Abstract
The sequence and gene organization of the van operons in vancomycin (MIC of >256 μg/ml)- and teicoplanin (MIC of ≥32 μg/ml)-resistant Paenibacillus thiaminolyticus PT-2B1 and Paenibacillus apiarius PA-B2B isolated from soil were determined. Both operons had regulatory (vanR and vanS), resistance (vanH, vanA, and vanX), and accessory (vanY, vanZ, and vanW) genes homologous to the corresponding genes in enterococcal vanA and vanB operons. The vanAPT operon in P. thiaminolyticus PT-2B1 had the same gene organization as that of vanA operons whereas vanAPA in P. apiarius PA-B2B resembled vanB operons due to the presence of vanW upstream from the vanHAX cluster but was closer to vanA operons in sequence. Reference P. apiarius strains NRRL B-4299 and NRRL B-4188 were found to harbor operons indistinguishable from vanAPA by PCR mapping, restriction fragment length polymorphism, and partial sequencing, suggesting that this operon was species specific. As in enterococci, resistance was inducible by glycopeptides and associated with the synthesis of pentadepsipeptide peptidoglycan precursors ending in d-Ala-d-Lac, as demonstrated by d,d-dipeptidase activities, high-pressure liquid chromatography, and mass spectrometry. The precursors differed from those in enterococci by the presence of diaminopimelic acid instead of lysine in the peptide chain. Altogether, the results are compatible with the notion that van operons in soil Paenibacillus strains and in enterococci have evolved from a common ancestor.
The glycopeptide antibiotics vancomycin and teicoplanin are drugs of primary importance for the treatment of hospital infections caused by multiresistant gram-positive bacteria such as Staphylococcus aureus, enterococci, and Clostridium difficile. Glycopeptides act by inhibiting cell wall synthesis (29), and resistance is due to the synthesis of peptidoglycan precursors with low affinity for these antibiotics (3, 32). Six types of van operons conferring glycopeptide resistance have been described in enterococci based on gene sequence and organization (25, 31). The various operons are designated according to the name of the gene, which encodes either a d-Ala:d-Lac (vanA, vanB, and vanD) or a d-Ala:d-Ser (vanC, vanE, and vanG) ligase for synthesis of peptidoglycan precursors with low affinity for glycopeptides. The operons coding for a d-Ala:d-Lac ligase contain genes for a two-component regulatory system (vanR and vanS), three resistance genes (vanH, vanA or vanB or vanD, and vanX), an accessory gene (vanY), and other genes with unknown functions (vanW or vanZ).
The most common glycopeptide resistance gene clusters in clinical enterococci are vanA and vanB. Both operons are associated with transposable elements, i.e., vanA with Tn1546 (2) and vanB with Tn1547 (28) and Tn1549-Tn5382 (7, 13). The recent acquisition of the vanA gene cluster by S. aureus (8) confirms that these genes are able to spread across bacterial genera. Previous indications of intergenic horizontal transfer of van operons were provided by the finding of vanA in Bacillus circulans (18), Oerskovia turbata, and Arcanobacterium haemolyticum (26) and of vanB in Streptococcus bovis (27) and anaerobic bacilli (4). All these bacteria were isolated from clinical specimens, mainly stools, suggesting that dissemination of glycopeptide resistance may occur in the intestinal microflora of patients.
It has been proposed that van operons originate from glycopeptide-producing organisms (14, 20, 23). This hypothesis is based on the observation that the glycopeptide producers contain resistance gene clusters homologous to those in human-pathogenic bacteria. However, these clusters have relatively low similarity with the vanA and vanB operons (from 54 to 64% predicted amino acid identity), lack the two-component regulatory systems, and do not appear to be transferable under laboratory conditions. Finding of the vanF operon in Paenibacillus popilliae (24) and the recent recovery of genes homologous to vanA in other Paenibacillus species (15) have raised interest about a possible role of this genus in the dissemination of glycopeptide resistance.
In this study, we have characterized the organization of the van operons in two glycopeptide-resistant Paenibacillus strains isolated from soil and known to harbor putative d-Ala:d-Lac ligase genes flanked by vanH- and vanX-like genes (15). The two operons were closely related to vanA on the basis of both gene sequence and organization. Glycopeptide resistance was inducible by vancomycin and teicoplanin and resulted from the synthesis of peptidoglycan precursors containing diaminopimelic acid and ending in d-Ala-d-Lac.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains Paenibacillus thiaminolyticus PT-2B1 and Paenibacillus apiarius PA-B2B were isolated from soil and identified by sequencing 16S rRNA genes (15). Reference strains P. apiarius NRRL B-4299 and NRRL B-4188 were isolated from dead bees and obtained from the collection of the National Center for Agricultural Utilization Research, U.S. Department of Agriculture. Strains were grown at 30°C in brain heart infusion (BHI) broth or on BHI agar (Difco Laboratories, Detroit, Mich.). The MICs of vancomycin and teicoplanin were determined by Etest (AB Biodisk, Solna, Sweden) after 48 h of incubation on Mueller-Hinton agar at 28°C.
TAIL PCR.
Thermal asymmetric interlaced (TAIL) PCR (19) was used to clone the 5′ and 3′ regions flanking the vanHAX clusters in strains PT-2B1 and PA-B2B and to determine their sequence. Three PCR steps were performed with a specific primer targeting the known sequence and arbitrary degenerate primer AD1 (Table 1 and Fig. 1). The target sequences of the anchor primers used in the second and third steps were selected at decreasing distance from the ends of the known sequence in order to obtain PCR products of slightly decreasing size. Total DNA obtained by use of the High Pure PCR template kit (Roche Diagnostics, Mannheim, Germany) was used as a template in the first PCR. The PCR mixture contained 0.15 μM specific primer (Table 1), 5 μM AD1 primer, 200 μM of each deoxynucleoside triphosphate, 1× PCR buffer with 22.5 mM MgCl2, and 2.5 U of the enzyme mix supplied by the Expand Long Template PCR system (Roche Diagnostics). PCR conditions were as previously described (9). The products obtained from the first and second PCRs were diluted 105 and 10 times, respectively, before being used as DNA templates in the following PCR. The DNA bands corresponding to the second and third PCRs, which had the length decrease expected from the positions of the specific primers, were purified using the QIAGEN PCR purification kit (QIAGEN S.A., Courtaboeuf, France) and sequenced.
TABLE 1.
Oligodeoxynucleotides used for TAIL and long PCR
| Primer | Sequence (5′ to 3′) | Positiona |
|---|---|---|
| Tpf1 | GTTTCAAAAGGATACGTGGC | 7419/7438 |
| Tpr1 | GCACATGACATCCAAATCCC | 5625/5606 |
| Tpf2 | CTTTATCGATTAGACACGGG | 7482/7501 |
| Tpr2 | ATGTCGCGCAGTACCTTGCC | 5548/5529 |
| Tpf3 | CAAAATCGCAGACGTTTGCG | 7587/7606 |
| Tpr3 | ATAATCGGCAACGCTATCCG | 5438/5419 |
| Ad1 | TGWGNAGWANCASAGA | NAb |
| Lpf1 | TCCAGAGAAGGATATGAC | 5017/5034 |
| Lpf2 | GAACTGGCATTTCGCAAGGC | 1998/2017 |
| Lpr1 | GCCCCCATTTCTTGGTAAAG | 8604/8585 |
| Lpf3 | GCTCCCATCATAGTCAAT | −190/−173 |
| Lpr2 | ACTGCGTTTTCAGAGCCTTT | 6839/6820 |
| Patf | TATCCTACGTGGATAAGCGG | 2948/2967 |
| Patr | GGGCCAAACTTGAGCACGAT | 9178/9159 |
Nucleotide numbering begins at the start site (+1) of the transposase gene preceding the vanAPT operon in P. thiaminolyticus PT-2B1 (accession no. DQ018710).
NA, not applicable.
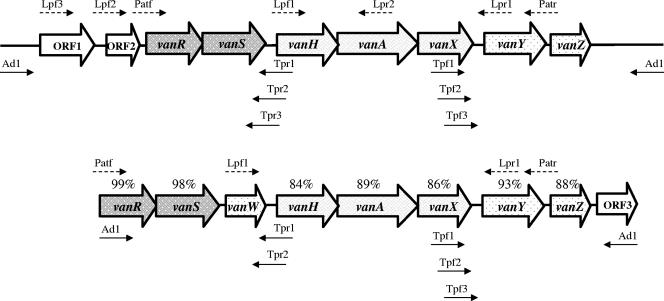
FIG. 1.
Schematic representation of the van operons in P. thiaminolyticus PT-2B1 (top) and P. apiarius PA-B2B (bottom) and of the PCR primers used for their characterization. The percentages refer to the levels of identity between the corresponding genes in the two operons. Open arrows represent coding sequences and indicate the direction of transcription. Solid and dashed small arrows indicate the primers used for TAIL and long PCR, respectively.
Cloning of the TAIL PCR products into Escherichia coli.
The purified PCR products were cloned in plasmid PCR2.1 into E. coli TOP10F′ using the TA cloning kit (Invitrogen Corporation, Carlsbad, CA). White colonies were isolated from BHI agar containing ampicillin (50 μg/ml) and 5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside (X-Gal; 80 μg/ml). Plasmid DNA was isolated according to the method of Birnboim and Doly (6) and digested with EcoRI (Invitrogen Corporation) to screen for the presence of an insert.
Nucleotide sequencing.
Plasmid DNA or PCR products were labeled using a dye-labeled ddNTP Terminator Cycle sequencing kit (Beckman Coulter UK Ltd.) and sequenced with a CEQ 2000 automated sequencer (Beckman). Sequences obtained from cloned TAIL PCR products were confirmed by sequencing PCR products obtained from total DNA using specific primers.
Computer analysis of sequences.
Sequences were aligned, translated, and analyzed using DNA Strider 1.3 (CEA/Saclay, Gif-sur-Yvette, France). Comparison with known genes and proteins was carried out using BlastN and BlastX, available at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST/). Nucleotide identities and fractional GC contents were calculated using EMBOSS Align (gap open 10 and gap extend 0.5) and EMBOSS Geecee, respectively (http://www.ebi.ac.uk/emboss/).
Restriction fragment length polymorphism (RFLP) analysis.
Specific primers were designed to amplify the entire operons or portions of them (Table 1; Fig. 1). Long PCR (Expand Long Template PCR system; Roche Diagnostics) was carried out using the following conditions: 2 min of denaturation at 94°C and 30 cycles of 10 s at 94°C, 30 s at 50°C, and 4 to 6 min at 68°C depending on the size of the product, followed by final extension at 68°C for 7 min. The long PCR products were partly sequenced and digested for 1 h at 37°C with NaeI (Biolabs, Saint Quentin Yvelines, France) and DdeI (Invitrogen Corporation) for RFLP analysis.
Mating experiments.
Transfer of glycopeptide resistance was attempted from Paenibacillus spp. to Enterococcus faecium BM4105 and J64/3 resistant to rifampin and fusidic acid. Donor and recipient strains were inoculated on BHI agar or on 0.45-μm-pore-size nitrocellulose filter (Millipore) placed on BHI agar. After 2 days of incubation at 28°C, bacteria were resuspended in 1 ml of BHI broth and plated on Slanetz agar (Oxoid) supplemented with vancomycin (10 μg/ml), rifampin (50 μg/ml), and fusidic acid (10 μg/ml). Longer mating time periods (up to 10 days) and selective enrichment in azide dextrose broth (Merck) supplemented with the three antibiotics were employed for enhancing detection of low-frequency transfer.
Assays of d,d-dipeptidase (VanX) and d,d-carboxypeptidase (VanY) activities.
The activities of VanX (cytoplasmic d,d-dipeptidase) and VanY (membrane-associated d,d-carboxypeptidase) were assayed by determining the amount of d-Ala released by hydrolysis of dipeptide d-Ala-d-Ala (6.56 mM) and pentapeptide UDP-MurNAc-l-Ala-γ-d-Glu-l-Lys-d-Ala-d-Ala (5 mM), respectively. Cytoplasmic and membrane extracts were obtained as previously described (9). Measurement was obtained through coupled indicator reactions using d-amino acid oxidase and horseradish peroxidase (1, 30). Specific activity was defined as the number of nanomoles of product formed at 37°C per minute per milligram of protein contained in the extract.
Peptidoglycan precursors.
UDP-MurNAc-l-Ala-γ-d-Glu-meso-A2pm-d-Ala-d-Ala (UDP-MurNAc-pentapeptide) was obtained as previously described (11). UDP-MurNAc-l-Ala-γ-d-Glu-meso-A2pm-d-Ala (UDP-MurNAc-tetrapeptide) was generated by removal of the C-terminal d-alanine of UDP-MurNAc-pentapeptide by action of the d,d-carboxypeptidase from Actinomura sp. strain R39 as described previously for the lysine-containing UDP-MurNAc-tetrapeptide (5).
Extraction and quantification of the UDP-MurNAc peptide precursors.
The UDP-MurNAc peptide precursors from mid-log-phase cells grown without and with vancomycin (32 μg/ml) were extracted according to the method of Reynolds et al. (33) and analyzed by high-pressure liquid chromatography (HPLC) as previously described (21).
Amino acid analysis.
Amino acid and amino sugar compositions were determined with a Hitachi model L8800 analyzer (ScienceTec, Les Ulis, France) equipped with a 2620MSC-PS column (80 × 4.6 mm). Prior to analysis samples were hydrolyzed in 6 M HCl at 95°C for 16 h.
Mass spectrometry analysis.
Matrix-assisted laser desorption ionization-time of flight mass spectra were recorded on a PerSeptive Voyager-DE STR instrument (Applied Biosystems, Foster City, CA) in the reflectron mode with delayed extraction. The samples were prepared as follows: 1 μl of a solution of compound in water at 20 pmol/μl was deposited on the plate and mixed with 1 μl of a 10-mg/ml solution of 2,5-dihydroxybenzoic acid in 0.1 M citric acid. After evaporation, desorption and ionization were obtained by pulses from a 337-nm nitrogen laser. Spectra were recorded in the negative ion mode at an acceleration voltage of −20 kV and an extraction delay time of 200 nanoseconds. A mixture of UDP-MurNAc, UDP-MurNAc-l-Ala-d-Glu, and UDP-MurNAc-pentapeptide was used as an external calibrant.
Nucleotide sequence accession numbers.
The sequences were submitted to GenBank and assigned the following accession numbers: DQ018710 (vanAPT operon in PT-2B1) and DQ018711 (vanAPA operon in PA-B2B).
RESULTS
Glycopeptide resistance phenotypes.
P. thiaminolyticus PT-2B1 and P. apiarius PA-B2B were resistant to high concentrations (MIC of >256 μg/ml) of vancomycin. An atypical growth response by PT-2B1 was observed with Etest, since the strain grew at high vancomycin concentrations (up to 256 μg/ml) but showed a thin inhibition zone at concentrations between 3 and 4 μg/ml. A similar observation was made by disk agar diffusion, the strain growing at the point of contact with the 30-μg vancomycin disk but displaying a thin zone of inhibition at a distance from the disk (Fig. 2). PA-B2B displayed reduced growth at vancomycin concentrations above 4 μg/ml in the Etest but grew in liquid medium containing higher concentrations of the antibiotic. The MIC of teicoplanin was higher for PT-2B1 (>256 μg/ml) than for PA-B2B (32 μg/ml).
FIG. 2.
Phenotypic characteristics of P. thiaminolyticus PT-2B1. A. Atypical inhibition zone centered on a 30-μg vancomycin disk. B. Inhibition of E. faecium BM4105 by a colony of PT-2B1.
Transfer of glycopeptide resistance from PT-2B1 or PA-B2B to E. faecium BM4105 and J64/3 could not be obtained, even when the mating mixtures were incubated for 10 days and selective enrichment was used for detection of transconjugants. Strain PT-2B1 inhibited the growth of E. faecium as indicated by the absence of growth around colonies of the donor strain (Fig. 2).
Organization of the van operon in PT-2B1.
Two TAIL PCR products of approximately 7 kb each were obtained from the upstream and downstream regions of the vanHAX cluster in PT-2B1 (Fig. 1). Sequencing revealed the same organization as in enterococcal vanA operons, with the vanHAX resistance gene cluster preceded by genes (vanR and vanS) for a two-component regulatory system and followed by a gene (vanY) coding for a putative d,d-carboxypeptidase. The percentage of identity to the corresponding genes in the vanA operon of Tn1546 in E. faecium BM4147 (2) varied between 83% and 94% (Fig. 3). Homology with the vanA operon of Tn1546 was also observed in the vanS-H (85%) and vanX-Y (74%) intergenic regions. As in Tn1546, vanS overlapped with vanR over 23 bp and vanA with vanH over 8 bp. Due to the high similarity with vanA operons in both gene sequence and organization, the cluster was designated vanAPT. All the genes had the same length as the corresponding genes in BM4147, except for vanSPT, which had a 6-bp insertion at the beginning of the gene, and vanYPT, which was shorter (885 versus 912 bp). A sequence homologous to vanZ (40% identity) was located downstream from vanYPT. The region upstream from vanRPT contained two open reading frames, ORF1 and ORF2. ORF1 (1,263 bp) encoded a hypothetical protein with 39% identity and 58% similarity to a putative transposase in Lactococcus lactis (accession no. AAC72261) (34). The predicted amino acid sequence encoded by ORF2 (405 bp) had 35% identity and 48% similarity with an acetyltransferase of the GCN5-related N-acetyltransferase (GNAT) superfamily (37) present in the genome of Bacillus cereus (accession no. AAP09041) and of other Bacillus spp.
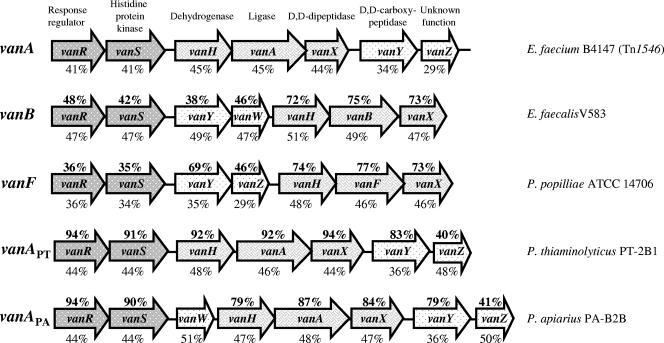
FIG. 3.
Organization of vanA operons (E. faecium B4147, accession no. M97297), vanB (E. faecalis V583, accession no. U35369), vanF (P. popilliae ATCC 14706, accession no. AF155139), vanAPT (P. thiaminolyticus PT-2B1, accession no. DQ018710), and vanAPA (P. apiarius PA-B2B, accession no. DQ018711). For every gene, identity to the corresponding gene of the vanA operon in Tn1546 and the GC content are indicated above and below the gene, respectively. Arrows indicate extent of the genes and direction of transcription.
Organization of the van operon in PA-B2B.
TAIL PCR products of approximately 3 and 4 kb were obtained from the regions upstream and downstream from the vanHAX cluster in PA-B2B (Fig. 2). The operon in this strain was a hybrid between vanA and vanB operons based on the relative gene organization: vanW was located upstream from vanHAX as in vanB operons whereas vanY was positioned downstream from vanHAX as in vanA operons (Fig. 3). However, the operon was designated vanAPA since the sequence of the genes was closer to that in vanA operons. The percentages of identity to the corresponding genes in the vanA operon of Tn1546 varied between 79% and 94% (Fig. 3). The vanWPA gene was 75% identical to vanW in the vanB operon of reference Enterococcus faecalis V583 (10). The typical overlaps between vanR and vanS and between vanH and vanA were also present in this operon. Similarly to vanSPT, a 12-bp insertion was found at the beginning of vanSPA relative to vanS in Tn1546, and vanYPA was followed by a putative vanZPA (Fig. 3). The vanRPA and vanSPA regulatory genes had sequences nearly identical to those of vanRPT and vanSPT whereas the remaining genes in vanAPA were less closely related to the corresponding genes in vanAPT (Fig. 1). The region downstream from vanYPA ended with a partial open reading frame (ORF3) homologous to btrU (79% identity), a gene in the aminoglycoside butirosin biosynthetic operon of B. circulans (accession no. CAD41946) (22).
PCR mapping and RFLP analysis of long PCR products.
PCR products of the expected size were obtained using specific primers (Table 1 and Fig. 1). Primers Patf and Patr allowed amplification of the entire operons in PT-2B1 and PA-B2B as well as in the two reference P. apiarius strains, NRRL B-4299 and NRRL B-4188. The van operons in the three P. apiarius strains had the same size and were indistinguishable based on RFLP analysis (data not shown) and partial sequencing of the long PCR products.
d,d-Peptidase activities.
Vancomycin and teicoplanin induced d,d-dipeptidase (VanX) and d,d-carboxypeptidase (VanY) activities in both strains (Table 2). No baseline enzymatic activity could be detected in the absence of antibiotic. There was good correlation between the d,d-peptidase and the nature of late peptidoglycan precursors. In particular, VanY activity was associated with an increase in UDP-MurNAC-tetrapeptide.
TABLE 2.
d,d-Dipeptidase (VanX) and d,d-carboxypeptidase (VanY) activities in cytoplamic and membrane extracts from Paenibacillus strainsa
| Strain and glycopeptide | Glycopeptide concn (μg/ml) | Activity (nmol/min/mg protein)
|
|
|---|---|---|---|
| Cytoplamic extract (VanX activity) | Membrane extract (VanY activity) | ||
| PA-B2B | |||
| Vancomycin | 4 | 108 ± 21 | 14 ± 4 |
| 64 | 142 ± 7 | 19 ± 9 | |
| Teicoplanin | 8 | 24 ± 2 | 1 ± 0.5 |
| 32 | 86 ± 9 | 5 ± 2 | |
| PT-2B1 | |||
| Vancomycin | 2 | 1 ± 0.5 | 37 ± 10 |
| 64 | 13 ± 4 | 81 ± 2 | |
| Teicoplanin | 4 | 1.5 ± 0.6 | 1 ± 0.5 |
| 32 | 22 ± 4 | 53 ± 8 | |
The reported mean values were calculated based on three separate measurements.
HPLC and mass spectrometry of peptidoglycan precursors.
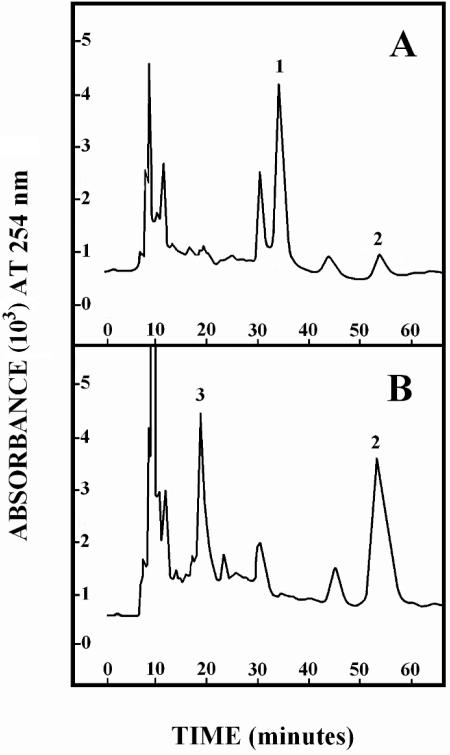
The crude cell wall extract from PT-2B1 was composed of ca. 25% A2pm-containing peptidoglycan. The main precursor peak detected in the HPLC profile of the cytoplasmic extract from untreated cells was eluted at 33 min (Fig. 4) and identified as A2pm-containing UDP-MurNAc-pentapeptide by its coelution with an authentic sample under two different HPLC conditions.
FIG. 4.
HPLC analysis of peptidoglycan precursors of P. thiaminolyticus PT-2B1 grown without (A) or with (B) vancomycin (32 μg/ml). Samples (one-fifth of the extracts) were applied to a μ-Bondapak C18 column (300 × 3.9 mm), and isocratic elution was performed with 0.05 M ammonium phosphate (pH 4.4) at a flow rate of 0.5 ml/min. The main peaks detected by absorbance at 254 nm were identified as UDP-MurNAc-pentapeptide (1), UDP-MurNAc-pentadepsipeptide (2), and UDP-MurNAc-tetrapeptide (3) and quantitated by their uridine content. Peak 1 in panel A, 1.2 nmol; peak 2 in panel A, 0.2 nmol; peak 2 in panel B, 1.5 nmol; peak 3 in panel B, 0.9 nmol.
In the extract from vancomycin-treated PT-2B1 cells, the UDP-MurNAc-(A2pm) pentapeptide peak was practically absent and replaced by two major peaks eluted at 18 and 50 min, respectively (Fig. 4). Both peaks were recovered and purified again by HPLC. The 18-min peak coeluted with an authentic sample of A2pm-containing UDP-MurNAc-tetrapeptide under two different HPLC conditions. This identification was confirmed by mass spectrometry which led to an [M-H]− ion with an m/z ratio of 1,121.13 in agreement with an A2pm-containing UDP-MurNAc-tetrapeptide, C38H60N8O27P2, having a monoisotopic mass of 1,122.30 g/mol.
Analysis of the 50-min peak (0.9 Mur, 1 A2pm, 1.1 Glu, and 1.5 Ala) was compatible with a precursor containing at least a MurNAc-(A2pm)tetrapeptide moiety. Mass spectrometry analysis led to an [M-H]− ion with an m/z ratio of 1,193.40, which was in agreement with a lactic acid-containing UDP-MurNAc-pentadepsipeptide, C41H64N8O29P2, having a monoisotopic mass of 1,194.32 g/mol. It was noteworthy that a low level of this UDP-MurNAc-pentadepsipeptide was detectable in cells grown without vancomycin (Fig. 4). Furthermore, both UDP-MurNAc-tetrapeptide and UDP-MurNAc-pentadepsipeptide were also found to be predominant peaks in an extract from vancomycin-treated PA-2B1.
DISCUSSION
This study shows that the genetic and biochemical basis of glycopeptide resistance in Paenibacillus from soil is the same as in enterococci and in other human-pathogenic bacteria. In particular, the glycopeptide resistance operons in Paenibacillus have primary sequences and gene organizations very similar to those of enterococcal vanA operons (Fig. 3). Furthermore, as in clinical isolates, resistance is inducible by glycopeptides (Table 2) and results from synthesis of peptidoglycan precursors terminating in d-Ala-d-Lac (Fig. 4). The pentadepsipeptide precursors differ from those in glycopeptide-resistant enterococci by the presence of diaminopimelic acid instead of lysine in the peptide chain. To the best of our knowledge, this is the first report of d-Ala-d-Lac-ending pentadepsipeptide precursors containing diaminopimelic acid. Diaminopimelic acid has been previously shown to be a normal constituent of peptidoglycan in various Paenibacillus species (17, 38, 39).
Occurrence of van operons in members of the genus Paenibacillus has been reported in the biopesticide P. popilliae ATCC 14706 (24). However, the level of identity with enterococcal operons is markedly lower than those reported in this study and the organization of the vanF operon in P. popilliae differs from those of vanA and vanB because of the presence of vanZ and vanY between the regulatory and the resistance genes (Fig. 3). In contrast, the vanAPT operon in P. thiaminolyticus had the same organization as the vanA operon (Fig. 3). Furthermore, as opposed to vanF, the similarity of vanAPT and vanAPA with enterococcal vanA operons was not limited to the resistance genes but extended to vanR and vanS with respect to both sequence and GC content (Fig. 3).
Irrespective of their sources and times of isolation, the three P. apiarius strains studied were resistant to glycopeptides and harbored indistinguishable operons. Based on current knowledge, it is unclear if glycopeptide resistance is acquired or intrinsic in Paenibacillus species. Unrelated P. popilliae isolates have been recently shown to be vancomycin resistant and to carry vanF operons (12). However, in a previous study (16), P. popilliae strains from Central and South America were reported to be susceptible to vancomycin and genetically divergent from North American isolates, which are vancomycin resistant and include the type strains for the species. The Paenibacillus genus has been only recently established (35), and thus, there is scarce information on the phylogenetic diversity within the species. Based on the results of this study it can be concluded that, if resistance is not intrinsic, it has been likely acquired a long time ago by certain genetic lineages, as indicated by the recovery of highly conserved genetic determinants among unrelated P. apiarius strains (NRRL B-4188, NRRL B-4299, and PA-B2B) isolated from different sample types (dead bees and soil), countries (United States, United Kingdom, and Denmark), and times (1973, 1975, and 2002).
The highly conserved structure of the van operons among various members of the same Paenibacillus species raises questions on the function of these gene clusters in environmental bacteria. The van operons could protect Paenibacillus from glycopeptides produced by actinomycetes in soil. However, it is not known whether glycopeptide production is common in soil and which concentrations can be achieved in situ. Alternatively, the van operons could be involved in another biological function and induced by physico-chemical factors other than glycopeptide antibiotics. Genome sequencing of various Bacillus species has revealed the presence of a cluster of genes homologous to vanR, vanS, and vanY in the chromosome of these bacteria (Integrated Genomics, www.ergo-light.com), but the role of these genes remains unknown. It has been proposed that the VanY-like d,d-carboxypeptidase in Bacillus spp. contributes to sporulation and germination (36). The predicted amino acid identities of putative VanR, VanS, and VanY in B. cereus ATCC 14579 (accession no. RZC01688) with the corresponding proteins in P. thiaminolyticus PT-2B1 are 43%, 32%, and 55%, respectively.
The close similarity of vanAPT and vanAPA operons with enterococcal vanA supports the hypothesis that these gene clusters have evolved from a common ancestor. Most likely, vanA operons originated in soil organisms and were subsequently acquired by enterococci. Two arguments suggest that vanA operons occurred in Paenibacillus before they occurred in enterococci: (i) glycopeptide resistance was first detected in enterococci in 1984 whereas the two P. apiarius reference strains studied were isolated in the early 1970s, when the use of glycopeptides was limited in clinical practice, and (ii) while glycopeptide resistance is associated with mobile genetic elements in enterococci, resistance in Paenibacillus is apparently chromosomal and intrinsic or at least acquired in very ancient times.
We do not provide any definitive evidence on the chromosomal location of the vanAPT and vanAPA operons. However, the open reading frames located upstream from vanAPT (ORF2) and downstream from vanAPA (ORF3) were homologous to chromosomal genes in other gram-positive bacteria. The chromosomal location of the operons is also supported by the fact that no plasmid of sufficient size to contain the operons was detected in the two strains (data not shown) and by lack of in vitro transfer of glycopeptide resistance.
Despite their similarity to enterococcal vanA and vanB operons, vanAPT and vanAPA were apparently not associated with any of the transposons previously described in enterococci of the VanA or VanB type (2, 7, 13, 28). Analysis of the region upstream from vanAPT revealed the presence of an open reading frame (ORF1) homologous to a putative transposase gene in L. lactis (34). Open reading frames encoding putative transposases have been described upstream from other van operons in gram-positive bacilli, such as vanF in P. popilliae (24) and vanABA in B. circulans (18). However, mobility of the genes has not been demonstrated under laboratory conditions. Thus, further investigation is needed to determine whether van operons in bacilli are associated with functional transposable elements and to investigate the possible mechanisms of transfer from bacilli to enterococci.
Acknowledgments
Luca Guardabassi was supported by grant no. 23-01-0170 from the Danish Agricultural and Veterinary Research Council.
REFERENCES
- 1.Arthur, M., F. Depardieu, P. Reynolds, and P. Courvalin. 1996. Quantitative analysis of the metabolism of soluble cytoplasmic peptidoglycan precursors of glycopeptide-resistant enterococci. Mol. Microbiol. 21:33-44. [DOI] [PubMed] [Google Scholar]
- 2.Arthur, M., C. Molinas, F. Depardieu, and P. Courvalin. 1993. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 175:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arthur, M., P. E. Reynolds, F. Depardieu, S. Evers, S. Dutka-Malen, R. Quintiliani, Jr., and P. Courvalin. 1996. Mechanisms of glycopeptide resistance in enterococci. J. Infect. 32:11-16. [DOI] [PubMed] [Google Scholar]
- 4.Ballard, S. A., E. A. Grabsch, P. D. Johnson, and M. L. Grayson. 2005. Comparison of three PCR primer sets for identification of vanB gene carriage in feces and correlation with carriage of vancomycin-resistant enterococci: interference by vanB-containing anaerobic bacilli. Antimicrob. Agents Chemother. 49:77-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Billot-Klein, D., L. Gutmann, S. Sable, E. Guittet, and J. van Heijenoort. 1994. Modification of peptidoglycan precursors is a common feature of the low-level vancomycin-resistant VANB-type enterococcus D366 and of the naturally glycopeptide-resistant species Lactobacillus casei, Pediococcus pentosaceus, Leuconostoc mesenteroides, and Enterococcus gallinarum. J. Bacteriol. 176:2398-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carias, L. L., S. D. Rudin, C. J. Donskey, and L. B. Rice. 1998. Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J. Bacteriol. 180:4426-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark, N. C., L. M. Weigel, J. B. Patel, and F. C. Tenover. 2005. Comparison of Tn1546-like elements in vancomycin-resistant Staphylococcus aureus isolates from Michigan and Pennsylvania. Antimicrob. Agents Chemother. 49:470-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Depardieu, F., M. G. Bonora, P. E. Reynolds, and P. Courvalin. 2003. The vanG glycopeptide resistance operon from Enterococcus faecalis revisited. Mol. Microbiol. 50:931-948. [DOI] [PubMed] [Google Scholar]
- 10.Evers, S., D. F. Sahm, and P. Courvalin. 1993. The vanB gene of vancomycin-resistant Enterococcus faecalis V583 is structurally related to genes encoding d-Ala:d-Ala ligases and glycopeptide-resistance proteins VanA and VanC. Gene 124:143-144. [DOI] [PubMed] [Google Scholar]
- 11.Flouret, B., D. Mengin-Lecreulx, and J. van Heijenoort. 1981. Reverse-phase high-pressure liquid chromatography of uridine diphosphate N-acetylmuramyl peptide precursors of bacterial cell wall peptidoglycan. Anal. Biochem. 114:59-63. [DOI] [PubMed] [Google Scholar]
- 12.Fraimow, H., C. Knob, I. A. Herrero, and R. Patel. 2005. Putative VanRS-like two-component regulatory system associated with the inducible glycopeptide resistance cluster of Paenibacillus popilliae. Antimicrob. Agents Chemother. 49:2625-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garnier, F., S. Taourit, P. Glaser, P. Courvalin, and M. Galimand. 2000. Characterization of transposon Tn1549, conferring VanB-type resistance in Enterococcus spp. Microbiology 146:1481-1489. [DOI] [PubMed] [Google Scholar]
- 14.Gholizadeh, Y., and P. Courvalin. 2000. Acquired and intrinsic glycopeptide resistance in enterococci. Int. J. Antimicrob. Agents 16(Suppl. 1):S11-S17. [DOI] [PubMed] [Google Scholar]
- 15.Guardabassi, L., H. Christensen, H. Hasman, and A. Dalsgaard. 2004. Members of the genera Paenibacillus and Rhodococcus harbor genes homologous to enterococcal glycopeptide resistance genes vanA and vanB. Antimicrob. Agents Chemother. 48:4915-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison, H., R. Patel, and A. A. Yousten. 2000. Paenibacillus associated with milky disease in Central and South American scarabs. J. Invertebr. Pathol. 76:169-175. [DOI] [PubMed] [Google Scholar]
- 17.Lee, J. S., K. C. Lee, Y. H. Chang, S. G. Hong, H. W. Oh, Y. R. Pyun, and K. S. Bae. 2002. Paenibacillus daejeonensis sp. nov., a novel alkaliphilic bacterium from soil. Int. J. Syst. Evol. Microbiol. 52:2107-2111. [DOI] [PubMed] [Google Scholar]
- 18.Ligozzi, M., G. Lo Cascio, and R. Fontana. 1998. vanA gene cluster in a vancomycin-resistant clinical isolate of Bacillus circulans. Antimicrob. Agents Chemother. 42:2055-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, Y. G., and R. F. Whittier. 1995. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25:674-681. [DOI] [PubMed] [Google Scholar]
- 20.Marshall, C. G., I. A. Lessard, I. Park, and G. D. Wright. 1998. Glycopeptide antibiotic resistance genes in glycopeptide-producing organisms. Antimicrob. Agents Chemother. 42:2215-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mengin-Lecreulx, D., B. Flouret, and J. van Heijenoort. 1982. Cytoplasmic steps of peptidoglycan synthesis in Escherichia coli. J. Bacteriol. 151:1109-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ota, Y., H. Tamegai, F. Kudo, H. Kuriki, A. Koike-Takeshita, T. Eguchi, and K. Kakinuma. 2000. Butirosin-biosynthetic gene cluster from Bacillus circulans. J. Antibiot. 53:1158-1167. [DOI] [PubMed] [Google Scholar]
- 23.Patel, R. 2000. Enterococcal-type glycopeptide resistance genes in non-enterococcal organisms. FEMS Microbiol. Lett. 185:1-7. [DOI] [PubMed] [Google Scholar]
- 24.Patel, R., K. Piper, F. R. Cockerill III, J. M. Steckelberg, and A. A. Yousten. 2000. The biopesticide Paenibacillus popilliae has a vancomycin resistance gene cluster homologous to the enterococcal VanA vancomycin resistance gene cluster. Antimicrob. Agents Chemother. 44:705-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perichon, B., and P. Courvalin. 2000. Update on vancomycin resistance. Int. J. Clin. Pract. 54:250-254. [PubMed] [Google Scholar]
- 26.Power, E. G., Y. H. Abdulla, H. G. Talsania, W. Spice, S. Aathithan, and G. L. French. 1995. vanA genes in vancomycin-resistant clinical isolates of Oerskovia turbata and Arcanobacterium (Corynebacterium) haemolyticum. J. Antimicrob. Chemother. 36:595-606. [DOI] [PubMed] [Google Scholar]
- 27.Poyart, C., C. Pierre, G. Quesne, B. Pron, P. Berche, and P. Trieu-Cuot. 1997. Emergence of vancomycin resistance in the genus Streptococcus: characterization of a vanB transferable determinant in Streptococcus bovis. Antimicrob. Agents Chemother. 41:24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quintiliani, R., Jr., and P. Courvalin. 1996. Characterization of Tn1547, a composite transposon flanked by the IS16 and IS256-like elements, that confers vancomycin resistance in Enterococcus faecalis BM4281. Gene 172:1-8. [DOI] [PubMed] [Google Scholar]
- 29.Reynolds, P. E. 1989. Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 8:943-950. [DOI] [PubMed] [Google Scholar]
- 30.Reynolds, P. E., O. H. Ambur, B. Casadewall, and P. Courvalin. 2001. The VanY(D) dd-carboxypeptidase of Enterococcus faecium BM4339 is a penicillin-binding protein. Microbiology 147:2571-2578. [DOI] [PubMed] [Google Scholar]
- 31.Reynolds, P. E., and P. Courvalin. 2005. Vancomycin resistance in enterococci due to synthesis of precursors terminating in d-alanyl-d-serine. Antimicrob. Agents Chemother. 49:21-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reynolds, P. E., F. Depardieu, S. Dutka-Malen, M. Arthur, and P. Courvalin. 1994. Glycopeptide resistance mediated by enterococcal transposon Tn1546 requires production of VanX for hydrolysis of d-alanyl-d-alanine. Mol. Microbiol. 13:1065-1070. [DOI] [PubMed] [Google Scholar]
- 33.Reynolds, P. E., H. A. Snaith, A. J. Maguire, S. Dutka-Malen, and P. Courvalin. 1994. Analysis of peptidoglycan precursors in vancomycin-resistant Enterococcus gallinarum BM4174. Biochem. J. 301:5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rince, A., A. Dufour, S. Le Pogam, D. Thuault, C. M. Bourgeois, and J. P. Le Pennec. 2000. Cloning, expression, and nucleotide sequence of genes involved in production of lactococcin DR, a bacteriocin from Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 60:1652-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shida, O., H. Takagi, K. Kadowaki, L. K. Nakamura, and K. Komagata. 1997. Transfer of Bacillus alginolyticus, Bacillus chondroitinus, Bacillus curdlanolyticus, Bacillus glucanolyticus, Bacillus kobensis, and Bacillus thiaminolyticus to the genus Paenibacillus and emended description of the genus Paenibacillus. Int. J. Syst. Bacteriol. 47:289-298. [DOI] [PubMed] [Google Scholar]
- 36.Sowell, M. O., and C. E. Buchanan. 1983. Changes in penicillin-binding proteins during sporulation of Bacillus subtilis. J. Bacteriol. 153:1331-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vetting, M. W., L. P. S. de Carvalho, M. Yu, S. S. Hegde, S. Magnet, S. L. Roderick, J. S. Blanchard, M. O. Sowell, and C. E. Buchanan. 2005. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 153:212-226. [DOI] [PubMed] [Google Scholar]
- 38.Yoon, J. H., W. T. Seo, Y. K. Shin, Y. H. Kho, K. H. Kang, and Y. H. Park. 2002. Paenibacillus chinjuensis sp. nov., a novel exopolysaccharide-producing bacterium. Int. J. Syst. Evol. Microbiol. 52:415-421. [DOI] [PubMed] [Google Scholar]
- 39.Yoon, J. H., D. K. Yim, J. S. Lee, K. S. Shin, H. H. Sato, S. T. Lee, Y. K. Park, and Y. H. Park. 1998. Paenibacillus campinasensis sp. nov., a cyclodextrin-producing bacterium isolated in Brazil. Int. J. Syst. Bacteriol. 48:833-837. [DOI] [PubMed] [Google Scholar]