Abstract
Salmonella enterica serovar Typhi is an important pathogen exclusively for humans and causes typhoid or enteric fever. It has been shown that type IVB pili, encoded by the S. enterica serovar Typhi pil operon located in Salmonella pathogenicity island 7, are important in the pathogenic process. In this study, by using both an adhesion-invasion assay and fluorescence quantitative PCR analysis, we demonstrated that the entry of type IVB piliated S. enterica serovar Typhi A21-6 (pil+ Kmr) into human THP-1 monocytic cells was greater than that of a nonpiliated S. enterica serovar Typhi pilS::Kmr (pil mutant) strain. We have applied a systematic evolution of ligands by exponential enrichment approach to select oligonucleotides (aptamers) as ligands that specifically bind to type IVB pili. Using this approach, we identified a high-affinity single-stranded RNA aptamer (S-PS8.4) as a type IVB pilus-specific ligand and further found that the selected aptamer (S-PS8.4) could significantly inhibit the entry of the piliated strain (but not that of the nonpiliated strain) into human THP-1 cells. The binding affinities between aptamers and pre-PilS (structural protein of type IVB pili) were determined by nitrocellulose filter-binding assays, and the Kd value was determined to be 8.56 nM for the S-PS8.4 aptamer alone. As an example of an aptamer against type IVB pili of S. enterica serovar Typhi, the aptamer S-PS8.4 can serve as a tool for analysis of bacterial type IVB pilus-host cell interactions and may yield information for the development of putative new drugs against S. enterica serovar Typhi bacterial infections, useful both in prevention of infection and in therapeutic treatment.
Of the more than 2,300 closely related Salmonella serovars identified, Salmonella enterica serovar Typhi is an important pathogen exclusively for humans and can be transmitted through contaminated food and water. It causes typhoid or enteric fever, which is a serious public health problem in developing countries. The genome of S. enterica serovar Typhi contains three large inserts (pathogenicity islands) (11), relative to the chromosome of Salmonella enterica serovar Typhimurium, which is normally noninvasive for humans. The type IVB pil operon of S. enterica serovar Typhi is located in Salmonella pathogenicity island 7 (18) and contains a pilS gene encoding the structural pilin (36, 37). It has been demonstrated that a pilS mutant of S. enterica serovar Typhi exhibited much-reduced adhesion to and invasion of human epithelial gastrointestinal cells in vitro and that purified soluble pre-PilS protein, retaining the signal sequence normally cleaved when the protein is excreted to form insoluble pili based on polymerized PilS, inhibited bacterial invasion (37). The structure of the N-terminally truncated type IVB structural pilin from S. enterica serovar Typhi was determined by nuclear magnetic resonance analysis (34). Type IVB pili, composed largely of polymerized PilS protein, also mediate bacterial self-association, but only when the presumptive minor pilus proteins PilV1 and PilV2 are not expressed (15). Bacterial self-association is an important virulence trait in enterotoxigenic strains of Escherichia coli and in Vibrio cholerae (1). These data indicated that the structural protein PilS of the type IVB pili might play important roles in the pathogenesis of S. enterica serovar Typhi in humans.
The SELEX (systematic evolution of ligands by exponential enrichment) method (28) is an oligonucleotide-based combinatorial library selection procedure that has been used extensively to isolate ligands (aptamers) that bind to proteins (3, 6, 9, 22, 32), cell surface epitopes (19, 21), and other targets (4, 12, 13, 17). Although in recent years SELEX has become increasingly important in the study of functions of proteins, as well as in the fields of drug discovery and identification of antagonists against many functional proteins, this in vitro selection strategy to generate inhibitors of the functions of bacterial proteins remains underutilized. Aptamers have several potential advantages over antibodies and antibiotics. Aptamers have high affinity and specificity for their targets and can be considered oligonucleotide analogs of antibodies. Being smaller than antibodies, aptamers are better candidates for cell penetration and blood clearance. A variety of chemical modifications, such as fluorescent probes, cross-linking reagents, and modifications to the backbone or specific bases by fluorine (2′-hydroxyl groups of the ribose moieties are replaced with fluorine) (7), can be introduced, thereby adding stability and functionality. Moreover, aptamers are nonimmunogenic and therefore do not cause side effects resulting from unwanted immune responses in hosts.
Both single-stranded DNA (ssDNA) and single-stranded RNA (ssRNA) are candidates for aptamers. Comparing ssRNA with ssDNA, ssRNA might have a more variable dimensional structure than ssDNA. As for the oligonucleotide structure, G-C, A-U, and G-U pairings can occur in ssRNA but only G-C and A-U pairings occur in ssDNA. So we chose ssRNA aptamers which had a richer spatial configuration and facilitated interaction with the PilS protein.
During natural Salmonella bacterial infections, monocytes/macrophages serve as key effector cells of the immune response. In order to further investigate the pathogenic roles of the PilS protein of the type IVB pili, we applied a SELEX approach to identify oligonucleotides (aptamers) as ligands that bind to type IVB pili and then investigated the effects of the aptamers on S. enterica serovar Typhi adhesion to and invasion of the human monocytic leukemia cell line THP-1. We have identified a single ssRNA aptamer (S-PS8.4) as a ligand of the S. enterica serovar Typhi type IVB pili and found it significantly inhibited the entry of the piliated strain (but not that of the nonpiliated strain) into human THP-1 cells.
MATERIALS AND METHODS
Materials.
E. coli DH5α and BL21(DE3)/plysS were used as described previously (37). S. enterica serovar Typhi A21-6 (pil+) (37) was generated by inserting a tac promoter between pilM and pilN with the 575-nucleotide pilM-pilN intergenic sequence removed. Therefore, transcription of the pilN through pilV genes of the pil operon was under the control of the tac promoter. This construct does not disrupt other genes, such as that for lipopolysaccharide, that are controlled by different promoters. The nonpiliated strain S. enterica serovar Typhi pilS::Kmr (37) was generated by inserting a kanamycin resistance gene into the pilS gene so that the destruction of the pilin operon did not influence other gene expression that was under the control of different promoters. All bacterial strains were grown in Luria-Bertani broth (LB) as previously described for 14 to 16 h at 30°C (37). Solid medium contained 1.5% (wt/vol) agar. Human acute monocytic leukemia cell line THP-1 cells were cultured in RPMI 1640 medium (Gibco) with 10% (vol/vol) fetal bovine serum (Gibco).
Expression and purification of pre-PilS-glutathione S-transferase (GST) fusion protein.
The PCR product of the pilS gene (which encodes the pre-PilS protein, in which the mature PilS protein is preceded by a signal sequence) was digested with EcoRI and BamHI, purified, and inserted into the EcoRI and BamHI sites of plasmid pGEX-2T to yield plasmid pGEXS5, in which pilS was fused in frame with the gene for GST. pGEXS5 was transformed into E. coli BL21(DE3)/plysS. After growth to logarithmic phase, the strain hosting the plasmid encoding the pre-PilS-GST fusion protein was induced with 1 mM IPTG for 4 h. Cells were sonicated in phosphate-buffered saline (PBS) and the GST-tagged protein purified through glutathione-Sepharose 4B (Amersham Biosciences).
Western blot analysis.
The purified pre-PilS-GST fusion protein and GST, as a control, were separated by 12% (wt/vol) sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes. Fat-free milk powder solution (10%, wt/vol) was used for blocking. The membrane was incubated with a primary anti-pre-PilS-GST polyclonal antibody (37) for 1 h at room temperature, washed three times, and then incubated with a 1:1,000 dilution of alkaline phosphatase-conjugated anti-mouse immunoglobulin G. Color was developed with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate.
SELEX.
SELEX procedures were generally performed as previously described (31, 33, 38). The oligonucleotide template was synthesized as a single-stranded 88-mer with the sequence 5′-GCGGAATTCTAATACGACTCACTATAGGGAACAGTCCGAGCC-N30-GGGTCAATGCGTCATA-3′, where the central N30 represents random oligonucleotides based on equal incorporation of A, G, C, and T at each position. The complementary strand was synthesized using the DNA polymerase I Klenow fragment with primer 2 (5′-GCGGGATCCTATGACGCATTGACCC-3′). The dsDNA molecules were generated by PCR amplification using primer 2 and primer 1 (5′-GCGGAATTCTAATACGACTCACTATAGGGAACAGTCCGAGCC-3′), which contains the T7 polymerase promoter (underlined). The initial RNA random library was prepared by incubation with T7 RNA polymerase for 2 h at 37°C in a 100-μl reaction mixture (2 to 5 μg DNA, 0.5 mM UTP, 0.5 mM ATP, 0.5 mM GTP, 0.5 mM CTP, 20 μl 5× transcription buffer [Promega], 10 mM dithiothreitol [DTT], 100 U RNasin, 40 U T7 RNA polymerase), and the transcribed RNAs were purified using an Rnaid Kit (Qbiogene). To initiate in vitro selection, purified random RNAs were incubated in binding buffer (25 mM Tris-HCl, 50 mM KCl, 200 mM NaCl, 0.2 mM EDTA, 5% [vol/vol] glycerol, 0.5 mM DTT) for 15 min at 20 to 25°C, with GST-pre-PilS protein bound to glutathione-Sepharose 4B. Following washing with at least 40 column volumes of binding buffer, bound RNAs were eluted with 1 M NaCl in binding buffer and purified using the Rnaid Kit. After reverse transcription (RT)-PCR amplification with primer 1 and primer 2, the resulting DNA was transcribed with T7 RNA polymerase for 2 h at 37°C in a 100-μl reaction mixture (2 μg to 5 μg DNA, 0.5 mM UTP, 0.5 mM ATP, 0.5 mM GTP, 0.5 mM CTP, 20 μl 5× transcription buffer [Promega], 10 mM DTT, 100 U RNasin, 40 U T7 RNA polymerase) and this RNA pool was used in the next round of selection. From round three to round eight, RNA pools were first bound to GST-Sepharose 4B to remove nonspecifically bound RNA and then bound to pre-PilS-GST-Sepharose 4B material.
In vitro transcription
The RNA products of the test application and of a control in which the T7 RNA polymerase was omitted were quantitated by liquid scintillation counting after 10 min, 30 min, and 60 min of incubation in 20-μl reaction volumes (1 μg DNA, 12.5 μM UTP, 0.5 mM ATP, 0.5 mM GTP, 0.5 mM CTP, 50 μCi [α-32P]UTP, 4 μl 5× transcription buffer [Promega], 10 mM DTT, 25 U RNasin, 20 U T7 RNA polymerase). Label incorporation into RNA of the test sample increased over time and exceeded that of the control. Label incorporation decreased slightly after 60 min, perhaps due to RNA degradation.
Cloning and sequencing.
After eight rounds of selection, RT-PCR products were digested with EcoRI and BamHI and then subcloned into pUC19. The bank was transformed into E. coli DH5α. Plasmid DNA was isolated from individual clones, purified, and analyzed by sequencing.
Adhesion and invasion assay using mixed infections.
Human acute monocytic leukemia THP-1 cells were maintained and prepared for bacterial adherence and invasion assays as previously described (23). THP-1 cells were grown in RPMI 1640 medium supplemented with 10% (vol/vol) fetal bovine serum to a density of 2 × 105 to 1 × 106/ml. Fresh cells (1 × 106/ml) were added to each well of a six-well plate using 1 ml/well. Then 1 ml of a dilution (in RPMI 1640 medium supplemented with 10% [vol/vol] fetal bovine serum) of an equal mixture of two S. enterica serovar Typhi strains, either S. enterica serovar Typhi pilS::Kmr or Typhi A21-6 (pil+ Kmr) and a wild-type strain of S. enterica serovar Typhi J341 (pil+), was added and mixed with the monocytes at a ratio of 30:1, 10:1, or 1:1 (bacterial cells to eukaryotic cells). Next, the mixtures were incubated at 37°C in a 5% (vol/vol) CO2 atmosphere for 2 h. Bacteria that did not enter the eukaryotic cells were removed by three washes with PBS or eliminated by incubation with gentamicin (200 μg/ml) for 1 h, followed by washing twice with PBS. Intracellular bacteria were recovered by eukaryotic cell lysis with 0.01% (wt/vol) Triton X-100 for 20 min and enumerated on both LB agar plates and LB agar plates supplemented with kanamycin (both the S. enterica serovar Typhi A21-6 wild-type strain and the S. enterica serovar Typhi pilS mutant were Kmr). Because mixtures of the wild-type and mutant strains S. enterica serovar Typhi A21-6 (pil+ Kmr) and S. enterica serovar Typhi pilS::Kmr were used, the procedure decreased the variability between independent trials.
Inhibition of cell invasion and adherence by aptamers.
THP-1 cells were grown in RPMI 1640 medium supplemented with 10% (vol/vol) fetal bovine serum to a density of 2 × 105 to 1 × 106/ml. S. enterica serovar Typhi strains (S. enterica serovar Typhi A21-6 and S. enterica serovar Typhi pilS::Kmr) were grown to stationary culture in LB for 14 to 16 h at 30°C to reach an optical density at 600 nm of 0.5 to 0.7. Prior to infection, the bacteria were incubated with 2 μg RNA aptamer S-PS8.4 in 100 μl diethyl pyrocarbonate-treated PBS at 20 to 25°C for 15 min. The treated bacteria were added to THP-1 cells (2.5 × 105 cells/well) in 24-well plates at a ratio of 12:1 (bacterial cells to eukaryotic cells). Following incubation at 37°C for 2 h in a 5% (vol/vol) CO2 atmosphere, cells were harvested by centrifugation at 900 × g for 10 min and washed thrice with PBS. Gentamicin (200 μg/ml) was added to kill residual extracellular bacteria. After further incubation for 1 h, the THP-1 cells were washed three more times and lysed with 0.01% (wt/vol) Triton X-100 to release intracellular bacteria. The bacteria were enumerated on LB agar plates with kanamycin. Basic controls for each experiment included bacteria only (no THP-1 cells) and THP-1 cells only. The percentage of bacterial adhesion and invasion inhibition afforded by the aptamer was determined as follows: percent inhibition = [(bacterial number with no S-PS8.4 − bacterial number with S-PS8.4)/bacterial number with no S-PS8.4] × 100%.
Real-time fluorescence quantitative PCR (FQ-PCR) assay.
Real-time FQ-PCR templates were prepared as follows. Experiments involving interaction of bacteria with THP-1 cells were performed basically as described above, except that cells were lysed with 0.1 M Tris-HCl (pH 8.0), 0.01 M EDTA, 1% (wt/vol) SDS, 0.04 M DTT, and 0.2 g/liter proteinase K at 55°C for 1 h after gentamicin treatment. Total DNA was purified from the cells, extracted with phenol-chloroform, and precipitated with 2 volumes of 100% ethanol. Quantification of intracellular bacteria was achieved using real-time quantitative PCR with the QuantiTect SYBR Green PCR Handbook Kit (QIAGEN). The DNA templates for real-time quantitative PCR in each sample were equal (35 ng). In this technique, the pilS gene was specifically amplified from S. enterica serovar Typhi A21-6 with primers PF1 (5′AATGGCTCGAGGAAACTGTGAAGCGA3′) and PR (5′TTCATCATGCCAATGGTGTTCGTGGC3′) or from S. enterica serovar Typhi pilS::Kmr with primers PF2 (5′TCGATAGATTGTCGCACCTGATTGCC3′) and PR, respectively. Forward primer PF2, for amplification of pilS::Kmr mutant DNA, lies in the Kmr gene and contains part of the kanamycin resistance gene sequence. The primers were designed to yield PCR products of similar sizes (each ca. 350 bp), and tests using mixtures of equal numbers of the two bacterial strains confirmed that the primer pairs were equally efficient in amplification (data not shown). The standard curves were generated as described previously (35). To determine the effects of aptamer S-PS8.4 on S. enterica serovar Typhi infection, the percentage of inhibition of bacterial intracellularization afforded by the aptamer was calculated as follows: percent inhibition = [(pilS gene concentration with no S-PS8.4 − pilS gene concentration with S-PS8.4)/pilS gene concentration with no S-PS8.4] × 100%.
Nitrocellulose filter-binding assay.
32P-labeled RNA was produced by in vitro transcription with 32P-labeled UTP, nucleoside triphosphates (GTP, CTP, UTP, and ATP), and T7 RNA polymerase. Each 20-μl reaction mixture containing 25 mM Tris-HCl (pH 7.5), 50 mM KCl, 200 mM NaCl, 0.2 mM EDTA, 5% (vol/vol) glycerol, 0.5 mM DTT, and 10 pmol of RNA was allowed to equilibrate with variable (excess) concentrations of protein at 20 to 25°C for 15 min. Samples were filtered through nitrocellulose disks prewetted with 25 mM Tris-HCl (pH 7.5) and immediately rinsed with 3 ml of the same solution. The disks were dried and counted by scintillation counting. Dissociation constants were determined as previously described (10, 24). The data points of binding curves were fitted to the equation F = P/(Kd + P) to determine the dissociation constants by Origin 6.0 (Microcal Software Inc.), where F is the fraction of RNA bound, P is the concentration of protein, and Kd is the dissociation constant.
RESULTS
Isolation of RNA aptamers binding to type IVB pili of S. enterica serovar Typhi.
Before the SELEX selection experiment (below), the random RNA pools obtained at various times after initiation of transcription were quantitated by liquid scintillation counting (Fig. 1). After 10 min, label incorporation into transcription products was detectable. The level of RNA product was significantly increased after 30 min, while label incorporation dropped slightly after 60 min, perhaps due to RNA degradation.
FIG. 1.
In vitro transcription in the SELEX procedure. Labeled RNA products were quantitated by liquid scintillation counting at 10 min, 30 min, and 60 min after initiation of transcription, respectively. Label incorporation into RNA products was increasingly higher than that of the control (no T7 RNA polymerase). The label incorporated into RNA decreased slightly after 30 min, perhaps due to RNA degradation.
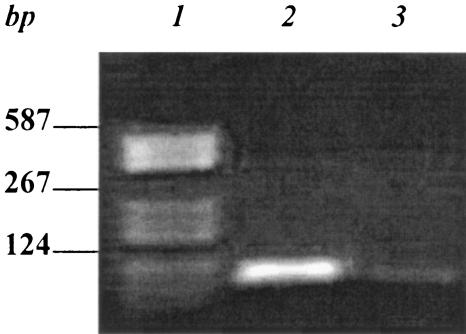
To isolate aptamers that specifically bind to type IVB pili of S. enterica serovar Typhi, we utilized the pilin structural protein, tagged with GST, as a selection target. The pre-PilS-GST fusion protein was expressed in and purified from E. coli (Fig. 2). After synthesis of the complementary strands from the ssDNA library, the dsDNA library was amplified by PCR. Then the dsDNAs were employed as templates for in vitro transcription. An RNA pool containing randomized 30-nucleotide inserts (>1012 molecules) was synthesized. The synthesized random RNA pool was added to pre-PilS-GST-Sepharose 4B beads to select aptamers which might bind to the pre-PilS protein. To ensure the isolation of aptamers of high affinity and specificity, preselections were used, from the third round, to remove RNAs binding to GST-Sepharose 4B. Eight rounds of selection were performed. The final (eighth) RNA pool was reverse transcribed by RT-PCR and subcloned into pUC19 (EcoRI and BamHI) prior to analysis by sequencing. The size of the RT-PCR product was 98 bp, which was the same as the size of the dsDNA (Fig. 3).
FIG. 2.
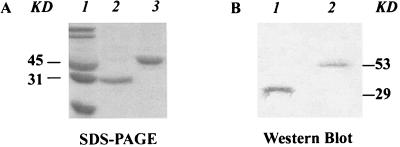
Expression and purification of pre-PilS-GST fusion protein. (A) SDS-polyacrylamide gel electrophoresis with Coomassie blue R-250 staining. Lane 1, molecular mass standard. Lane 2, GST protein (29 kDa). Lane 3, pre-PilS-GST fusion protein (53 kDa). (B) Western blot assay with anti-pre-PilS-GST antibody. Lane 1, GST protein (29 kDa). Lane 2, pre-PilS-GST fusion protein (53 kDa). KD, kilodaltons.
FIG. 3.
Products from the eighth and final round of RT-PCR in the SELEX procedure. Lane 1, DNA molecular weight marker. Lane 2, eighth-round RT-PCR products (98 bp). Lane 3, negative control (no reverse transcriptase).
Aptamer S-PS8.4 secondary-structure prediction.
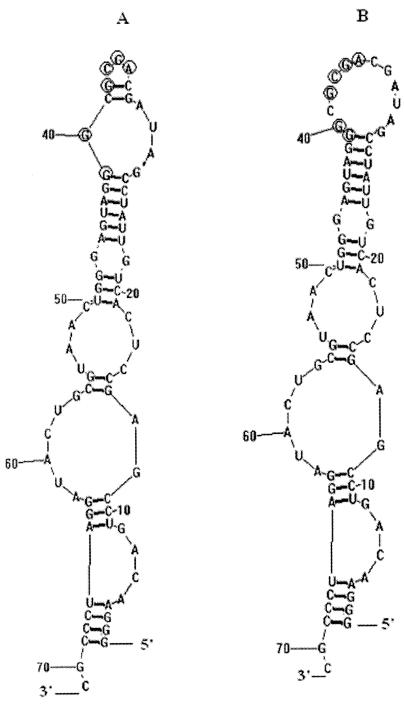
After eight rounds of selection, 14 individual clones that bound to the pre-PilS-GST-Sepharose 4B beads were selected and sequenced. Among them, nine clones were unique and all 14 individual RNAs were aligned using the Clustal software package (Table 1). A consensus sequence (underlined and present eight times in the 14 clones) within individual RNA aptamers was AGCG-(X)-GG. The six RNA aptamers that had no consensus regions (Table 1) perhaps belonged to other small aptamer families. It is interesting that clone S-PS8.4 appeared five times in the RNA aptamer pool. The secondary structure of the S-PS8.4 RNA aptamer was predicted with the RNA structure program (Fig. 4) (version 4.2; D. H. Mathews, University of Rochester Medical Center [http://www.rna.urmc.Rochester.edu/RNAstructure.zip]). The predicted secondary structure of S-PS8.4 shows that the consensus AGCG-(X)-GG sequence occurs in the terminal loop of a stem-loop structure, suggesting that this stem-loop might be the site of binding to the target protein. There are two possible secondary structures (Fig. 4A and B) for the S-PS8.4 RNA aptamer. These two predicted secondary structures are equally likely from the sequence analysis of aptamer S-PS8.4, and the free energies of the two predicted secondary structures are very similar. The only difference between the two predicted structures lies in their top stem-loops. In Fig. 4B, 14 nucleotides are connected to form the top stem-loop, but in Fig. 4A, the same 14 nucleotides form the two stem-loops, 5 nucleotides connecting to form the small top loop and the other 9 nucleotides forming the larger one.
TABLE 1.
Aptamers selected by the SELEX procedure
| Aptamer | Frequency | RNA aptamer sequence (N30)a |
|---|---|---|
| S-PS8.4 | 5/14 | UCACUGUUAUCCGAUAGCAGCGCGGGAUGA |
| S-PS8.3 | 2/14 | AUUACCUAGAGCGGGAUAAAGUAUAGGUU |
| S-PS8.2 | 1/14 | CAUCGAGGAGGCGGGAUAUUCGAUGAGUU |
| S-PS8.5 | 1/14 | GAGUACGAGCGGGAUAGUAAUCGGGUGAU |
| S-PS8.6 | 1/14 | GAAGGGGUCCGGCUCGGGUGAGGGUCGGGU |
| S-PS8.9 | 1/14 | AGCUAGCGGGGGGGCUCGACGGUGGUGGGUU |
| S-PS8.7 | 1/14 | UAGCGGGAGCUUGGACCUGGUGGUCGCGGC |
| S-PS8.1 | 1/14 | UUGGGAUCAGCUCGGCGCUGGAGGAGGGGC |
| S-PS8.8 | 1/14 | AACUGUACAUGGGCGCAACAGGGAGUUAGC |
Consensus regions within the 14 individual aptamers are underlined.
FIG. 4.
Prediction of aptamer S-PS8.4 secondary structures. After eight rounds of SELEX selection, clone S-PS8.4, which represented the most frequent RNA isolate, was selected. The secondary structures of S-PS8.4 (both panels A and B) were predicted by using the RNA structure program. The predicted secondary structures of S-PS8.4 show that the consensus AGCG-(X)-GG sequence occurs in the terminal loop of a stem-loop structure. (A) The consensus AGCG-(X)-GG sequence encircled with rings occurs in the two terminal loops of a stem-loop structure. (B) The consensus AGCG-(X)-GG sequence encircled with rings occurs in the one terminal loop of a stem-loop structure.
A pool of RNA aptamers significantly inhibited S. enterica serovar Typhi invasion of THP-1 cells.
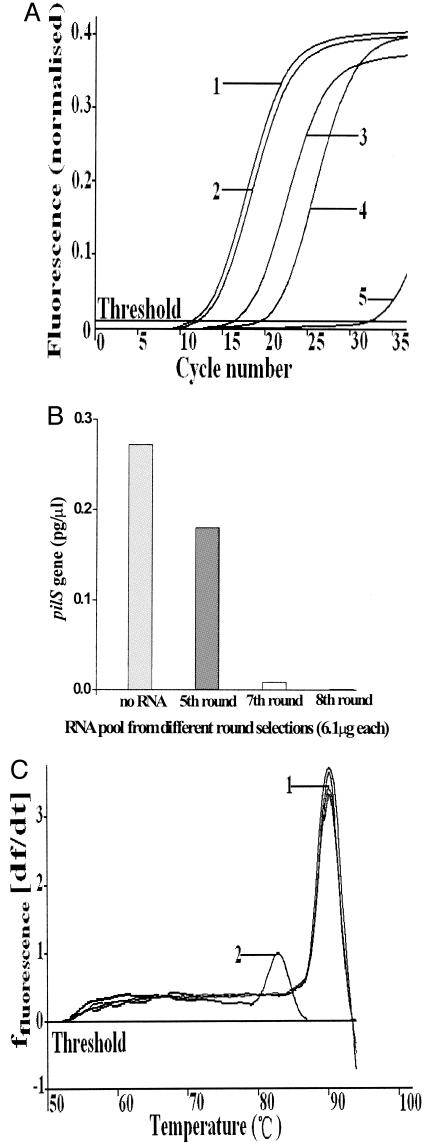
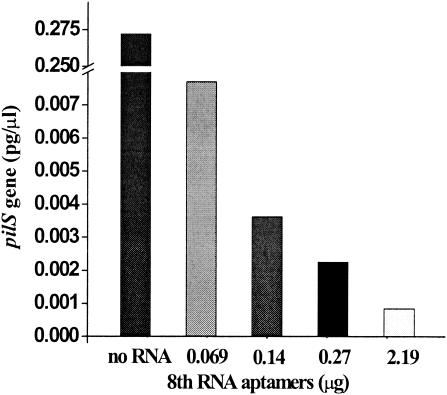
Based on previous reports, an obvious improvement in aptamer-protein binding was usually apparent after five rounds of amplification (6, 12). In this study, we decided to examine the biological activities of aptamers after the fifth, seventh, and eighth rounds of selection. Compared with a control containing no RNA, addition of the RNA aptamer pools caused significant inhibition of bacterial infection in the real-time FQ-PCR assay (Fig. 5A and B). The inhibitory effects of the RNA aptamer pool increased with advancing cycles (Fig. 5A and B). Moreover, the inhibitory effect of the eighth-round aptamer pool was concentration dependent (Fig. 6). With the addition of 6.1 μg of the RNA aptamer pool from the fifth, seventh, or eighth amplification round, cell invasion by S. enterica serovar Typhi A21-6 decreased to 66%, 3%, and 0.3%, respectively, compared to the cell invasion level seen in the RNA-free control (Fig. 5B). The melting curves of the PCR products suggested that there were no nonspecific PCR products in the PCRs (Fig. 5C). After the eighth round of selection, high-affinity RNA aptamers were obtained and almost completely inhibited bacterial infection (Fig. 6).
FIG. 5.
Inhibition of bacterial adhesion and invasion by the RNA pools obtained at various intervals during the SELEX procedure. The assay of inhibition of bacterial adherence and invasion was performed as described in Materials and Methods. (A) Inhibition of adhesion and invasion by various RNA pools as shown by real-time FQ-PCR amplification curves. The pil+ strain S. enterica serovar Typhi A21-6 was used at a 12:1 ratio of bacterial cells to eukaryotic cells. Curves: 1, S. enterica serovar Typhi A21-6 control (no RNA); 2, the fifth RNA pool; 3, the seventh RNA pool; 4, the eighth RNA pool; 5, control (THP-1 cells only). (B) Inhibition of adhesion and invasion by the various RNA pools as shown by quantitation of the cell-associated pilS gene PCR product (pg/μl), calculated with data from the real-time quantitative PCR method. (C) Melting curves of PCR products suggesting that no unspecific PCR products were formed in the PCRs. Curves: 1, real-time melting temperatures of RNA pools from the fifth, seventh, and eighth rounds of amplification; 2, melting temperature of PCR products from the control (THP-1 cells only) in the real-time FQ-PCR.
FIG. 6.
Inhibition of adhesion and invasion by the eighth-round RNA pool as measured by levels of the intracellular pilS gene PCR product (pg/μl), calculated from data obtained using the real-time quantitative PCR method.
Selected RNA aptamer S-PS8.4 significantly inhibited S. enterica serovar Typhi infection of human monocytes.
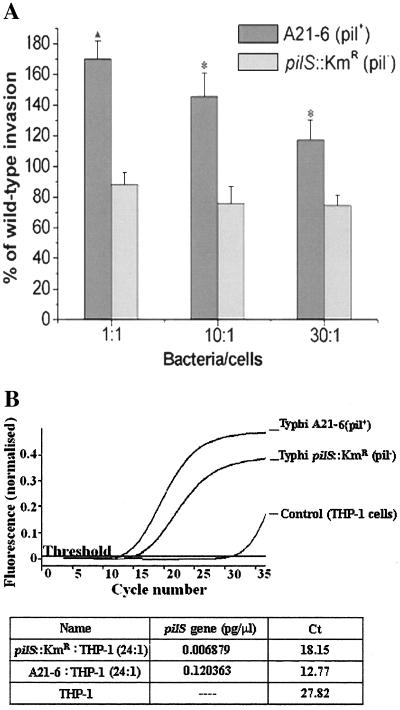
The mixed-infection assay (Fig. 7A) and the FQ-PCR assay (Fig. 7B) showed that S. enterica serovar Typhi A21-6 (pil+) entered THP-1 cells to an extent greater than that of S. enterica serovar Typhi pilS::Kmr. PCR products were quantitated by measurement of the density levels of strain-specific PCR products after agarose gel electrophoresis, and the results were the same (data not shown). By the mixed-infection assay, the entry level of the pilS mutant was about half that of the pil+ strain. In the FQ-PCR assay, however, the entry of the pilS mutant was reduced to a greater extent. Thus, 19 rounds of PCR were required to achieve a fluorescence level of 0.2 for the pil+ strain but 23 rounds were needed to attain the same fluorescence level for the pilS mutant. The data suggest that the intracellular pilS DNA concentration of the pilS mutant was present at a level only 6 to 7% of that of the pil+ strain. There are two possibilities leading to this result. One is that the invasion of the pil+ strain induced cell ruffles neighboring the pilS mutant bacteria. As the type IVB pili are known to mediate bacterial self-association (15), another possibility is that enmeshment of the pilS mutant by the accompanying pil+ strain in the mixed-infection assay yields artificially high levels of pilS mutant entry levels. The results of experiments comparing mixed and individual infections using a gentamicin assay also indicated that the pilS mutant could accompany the pilS+ strain in the mixed-infection assay (data not shown). This concern is absent with the FQ-PCR assay, where single bacterial strains are used.
FIG. 7.
Adhesion to and invasion of THP-1 cells by S. enterica serovar Typhi A21-6 (pil+) and S. enterica serovar Typhi pilS::Kmr using both mixed-infection and real-time quantitative PCR assays. (A) Adhesion to and invasion of THP-1 cells by S. enterica serovar Typhi A21-6 (pil+) and S. enterica serovar Typhi pilS::Kmr using the mixed-infection assay. A21-6 (pil+) versus pilS::Kmr (pil) ▴, P < 0.01 (bacterium/cell ratio: 1:1). *, P < 0.05 (bacterium/cell ratios: 10:1 and 30:1). (B) Adhesion to and invasion of THP-1 cells by S. enterica serovar Typhi A21-6 (pil+) and S. enterica serovar Typhi pilS::Kmr using the real-time quantitative PCR assay. The threshold cycle (Ct) is the cycle at which there is a significant increase in fluorescence, and this value is associated with exponential growth of the PCR product during the log-linear phase. Generally, the more original the DNA template is, the fewer the threshold cycles required.
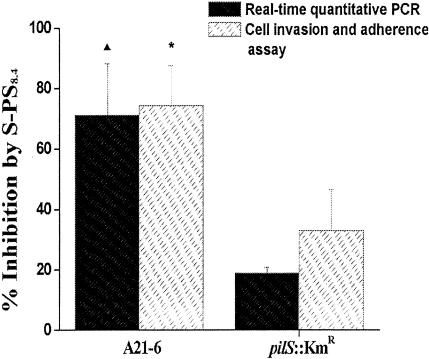
The effects of the selected RNA aptamer S-PS8.4 alone on S. enterica serovar Typhi A21-6 and S. enterica serovar Typhi pilS::Kmr cell infection were determined with real-time quantitative PCR and the mixed-infection assays (Fig. 8). The results showed that 2.0 μg of RNA aptamer S-PS8.4 effected ca. 71% inhibition of cell invasion by pil+ S. enterica serovar Typhi A21-6 but only ca. 19% inhibition in the case of the pilS::Kmr mutant. These results suggested that the selected aptamer, S-PS8.4, alone inhibited S. enterica serovar Typhi invasion of monocytes by specifically binding to type IVB pili. However, S-PS8.4 still showed a mild inhibitory effect on the entry of the pilS::Kmr mutant strain. It is possible that S-PS8.4 had a low affinity for an unknown component in the pilS::Kmr strain and therefore caused a mild reduction in cell adhesion and invasion. Results from both real-time quantitative PCR analysis and the mixed-infection assay were consistent, and we conclude that the real-time quantitative PCR analysis fairly reflects the effects of the aptamer on S. enterica serovar Typhi invasion.
FIG. 8.
S-PS8.4 inhibits cell invasion by S. enterica serovar Typhi, as shown by both real-time quantitative PCR and cell invasion and adherence assays. S-PS8.4 inhibited THP-1 cell invasion by S. enterica serovar Typhi A21-6 by 71% (real-time quantitative PCR) or 74% (cell invasion and adherence assay) and THP-1 cell invasion by the S. enterica serovar Typhi pilS::Kmr strain by 19% or 33%, respectively. In each assay, three experiments were carried out and each was performed in triplicate. The ratio of inhibition by S-PS8.4 between A21-6 (pil+) and pilS::Kmr (pil) was statistically significant (▴, P < 0.01 in the real-time quantitative PCR; *, P < 0.05 in the cell invasion and adherence assay).
Determination of the affinity of binding of RNA aptamers to target protein.
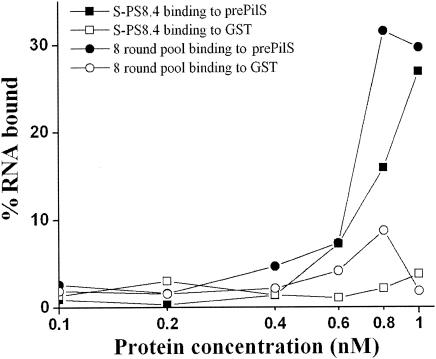
In order to determine the binding affinities of aptamers, nitrocellulose filter-binding assays were performed. Binding reactions were carried out with a constant concentration of RNA aptamer, and the concentration of pre-PilS-GST or GST protein was titrated from 0.1 nM to 1 nM (Fig. 9). Binding curves of selected aptamers and pre-PilS showed that affinities were increased in a pre-PilS protein concentration-dependent manner and that the affinity of RNA binding to pre-PilS was much higher than the affinity of binding to GST (Fig. 9). The Kd values of binding to pre-PilS of the eighth-round RNA aptamer pool and of clone S-PS8.4 alone were 6.08 nM and 8.56 nM, respectively, while the Kd values of binding to GST were 98.13 nM and 119.28 nM, respectively. Our experimental results have provided evidence of strong binding and interaction between the aptamers and PilS protein and very weak binding and interaction between the aptamers and GST protein (Fig. 9). In addition, the binding affinity of the eighth-round RNA pool was slightly higher than that of the S-PS8.4 aptamer alone (Fig. 9).
FIG. 9.
Pre-PilS-binding curves of selected aptamers. Proteins were incubated with labeled RNA aptamer S-PS8.4 or the eighth-round labeled RNA pool. Bound aptamers were quantitated by scintillation counting. The levels of RNA bound by the pre-PilS protein were determined by subtracting the levels of RNA bound by GST protein from the levels of RNA bound by the pre-PilS-GST fusion protein. Both S-PS8.4 and the eighth-round pool bound strongly to pre-PilS (the Kd values were 8.56 nM and 6.08 nM, respectively) but bound weakly to GST (the Kd values were 98.13 nM and 119.28 nM, respectively).
DISCUSSION
S. enterica serovar Typhi contains a type IVB pil operon that is absent in S. enterica serovar Typhimurium. Moreover, the type IVB pilus operon is confined to S. enterica serovar Typhi and a few other human-invasive strains such as S. enterica serovars Paratyphi C and Dublin (14, 26). The type IVB pilus-mediated events may be important in the mediation of enteric fever in humans as elements of pathogenicity required for the development of epidemics of typhoid fever.
RNA aptamers could be used as potential agents against bacterial invasion and pathogenesis. SELEX approaches have been used in the identification of RNA aptamers used to determine the toxicity of bacteria to host cells (20, 25). In addition, in vitro selection of RNA ligands that block adhesion to and invasion of monkey kidney epithelial cells by Trypanosoma cruzi has also been described (30) and the aptamers were successfully used to inhibit invasion by the parasite. However, selection of nucleic acids directly by affinity for bacterial proteins involved in bacterial invasion has not been reported. S. enterica serovar Typhi remains epidemic in developing countries, with an estimated incidence of 33 million cases each year (27). For patients with typhoid fever, administration of an effective antibiotic is essential. However, the development of bacterial resistance to antibiotics such as chloramphenicol, ampicillin, and trimethoprim in epidemic strains is a major concern (27). RNA aptamers against type IVB pili of S. enterica serovar Typhi generated from SELEX selection may be further developed into new antibacterial agents with the potential to overcome resistance issues, since these RNA aptamers are different from antibiotics in terms of structure and function. In addition, these molecules may inhibit bacterial pathogenesis prior to eukaryotic cell invasion, perhaps by interfering with bacterial aggregation or directly by prohibiting bacterial entry.
Since the first description of the in vitro selection of novel ligands from combinational nucleic acid libraries by SELEX, much effort has been devoted to optimization of the methodology. In this study, we used Sepharose 4B instead of nitrocellulose as the selection matrix. In order to increase the specificity of selected RNA aptamers, we have utilized a combinatorial library approach by pretreating RNA aptamers with GST-Sepharose 4B before each round of SELEX selection, from the third round to the eighth and final round. Although this combinatorial library approach has been successfully used to isolate aptamers blocking ligand-receptor interactions between eukaryotic cells and viruses (2, 5, 8) or between eukaryotic cells themselves (17, 22, 29), the technique has not been applied to the selection of ligands blocking an interaction between bacterial and eukaryotic cells. Our studies indicate that the SELEX approach is a useful tool to study the roles of bacterial proteins in pathogenesis and invasion of host cells, as well as to provide insights into the mechanisms of pathogen-host cell interactions.
Our data show that the selected aptamer, S-PS8.4, had the highest ratio from eight rounds of selection and a sequence that was identical to that of 9 of 14 individual clones. These findings suggested that aptamer S-PS8.4 should have a high affinity for the type IVB pilus protein. In the selection procedures, the RNA aptamers bound to pre-PilS competitively. Only those high-affinity aptamers had competitive advantages and survived in the final RNA pool. The aptamer with the highest frequency of occurrence in the final selected RNA pool may potentially bind to pre-PilS protein the most tightly. From our experimental results (Fig. 9), total RNA pools which contained an aptamer mixture had more binding affinity than aptamer S-PS8.4 alone. (The Kd values of binding to pre-PilS of the eighth-round RNA aptamer pool and of clone S-PS8.4 alone were 6.08 nM and 8.56 nM, respectively.) These results suggested that, except for aptamer S-PS8.4 alone, the other aptamers from the eighth-round selection might also have some inhibition efficiency but should have less inhibition efficiency than aptamer S-PS8.4.
A consensus sequence (present eight times in the 14 clones) within individual RNA aptamers was AGCG-(X)-GG, and this consensus region is located in the terminal loop of the predicted S-PS8.4 secondary structure, suggesting that this stem-loop might be the site of binding to the target protein. Several of the other six aptamers (Table 1) without the AGCG(X)GG sequence also have stem-loop structures in predicted secondary structures; however, these stem-loops appeared to occur only once and were not present in other aptamers and therefore these stem-loops might have a lower affinity for the pre-PilS protein. In our work, we focused on aptamer S-PS8.4, which had the highest frequency of occurrence because of its high affinity for pre-PilS protein.
To identify the biological activities, and substrate affinity, of an RNA aptamer, a real-time quantitative PCR assay for quantification of bacterial adhesion and invasion was used in this work. After interaction with the RNA aptamer, a reduction of cell invasion resulted. Intracellular bacteria were quantitated by real-time PCR. Although killed extracellular bacteria may not have been completely removed, the technique is useful to measure and quantify cell invasion effects and eliminates the need for microscopic enumeration of adherent and invasive bacteria. In our studies, the results from PCR assays were consistent with those from the classical cell invasion and adherence assay. Naguleswaran et al. (16) compared the real-time quantitative PCR assay with other adhesion-invasion assays, and those results also showed the reliability of the method.
Two or three of the most efficient aptamers together to inhibit cell invasion by S. enterica serovar Typhi perhaps might be the perfect therapeutic agents. However, selecting several of the most efficient aptamers requires further affinity assays and biological function assays, and we have found dramatically efficient inhibition of cell invasion by S. enterica serovar Typhi in the high-affinity aptamer pool (the final selected RNA pool). In our study, aptamer S-PS8.4 was found to inhibit Salmonella invasion of human monocytic cells significantly.
In summary, our results suggest that aptamer S-PS8.4, selected here for affinity for type IVB pili, has strong potential both as an antagonist against S. enterica serovar Typhi invasion of human monocytic cells and as a tool to analyze the interactions of bacterial type IVB pili with host cells and examine important aspects of bacterial pathogenesis.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (30270076) and the Ministry of Education Scientific Research Foundation for Returned Overseas Chinese Scholars (301153033), Outstanding Youth Scholars grants from Hubei Province (2002AC017 and 2002AA301B15), and grants from Wuhan University (301270055 and 301276010).
REFERENCES
- 1.Bieber, D., S. W. Ramer, C. Y. Wu, W. J. Murray, T. Tobe, R. Fernandez, and G. K. Schoolnik. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280:2114-2118. [DOI] [PubMed] [Google Scholar]
- 2.Biroccio, A., J. Hamm, I. Incitti, R. De Francesco, and L. Tomei. 2002. Selection of RNA aptamers that are specific and high-affinity ligands of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 76:3688-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, C. H., G. A. Chernis, V. Q. Hoang, and R. Landgraf. 2003. Inhibition of heregulin signaling by an aptamer that preferentially binds to the oligomeric form of human epidermal growth factor receptor-3. Proc. Natl. Acad. Sci. USA 100:9226-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daniels, D. A., H. Chen, B. J. Hicke, K. M. Swiderek, and L. Gold. 2003. A tenascin-C aptamer identified by tumor cell SELEX: systematic evolution of ligands by exponential enrichment. Proc. Natl. Acad. Sci. USA 100:15416-15421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukuda, K., D. Vishnuvardhan, S. Sekiya, J. Hwang, N. Kakiuchi, K. Taira, K. Shimotohno, P. K. Kumar, and S. Nishikawa. 2000. Isolation and characterization of RNA aptamers specific for the hepatitis C virus nonstructural protein 3 protease. Eur. J. Biochem. 267:3685-3694. [DOI] [PubMed] [Google Scholar]
- 6.Hicke, B. J., C. Marion, Y. F. Chang, T. Gould, C. K. Lynoct, D. Parma, P. G. Schmidt, and S. Warren. 2001. Tenascin-C aptamers are generated using tumor cells and purified protein. J. Biol. Chem. 276:48644-48654. [DOI] [PubMed] [Google Scholar]
- 7.Jellinek, D., L. S. Green, C. Bell, C. K. Lynott, N. Gill, C. Vargeese, G. Kirschenheuter, D. P. McGee, P. Abesinghe, W. A. Pieken, R. Shapiro, D. B. Rifkin, D. Moscatelli, and N. Janjc. 1995. Potent 2′-amino-2′-deoxypyrimidine RNA inhibitors of basic fibroblast growth factor. Biochemistry 34:11363-11372. [DOI] [PubMed] [Google Scholar]
- 8.Joshi, P., and V. R. Prasad. 2002. Potent inhibition of human immunodeficiency virus type 1 replication by template analog reverse transcriptase inhibitors derived by SELEX (systematic evolution of ligands by exponential enrichment). J. Virol. 76:6545-6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimoto, M., K. Sakamoto, M. Shirouzu, I. Hirao, and S. Yokoyama. 1998. RNA aptamers that specifically bind to the Ras-binding domain of Raf-1. FEBS Lett. 441:322-326. [DOI] [PubMed] [Google Scholar]
- 10.Lim, F., and D. S. Peabody. 2002. RNA recognition site of PP7 coat protein. Nucleic Acids Res. 30:4138-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu, S. L., and K. E. Sanderson. 1995. Rearrangements in the genome of the bacterium Salmonella typhi. Pro. Natl. Acad. Sci. USA 92:1018-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meli, M., J. Vergne, J. L. Decout, and M. C. Maurel. 2002. Adenine-aptamer complexes: a bipartite RNA site that binds the adenine nucleic base. J. Biol. Chem. 277:2104-2111. [DOI] [PubMed] [Google Scholar]
- 13.Morris, K. N., K. B. Jensen, C. M. Julin, M. Weil, and L. Gold. 1998. High affinity ligands from in vitro selection: complex targets. Proc. Natl. Acad. Sci. USA 95:2902-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris, C., C. K. Tam, T. S. Wallis, P. W. Jones, and J. Hackett. 2003. Salmonella enterica serovar Dublin strains which are Vi antigen-positive use type IVB pili for bacterial self-association and human intestinal cell entry. Microb. Pathog. 35:279-284. [DOI] [PubMed] [Google Scholar]
- 15.Morris, C., C. M. Yip, I. S. Tsui, D. K. Wong, and J. Hackett. 2003. The shufflon of Salmonella enterica serovar typhi regulates type IVB pilus-mediated bacterial self-association. Infect. Immun. 71:1141-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naguleswaran, A., N. Muller, and A. Hemphill. 2003. Neospora caninum and Toxoplasma gondii: a novel adhesion/invasion assay reveals distinct differences in tachyzoite-host cell interactions. Exp. Parasitol. 104:149-158. [DOI] [PubMed] [Google Scholar]
- 17.O'Connell, D., A. Koenig, S. Jennings, B. Hicke, H. L. Han, T. Fitwater, Y. F. Chang, N. Varki, D. Parma, and A. Varki. 1996. Calcium-dependent oligonucleotide antagonists specific for l-selectin. Proc. Natl. Acad. Sci. USA 93:5883-5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pickard, D., J. Wain, S. Baker, A. Line, S. Chohan, M. Fookes, A. Barron, P. O. Gaora, J. A. Chabalgoity, N. Thanky, C. Scholes, N. Thomson, M. Quail, J. Parkhill, and G. Dougan. 2003. Composition, acquisition, and distribution of the Vi exopolysaccharide-encoding Salmonella enterica pathogenicity island SPI-7. J. Bacteriol. 185:5055-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Proske, D., M. Hofliger, R. M. Soll, A. G. Beck-Sickinger, and M. Famulok. 2002. A Y2 receptor mimetic aptamer directed against neuropeptide Y. J. Biol. Chem. 277:11416-11422. [DOI] [PubMed] [Google Scholar]
- 20.Qian, Y., J. H. Lee, and R. K. Holmes. 2002. Identification of a DtxR-regulated operon that is essential for siderophore-dependent iron uptake in Corynebacterium diphtheriae. J. Bacteriol. 184:4846-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhodes, A., A. Deakin, J. Spaull, B. Coomber, A. Aitken, P. Life, and S. Rees. 2000. The generation and characterization of antagonist RNA aptamers to human oncostatin M. J. Biol. Chem. 275:28555-28561. [DOI] [PubMed] [Google Scholar]
- 22.Rhodes, A., N. Smithers, T. Chapman, S. Parsons, and S. Rees. 2001. The generation and characterization of antagonist RNA aptamers to MCP-1. FEBS Lett. 506:85-90. [DOI] [PubMed] [Google Scholar]
- 23.Schmitt, C. K., J. S. Ikeda, S. C. Darnell, P. R. Watson, J. Bispham, T. S. Wallis, D. L. Weinstein, E. S. Metcalf, and A. D. O'Brien. 2001. Absence of all components of the flagellar export and synthesis machinery differentially alters virulence of Salmonella enterica serovar Typhimurium in models of typhoid fever, survival in macrophages, tissue culture invasiveness, and calf enterocolitis. Infect. Immun. 69:5619-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider, D., C. Tuerk, and L. Gold. 1992. Selection of high affinity RNA ligands to the bacteriophage R17 coat protein. J. Mol. Biol. 228:862-869. [DOI] [PubMed] [Google Scholar]
- 25.Sterba, K. M., S. G. Mackintosh, J. S. Blevins, B. K. Hurlburt, and M. S. Smeltzer. 2003. Characterization of Staphylococcus aureus SarA binding sites. J. Bacteriol. 185:4410-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tam, C. K., J. Hackett, and C. Morris. 2004. Salmonella enterica serovar Paratyphi C carries an inactive shufflon. Infect. Immun. 72:22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Threlfall, E. J. 2002. Antimicrobial drug resistance in Salmonella: problems and perspectives in food- and water-borne infections. FEMS Microbiol. Rev. 26:141-148. [DOI] [PubMed] [Google Scholar]
- 28.Tuerk, C., and L. Gold. 1990. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249:505-510. [DOI] [PubMed] [Google Scholar]
- 29.Ulrich, H., J. E. Ippolito, O. R. Pagan, V. A. Eterovic, R. M. Hann, H. Shi, J. T. Lis, M. E. Eldefrawi, and G. P. Hess. 1998. In vitro selection of RNA molecules that displace cocaine from the membrane-bound nicotinic acetylcholine receptor. Proc. Natl. Acad. Sci. USA 95:14051-14056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ulrich, H., M. H. Magdesian, M. J. Alves, and W. Colli. 2002. In vitro selection of RNA aptamers that bind to cell adhesion receptors of Trypanosoma cruzi and inhibit cell invasion. J. Biol. Chem. 277:20756-20762. [DOI] [PubMed] [Google Scholar]
- 31.Venables, J. P., M. Ruggiu, and H. J. Cooke. 2001. The RNA-binding specificity of the mouse Dazl protein. Nucleic Acids Res. 29:2479-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White, R. R., S. Shan, C. P. Rusconi, G. Shetty, M. W. Dewhirst, C. D. Kontos, and B. A. Sullenger. 2003. Inhibition of rat corneal angiogenesis by a nuclease-resistant RNA aptamer specific for angiopoietin-2. Proc. Natl. Acad. Sci. USA 100:5028-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu, A., and K. Y. Chen. 2001. Hypusine is required for a sequence-specific interaction of eukaryotic initiation factor 5A with postsystematic evolution of ligands by exponential enrichment RNA. J. Biol. Chem. 276:2555-2561. [DOI] [PubMed] [Google Scholar]
- 34.Xu, X. F., Y. W. Tan, L. Lam, J. Hackett, M. Zhang, and Y. K. Mok. 2004. NMR structure of a type IVB pilin from Salmonella typhi and its assembly into pilus. J. Biol. Chem. 279:31599-31605. [DOI] [PubMed] [Google Scholar]
- 35.Yukiue, H., H. Sasaki, Y. Kobayashi, Y. Nakashima, S. Moriyama, M. Yano, M. Kaji, M. Kiriyama, I. Fukai, Y. Yamakawa, and Y. Fujii. 2003. Clinical significance of tissue inhibitor of metalloproteinase and matrix metalloproteinase mRNA expression in thymoma. J. Surg. Res. 109:86-91. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, X. L., C. Morris, and J. Hackett. 1997. Molecular cloning, nucleotide sequence, and function of a site-specific recombinase encoded in the major ‘pathogenicity island’ of Salmonella typhi. Gene 202:139-146. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, X. L., I. S. Tsui, C. M. Yip, A. W. Fung, D. K. Wong, X. Dai, Y. Yang, J. Hackett, and C. Morris. 2000. Salmonella enterica serovar typhi uses type IVB pili to enter human intestinal epithelial cells. Infect. Immun. 68:3067-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zolotukhin, A. S., D. Michalowski, S. Smulevitch, and B. K. Felber. 2001. Retroviral constitutive transport element evolved from cellular TAP(NXF1)-binding sequences. J. Virol. 75:5567-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]