kit as recommended by the manufacturer (Boehringer Mannheim, GmbH) (19). The sequencing method was dye terminator cycle sequencing with the ABI Prism BigDye Terminator kit (Applied Biosystems, Foster City, Calif.).
VIM-5 was purified through a Q-Sepharose column (HiTrap Q; Amersham) and later polished through a Sephacryl S-100 HR column (packing dimensions, 10 by 300 mm). Protein contents were measured by the Bio-Rad protein assay (Richmond, CA). The relative molecular mass of VIM-5 was estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis. The kcat and Km values were determined by analyzing β-lactam hydrolysis under initial rate conditions with a UV spectrophotometer (UV 1601; Schimadzu) by at least three independent measurements. One unit of activity was defined as the amount of enzyme that hydrolyzed 1 μmol of imipenem per min. The 50% inhibitory concentrations of EDTA were determined with 100 μM imipenem.
Analytical isoelectric focusing (IEF) was performed with a Model 111 Mini IEF Cell (Bio-Rad Laboratories) (18).
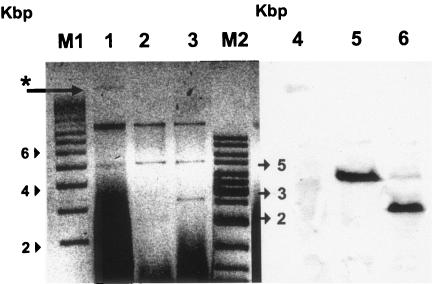
E. cloacae EDV/1 was identified with reduced susceptibility to β-lactam antibiotics in a set of clinical isolates stored before 2002 in a university hospital located in southeast Turkey. The crude enzyme extract of EDV/1 showed carbapenemase activity typical of metalloenzymes. A PCR screen with consensus primers of IMP and VIM family metallo-β-lactamases was performed (data not shown). Upon detection of a VIM allele with PCR, a DIG-dUTP-labeled probe was generated. Hybridization experiments with this probe detected blaVIM on an approximately 3,900-bp (BamHI) or 2,712-bp (BamHI-HindIII) fragment of an approximately 23-kbp plasmid, pEDV5 (Fig. 1), which seems to be nonconjugative. Sequence analysis of the bla gene was identical to that of VIM-5 (GenBank accession no. AY910754).
FIG. 1.
Plasmids, their restriction fragments, and hybridization with a DIG-dUTP-labeled VIM-5 probe of E. cloacae EDV/1. Lanes: M1, supercoiled DNA ladder (Sigma); 1, plasmids of EDV/1 (*, pEDV5, a 23-kbp plasmid carrying VIM-5); 2, BamHI-digested plasmid; 3, BamHI-HindIII-digested plasmid; M2, gene ruler DNA ladder (Fermentas); 4, 5, and 6, corresponding lanes to lanes 1, 2, and 3 hybridized with the VIM-5 probe on a nylon membrane.
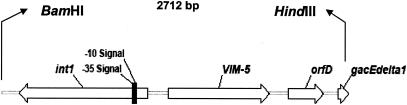
The BamHI fragment of the plasmid (pEDV5) shown to carry blaVIM-5 was successfully inserted into the vector pACYCDuet-1 (pACDV5/4). A BamHI-HindIII double digestion of this plasmid yielded two fragments from the insert. These fragments were inserted into pUC19 as pUCSCV5/1 (2,712 bp) and pUCSCV5/2 (approximately 1,100 bp). The VIM-5 gene was located on the plasmid pUCSCV5/1. Sequence analysis of this insert showed that blaVIM-5 is associated with a class 1 integron as a single antibiotic resistance gene cassette, followed by orfD and qacEdelta1 genes (Fig. 2) (GenBank accession no. DQ023222).
FIG. 2.
Schematic map of the BamHI-HindIII insert carrying a class 1 integron with the VIM-5 cassette. The coding regions are shown as arrows indicating the direction of transcription.
VIM-1 was first reported from Italy in 1999 (6) and is now prevalent in the northern part of the same country (2, 8). However, a recent retrospective study determined that VIM-1 was present in a P. aeruginosa isolate from a collection of strains stored in 1996 in a Greek hospital (17). Likewise, VIM-4 was first detected in Greece (14) and was then found in Poland (12), Hungary (7), and Italy (9). In Hungary the VIM-4 outbreak was related to a Greek patient visiting the country. Current knowledge, therefore, raises the probability that VIM-1 and later VIM-4 evolved in Greek hospitals and was disseminated by carriers (17). The present study shows that VIM-5 existed before 2002 and is now scattered around hospitals in different parts of Turkey. These evolutionarily related VIMs occurred in countries geographically close to one another. However, whether this figure represents a unifocal epidemic of VIM-1, VIM-4, and VIM-5 has not been documented so far. Moreover, we do not have data for the dissemination of VIM alleles in countries bordering the eastern part of Turkey.
Although the crude enzyme extract failed, purified VIM-5 was visible at a pI of approximately 4.5 on the IEF gel. According to sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis, the molecular mass of VIM-5 was calculated as 28 kDa and the purity of the protein was >90%. The purification steps are summarized in Table 1. Kinetic parameters kcat, Km, and kcat/Km were determined with clinically significant β-lactam antibiotics. All β-lactams with significant differences in kcat and Km values were hydrolyzed by VIM-5, with the exception of aztreonam (Table 2). Notably, ceftazidime and cefepime were less effectively hydrolyzed by VIM-5 than VIM-1 (3) and VIM-2 (13), while cefotaxime was hydrolyzed at a comparable efficacy to that of VIM-2. The behavior of VIM-5 against carbapenems was similar to that of VIM-1 and VIM-2, with greater efficiency on imipenem than meropenem. VIM-5 was inhibited by EDTA (50% inhibitory concentration, 20 μM EDTA).
TABLE 1.
Summary of the purification steps of VIM-5 enzyme
| Step | Volume (ml) | Total acta (U) | Total protein (mg) | Sp act (U/mg) | Purifi- cation (fold) | Recovery (%) |
|---|---|---|---|---|---|---|
| Crude extract | 4 | 21 | 8.99 | 2 | 100 | |
| Anion exchange (Q-sepharose) | 15 | 10 | 0.45 | 23 | 10 | 48 |
| Gel filtration (Sephacryl S-100 HR) | 6 | 6 | 0.22 | 26 | 11 | 28 |
Total activity was determined by monitoring imipenem hydrolysis. One unit of activity was defined as the amount of enzyme that hydrolyzes 1 μmol of imipenem per min.
TABLE 2.
Kinetic parameters of VIM-5 compared to VIM-1 and VIM-2 enzymes
| Drug |
kcat (s−1)
|
Km (μM)
|
kcat/Km (μM−1 · s−1)
|
Relativeakcat/Km
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VIM-1b | VIM-2 | VIM-5 | VIM-1 | VIM-2 | VIM-5 | VIM-1 | VIM-2 | VIM-5 | VIM-1 | VIM-2 | VIM-5 | |
| Penicillin | 29 | 55.8 | 29 | 841 | 49 | 113 | 0.034 | 1.14 | 0.26 | 100 | 100 | 100 |
| Ampicillin | 37 | 14 | 917 | 125 | 0.04 | 0.11 | 117.6 | 44.7 | ||||
| Piperacillin | 1,860 | 32.7 | 47 | 3,500 | 72 | 1,753 | 0.53 | 0.45 | 0.03 | 1,558.8 | 39.5 | 10.6 |
| Ceftazidime | 60 | 89 | 0.2 | 794 | 98 | 149 | 0.076 | 0.9 | 0.001 | 223.5 | 78.9 | 0.5 |
| Cefepime | 549 | 4.7 | 0.1 | 145 | 184 | 76 | 3.8 | 0.03 | 0.001 | 11,176.5 | 2.6 | 0.4 |
| Cefotaxime | 169 | 27.5 | 9.2 | 247 | 32 | 101 | 0.68 | 0.86 | 0.09 | 2,000 | 75.4 | 35.7 |
| Imipenem | 2 | 9.9 | 3.5 | 1.5 | 10 | 12 | 1.3 | 0.99 | 0.29 | 3,823.5 | 86.8 | 114.2 |
| Meropenem | 13 | 1.4 | 2.4 | 48 | 5 | 49 | 0.27 | 0.28 | 0.05 | 794.1 | 24.6 | 18.9 |
| Aztreonam | <0.01 | <0.5 | <0.01 | >1,000 | ND | NDc | <10-5 | ND | ND | NC | NC | NC |
MICs for the wild-type strain E. cloacae EDV/1 and the clones are presented in Table 3. The MICs of cefepime and carbapenems for both the wild-type strain and clones were in the susceptible breakpoint range. Although these antibiotics are hydrolyzed by VIM-5, the genotypic resistance is masked in susceptibility tests when the recommended inoculum size is applied. However, significant differences were observed between the MICs in independent experiments. Slight changes in inoculum (not more than 10-fold) caused significant changes in MICs (2 μg/ml versus 16 μg/ml for imipenem; 4 μg/ml versus 32 μg/ml for meropenem). Such striking discrepancies were observed with VIM-1 and VIM-4 as well (5, 9).
TABLE 3.
MICs of antibiotics for E. cloacae (EDV/1) and E. coli ER2267 harboring recombinant plasmids and reference strains
| Ab(s)a | E. cloacae EDV/1 | VIM-5+ clonesb
|
VIM-5− clone
|
Control strains
|
|||
|---|---|---|---|---|---|---|---|
| pACDV5/4 | pUCSCV5/1 | pUCSCV5/2 | E. coli ER2267 | E. coli pACYC184c | E. coli ATCC 25922 | ||
| Ampicillin | >128 | >128 | >128 | >128 | <1 | <1 | 4 |
| Piperacillin | 128 | 32 | >128 | >128 | <1 | <1 | 2 |
| Piperacillin-TZ | 128 | 32 | 64 | 32 | <1 | <1 | 1 |
| Ceftazidime | 32 | 16 | 16 | <0.5 | <0.5 | <0.5 | <0.5 |
| Ceftazidime-CLV | 32 | 16 | 32 | <0.5 | <0.5 | <0.5 | 0.5 |
| Cefotaxime | >32 | 16 | 16 | <0.5 | <0.5 | <0.5 | <0.5 |
| Cefepime | 0.5 | 0.25 | <025 | <0.25 | <0.25 | <0.25 | <0.25 |
| Aztreonam | 4 | 0.06 | <0.06 | 0.06 | 0.06 | 0.06 | <0.06 |
| Imipenem | 2 | 1 | 0.5 | <0.5 | <0.5 | <0.5 | <0.5 |
| Meropenem | 4 | 0.5 | 0.5 | <0.5 | <0.5 | <0.5 | <0.5 |
| Ciprofloxacin | <0.25 | <0.25 | <0.25 | <0.25 | <0.25 | <0.25 | <0.25 |
| Gentamycin | 1 | 1 | 0.25 | 0.5 | 0.5 | 0.5 | 0.5 |
| Amikacin | 2 | 1 | 1 | 1 | 0.5 | 2 | 4 |
Ab(s), antibiotic(s); piperacillin-TZ, tazobactam with a ratio of 2:1 to piperacillin; ceftazidime-CLV, clavulanate with a fixed ratio of 4 μg/ml.
VIM-5+ clones of ER2267 carrying recombinant plasmids; pACDV5/4, recombinant pACYCDuet-1 with a complete insert; pUCSCV5/1, recombinant pUC19 with the subclone carrying VIM-5.
E. coli K-12 strain ER2420 carrying the vector pACYC184.
Finally, the present data show that VIM-5 entered Turkish hospitals before 2002, and it is now found in distinct centers.
Nucleotide sequence accession numbers. The sequences determined in the course of this work have been deposited in GenBank under accession numbers AY910754 and DQ023222.
Acknowledgments
We are grateful to Sinem Torol and Yoruk Divanoglu for their excellent technical assistance.
REFERENCES
- 1.Bahar, G., A. Mazzariol, R. Koncan, A. Mert, R. Fontana, G. M. Rossolini, and G. Cornaglia. 2004. Detection of VIM-5 metallo-beta-lactamase in a Pseudomonas aeruginosa clinical isolate from Turkey. J. Antimicrob. Chemother. 54:282-283. [DOI] [PubMed] [Google Scholar]
- 2.Cornaglia, G., A. Mazzariol, L. Lauretti, G. M. Rossolini, and R. Fontana. 2000. Hospital outbreak of carbapenem-resistant Pseudomonas aeruginosa producing VIM-1, a novel transferable metallo-beta-lactamase. Clin. Infect. Dis. 31:1119-1125. [DOI] [PubMed] [Google Scholar]
- 3.Franceschini, N., B. Caravelli, J. D. Docquier, M. Galleni, J. M. Frere, G. Amicosante, and G. M. Rossolini. 2000. Purification and biochemical characterization of the VIM-1 metallo-β-lactamase. Antimicrob. Agents Chemother. 44:3003-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galani, I., M. Souli, Z. Chryssouli, K. Orlandou, and H. Giamarellou. 2005. Characterization of a new integron containing blaVIM-1 and aac(6′)-IIc in an Enterobacter cloacae clinical isolate from Greece. J. Antimicrob. Chemother. 55:634-638. [DOI] [PubMed] [Google Scholar]
- 5.Giakkoupi, P., L. S. Tzouvelekis, G. L. Daikos, V. Miriagou, G. Petrikkos, N. J. Legakis, and A. C. Vatopoulos. 2005. Discrepancies and interpretation problems in susceptibility testing of VIM-1-producing Klebsiella pneumoniae isolates. J. Clin. Microbiol. 43:494-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lauretti, L., M. L. Riccio, A. Mazzariol, G. Cornaglia, G. Amicosante, R. Fontana, and G. M. Rossolini. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Libisch, B., M. Gacs, K. Csiszar, M. Muzslay, L. Rokusz, and M. Fuzi. 2004. Isolation of an integron-borne blaVIM-4 type metallo-β-lactamase gene from a carbapenem-resistant Pseudomonas aeruginosa clinical isolate in Hungary. Antimicrob. Agents Chemother. 48:3576-3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lombardi, G., F. Luzzaro, J. D. Docquier, M. L. Riccio, M. Perilli, A. Coli, G. Amicosante, G. M. Rossolini, and A. Toniolo. 2002. Nosocomial infections caused by multidrug-resistant isolates of Pseudomonas putida producing VIM-1 metallo-β-lactamase. J. Clin. Microbiol. 40:4051-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luzzaro, F., J. D. Docquier, C. Colinon, A. Endimiani, G. Lombardi, G. Amicosante, G. M. Rossolini, and A. Toniolo. 2004. Emergence in Klebsiella pneumoniae and Enterobacter cloacae clinical isolates of the VIM-4 metallo-β-lactamase encoded by a conjugative plasmid. Antimicrob. Agents Chemother. 48:648-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Midilli, K., G. Aygün, M. Kuskucu, et al. 2003. A novel metallo-β-lactamase (VIM-5) from a clinical isolate of Klebsiella pneumoniae, p. 275. In H. Eraksoy (ed.), Proceedings of the KLIMIK Congress. Tavasli Matbaasi, Istanbul, Turkey.
- 11.Miriagou, V., E. Tzelepi, D. Gianneli, and L. S. Tzouvelekis. 2003. Escherichia coli with a self-transferable, multiresistant plasmid coding for metallo-β-lactamase VIM-1. Antimicrob. Agents Chemother. 47:395-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patzer, J., M. A. Toleman, L. M. Deshpande, W. Kaminska, D. Dzierzanowska, P. M. Bennett, R. N. Jones, and T. R. Walsh. 2004. Pseudomonas aeruginosa strains harbouring an unusual blaVIM-4 gene cassette isolated from hospitalized children in Poland (1998-2001). J. Antimicrob. Chemother. 53:451-456. [DOI] [PubMed] [Google Scholar]
- 13.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J. D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pournaras, S., A. Tsakris, M. Maniati, L. S. Tzouvelekis, and A. N. Maniatis. 2002. Novel variant (blaVIM-4) of the metallo-β-lactamase gene blaVIM-1 in a clinical strain of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 46:4026-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sader, H. S., M. Castanheira, R. E. Mendes, M. Toleman, T. R. Walsh, and R. N. Jones. 2005. Dissemination and diversity of metallo-beta-lactamases in Latin America: report from the SENTRY Antimicrobial Surveillance Program. Int. J. Antimicrob. Agents. 25:57-61. [DOI] [PubMed] [Google Scholar]
- 16.Toleman, M. A., D. Biedenbach, D. M. Bennett, R. N. Jones, and T. R. Walsh. 2005. Italian metallo-beta-lactamases: a national problem? Report from the SENTRY Antimicrobial Surveillance Programme. J. Antimicrob. Chemother. 55:61-70. [DOI] [PubMed] [Google Scholar]
- 17.Tsakris, A., S. Pournaras, N. Woodford, M. F. Palepou, G. S. Babini, J. Douboyas, and D. M. Livermore. 2000. Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J. Clin. Microbiol. 38:1290-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vahaboglu, H., S. Dodanli, C. Eroglu, R. Ozturk, G. Soyletir, I. Yildirim, and V. Avkan. 1996. Characterization of multiple-antibiotic-resistant Salmonella typhimurium strains: molecular epidemiology of PER-1-producing isolates and evidence for nosocomial plasmid exchange by a clone. J. Clin. Microbiol. 34:2942-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vahaboglu, H., R. Ozturk, G. Aygun, F. Coskunkan, A. Yaman, A. Kaygusuz, H. Leblebicioglu, I. Balik, K. Aydin, and M. Otkun. 1997. Widespread detection of PER-1-type extended-spectrum beta-lactamases among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nationwide multicenter study. Antimicrob. Agents Chemother. 41:2265-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]