Abstract
Compounds A-782759 (an N-1-aza-4-hydroxyquinolone benzothiadiazine) and BILN-2061 are specific anti-hepatitis C virus (HCV) agents that inhibit the RNA-dependent RNA polymerase and the NS3 serine protease, respectively. Both compounds display potent activity against HCV replicons in tissue culture. In order to characterize the development of resistance to these anti-HCV agents, HCV subgenomic 1b-N replicon cells were cultured with A-782759 alone or in combination with BILN-2061 at concentrations 10 times above their corresponding 50% inhibitory concentrations in the presence of neomycin. Single substitutions in the NS5B polymerase gene (H95Q, N411S, M414L, M414T, or Y448H) resulted in substantial decreases in susceptibility to A-782759. Similarly, replicons containing mutations in the NS5B polymerase gene (M414L or M414T), together with single mutations in the NS3 protease gene (A156V or D168V), conferred high levels of resistance to both A-782759 and BILN-2061. However, the A-782759-resistant mutants remained susceptible to nucleoside and two other classes of nonnucleoside NS5B polymerase inhibitors, as well as interferon. In addition, we found that the frequency of replicons resistant to both compounds was significantly lower than the frequency of resistance to the single compound. Furthermore, the dually resistant mutants displayed significantly reduced replication capacities compared to the wild-type replicon. These findings provide strategic guidance for the future treatment of HCV infection.
Hepatitis C virus (HCV) is a leading cause of chronic liver disease, affecting over 4 million Americans and about 170 million people worldwide. The current standard of care for chronic HCV infection involves extended dosing with alpha interferon (IFN-α) and ribavirin (10, 14, 18). However, these regimens have limited clinical benefit due to poor tolerability and limited efficacy (only approximately half of genotype 1 HCV-infected individuals have a sustained virological response, whereas the response rate improves significantly [∼80%] when genotypes 2 and 3 are treated) (9, 11, 35). Therefore, development of inhibitors against virally encoded targets is urgently needed.
The HCV genome is a 9.6-kb single-stranded RNA of positive polarity encoding a large polyprotein, which is the precursor of at least 10 mature viral proteins: C, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B (2). The HCV polymerase encoded by nonstructural protein 5B (NS5B) is responsible for HCV RNA-dependent RNA polymerase (RdRp) and terminal transferase activities (4, 31, 32). The N-terminal domain (approximately 180 amino acids) of NS3 and the small hydrophobic NS4A protein compose a heterodimeric enzyme catalyzing the posttranslational processing of the HCV NS proteins (1, 21). Both NS5B RdRp and NS3 serine protease are believed to be components of the HCV replication complex, responsible for viral RNA replication, and have been shown to be indispensable for HCV replication in chimpanzees (26).
To date, a number of distinct classes of NS5B RdRp inhibitors, including nucleoside analogs, pyrophosphate mimics, and nonnucleoside analogs, have been reported in the scientific and patent literature (12, 13, 20, 40, 50, 51). The nucleoside analogs act as chain terminators by competing with the substrate of RdRp (40). This class of compounds has been shown to inhibit HCV replication in both subgenomic replicons and HCV-infected chimpanzees (5, 19, 41, 49). The pyrophosphate mimics, including dihydroxypyrimidine carboxylic acids and diketoacid derivatives, are believed to inhibit RdRp activity through an interaction with the catalytic metal ions in the enzyme active site (42, 50, 51). For nonnucleoside analogs, several structurally distinct series have been identified, including benzothiadiazines, benzimidazoles/diamides, substituted pyranones, and disubstituted phenylalanine/thiophene amides (13, 22, 25, 33, 47, 52, 57). Crystal structures of NS5B complexed with substituted pyranone derivatives and phenylalanine amides demonstrated that the inhibitors were allosterically bound on the protein surface within a narrow cleft at the base of the thumb domain, ∼30 and 35 Å from the active site, respectively (see Fig. 3) (27, 33, 57). Recently, X-ray crystal structures of several benzothiadiazines bound to HCV NS5B RdRp revealed that they bind to a second allosteric site in close proximity to the interface of the palm and thumb domains of NS5B (15). Furthermore, resistance and nuclear magnetic resonance studies of the benzimidazoles/diamides suggested that this class of inhibitors binds to a third allosteric site of the NS5B RdRp protein, which is located in the upper thumb domain of the enzyme (3, 8, 52). Since structurally distinct nonnucleoside analogs bind to three different allosteric binding sites, it is of interest to examine whether they have nonoverlapping resistances so that combination therapy regimens based on the use of multiple nonnucleoside inhibitors may be considered as a future option for HCV treatment.
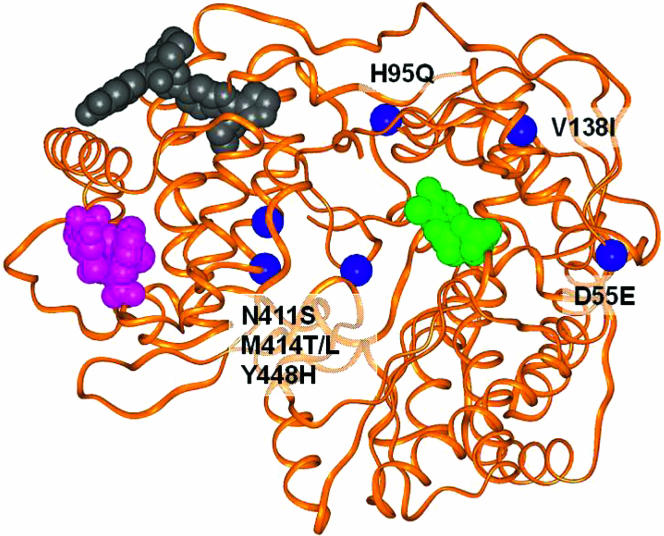
FIG. 3.
Locations of mutations and inhibitor binding sites for HCV polymerase enzyme. A ribbon representation (orange) of the protein is shown for Protein Database entry 1C2P (27). Mutations are shown as blue balls at the alpha carbons of the specified residues. Space-filling representations of various inhibitors are also shown: phenylalanine-amide (purple) (57), diamide (gray) (8), and nucleoside (green) (24).
Despite the shallow substrate binding site of the HCV NS3 serine protease, a number of inhibitors of this enzyme have been identified (17, 43, 55, 59). Generally, these inhibitors are substrate-based peptidomimetic compounds. They bind to the active site of HCV serine protease and competitively inhibit enzymatic function. Among these inhibitors, BILN-2061 is the first serine protease inhibitor in clinical trials for HCV (23). In both IFN treatment-experienced and -naïve patients with genotype 1 infections, twice-daily administration of 25-mg, 200-mg, or 500-mg doses of BILN-2061 resulted in a 2- to 3-log-unit reduction of HCV RNA within 48 h after treatment (23). This provided the first proof-of-concept evidence that HCV serine protease inhibitors could be efficacious in treating HCV infection in vivo.
Resistance to specific antiretroviral drugs has limited the efficacy of human immunodeficiency virus (HIV) protease and reverse transcriptase inhibitors due to the error-prone nature of the HIV reverse transcriptase, together with the high rate of viral replication (37, 38, 48). Like HIV, HCV also has a high replication rate in vivo, and its polymerase also has poor fidelity; thus, it is highly likely that drug-resistant HCV variants will emerge in patients treated with HCV polymerase or serine protease inhibitors as well. In fact, in vitro selection of HCV replicons resistant to nonnucleoside benzothiadiazine analogs (NS5B RdRp inhibitors) has been reported (39, 52, 53) and was found to be due to selection of a specific mutation(s) (M414T, C451R, G558R, or H95R) in the NS5B gene that led to decreased susceptibility to benzothiadiazines (for example, Fig. 1, Glaxo SmithKline [GSK], 6). Similarly, characterization of drug-resistant HCV replicons revealed that a single S282T mutation in NS5B polymerase conferred resistance to 2′-C Me-adenosine nucleosides (36). Furthermore, resistance to the NS3/4A serine protease inhibitors, including VX-950, BILN-2061, and another acyclic tripeptide inhibitor, was also reported (28, 29, 34, 54). Single amino acid substitutions (R155Q, A156S, A156T, D168V, D168A, and D168Y) in the NS3 protease gene mediated significant HCV replicon resistance to these compounds.
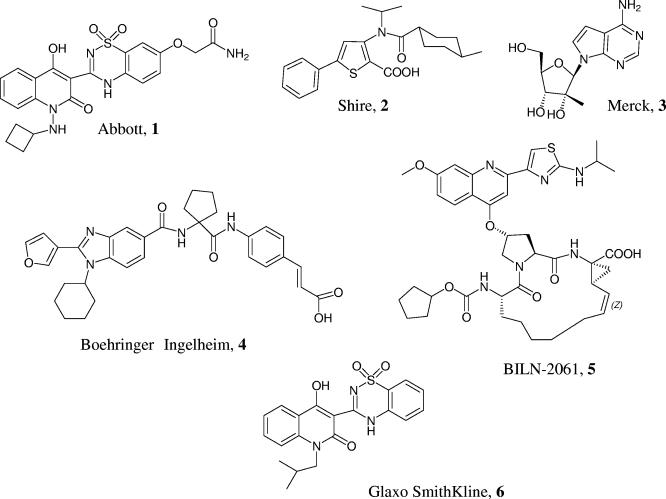
FIG. 1.
Chemical structures of the HCV polymerase inhibitors from Abbott, 1 (A-782759); Shire, 2; Merck, 3; Boehringer Ingelheim, 4; and Glaxo Smithkline, 6, as well as the HCV protease inhibitor BILN-2061, 5.
Our efforts at optimization of the benzothiadiazine series through chemistry have led us to identify a compound with significantly improved potency against HCV replicons compared to the benzothiadiazines previously reported (39, 53). In addition, the side chain substitutions of this compound differ from other benzothiadiazines, and this compound has a favorable pharmacokinetic profile in animals (45). Because of these features, it is of interest to map the binding site and investigate the resistance profile of this potent inhibitor. In addition, exploring the effect of the combination treatment on in vitro resistance selection may provide insight into future HCV treatment strategies. In this report, we describe the in vitro selection of HCV replicons with decreased susceptibility to a potent polymerase inhibitor, A-782759, alone and in combination with a protease inhibitor, BILN-2061. In addition, we have defined the genotypic basis for the observed resistance phenotype. Finally, the replication capacities and the susceptibilities of the mutants to other classes of polymerase inhibitors were analyzed.
MATERIALS AND METHODS
Inhibitors.
The details of the synthesis of A-782759, an Abbott compound, 2-[3-(1-cyclobutylamino-4-hydroxy-2-oxo-1,2-dihydro-quinolin-3-yl)-1,1-dioxo-1,4-dihydro-1l6-benzo(1,2,4)thiadiazin-7-yloxy]-acetamide (Fig. 1, Abbott, 1), has been described and will be published elsewhere (45). Polymerase inhibitors, 3-[isopropyl-(4-methyl-cyclohexanecarbonyl)-amino]-5-phenyl-thiophene- 2-carboxylic acid (Fig. 1, Shire, 2), the nucleoside 2-(4-amino-pyrrolo[2,3-d] pyrimidin-7-yl)-5-hydroxymethyl-3-methyl-tetrahydro-furan-3,4diol (Fig. 1, Merck, 3), 3-[4-({1-[(1-cyclohexyl-2-furan-3-yl-1H-benzoimidazole-5-carbonyl)-amino]-cyclopentanecarbonyl}-amino)-phenyl]-acrylic acid (Fig. 1, Boehringer Ingelheim, 4), and BILN-2061, 14-cyclopentyloxycarbonylamino-18-[2-(2-isopropylamino-thiazol-4-yl)-7-methoxy-quinolin-4-yloxy]-2,15-dioxo-3,16-diazatricyclo [14.3.0.04,6]nonadec-7-ene-4-carboxylic acid (Fig. 1, BILN-2061, 5), were synthesized according to procedures described previously (3, 6, 16, 30). Recombinant IFN-α was purchased from PBL Biomedical Laboratories, Piscataway, NJ.
Drug susceptibility assay.
HCV genotype 1b strain N (1b-N) subgenomic replicon cell lines were obtained and licensed from Stanley Lemon, University of Texas Medical Branch. The cells were grown in Dulbecco's modified Eagle medium (Invitrogen) supplemented with 10% fetal bovine serum (Atlanta Biologicals) and 400 μg/ml G418 (Invitrogen). Replicon cells containing the secreted alkaline phosphatase (SEAP) reporter were also passaged in the presence of blasticidin at 2 μg/ml. The inhibitory effects of compounds against HCV 1b-N replicon and resistant colonies were determined by monitoring the levels of HCV RNA and/or SEAP reporter as described previously (58). Briefly, 3,000 replicon-containing cells were seeded into each well of 96-well plates. The next day, the compound was initially diluted in dimethyl sulfoxide (DMSO) to generate a 200× stock for each dilution, while the human IFN-α was diluted in cell culture medium, not in DMSO. The medium from overnight cell culture plates was removed and replaced with fresh medium containing a 0.5% DMSO solution of inhibitor in a series of half-log dilutions in medium. Replicon cells were incubated for 3 days with compound, after which total cellular RNA was extracted using RNeasy-96 (QIAGEN), and the copy number of the HCV RNA was determined by a quantitative real-time reverse transcription (RT)-PCR (Taqman) assay. Alternatively, the activity of SEAP was measured in each culture supernatant after 4 days of incubation with compound according to the manufacturer's instructions. Cellular toxicity was determined on the remaining cells by the 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide colorimetric assay (44). Each assay was performed in either duplicate or triplicate. The 50% inhibitory concentration (IC50) was then calculated by nonlinear regression analysis using Prism (GraphPad Software, Inc.).
In vitro selection of resistant colonies.
The selection protocol was performed as described by Lu et al. with minor modifications (34). Genotype 1b replicon cells (2 × 104 or 2 × 105) were plated in 10-cm- or 15-cm-diameter tissue culture dishes, respectively, and cultured in the presence of selection medium containing 10% fetal bovine serum plus 400 μg/ml G418, 2 μg/ml blasticidin, and 350 nM or 700 nM (5 or 10 times the IC50) of A-782759 and/or 20 or 40 nM (5 or 10 times the IC50) BILN-2061. The reasons for using 5 and 10 times the IC50 were (i) both A782759 and BILN-2061 at 5 and 10 times the IC50 inhibit HCV RNA by at least 90% and (ii) 5 to 10 times is considered the target ratio of plasma concentration versus serum-adjusted IC50 that potentially produces antiviral activity in vivo. Under these conditions, cells containing drug-susceptible replicons were killed and cells harboring drug-resistant replicons formed colonies after approximately 2 to 3 weeks. Some of these resistant colonies were randomly picked, grown in the presence of inhibitor(s) for approximately 2 weeks, and subsequently characterized. Replicon cells incubated in the selection medium without inhibitor were used for the parental-cell control.
Sequence analysis of the NS3 and/or NS5B genes of the individual colonies.
Total cellular RNA was extracted using the RNeasy minikit (QIAGEN) according to the manufacturer's instructions and subjected to RT-PCR using primers SNS3-5-s (5′-GATAATACCATGGCGCCCATCACGGCCTAC-3′ [the underlined portion corresponds to nucleotides 3423 to 3440 of accession number AF139594]) and SNS3-5-as (5′-GGAAATGGCCTATTGGCCTGGAGTGTTTAGCTC-3′ [9395 to 9427 of accession number AF139594]). At least three independent PCRs were performed for each sample, and the PCR products were pooled. The amplified DNA fragments were purified using the QIAquick PCR purification kit (QIAGEN), and nucleotide sequences were determined by automated sequencing using BigDye terminator v. 3.1 (Applied Biosystems).
Construction of molecular clones containing NS3 or NS5B mutants.
The replicon obtained from the University of Texas Medical Branch has HIV tat followed by the gene encoding the 2A protease of foot-and-mouth disease virus fused to the 5′ end of the neomycin resistance gene (tat-2A-Neo) (58). To generate a replicon construct that can be used in transient-transfection assays, the tat-2A-Neo region was replaced with a sequence encoding the N-terminal 12 amino acids of the HCV core fused to the firefly luciferase gene. This replicon is referred to as 1b-N-Luc. For introducing the NS3 or NS5B mutations, the AvrII-AfeI and AfeI-ClaI fragments, containing the NS3-NS4B and NS5A-5B regions, respectively, were separately cloned into the vector pCR2.1TOPO (Invitrogen). All mutations were engineered using 5′-phosphorylated sense-strand synthetic oligonucleotides (Integrated DNA Technologies) and the Multi Site Directed Mutagenesis Kit (Stratagene) according to the manufacturers' instructions. The mutagenized fragments (AvrII-AfeI for the NS3-NS4B region and AfeI-ClaI for the NS5A-5B region) were reconstructed back into the 1b-N-Luc replicon construct after the sequence was confirmed.
RNA transcription and RNA transient-transfection assay.
The transcripts of HCV mutant replicons were generated using XbaI-linearized replicon plasmids and the Megascript T7 kit (Ambion) according to the manufacturer's instructions. Transient-transfection assays were performed as described previously (34). Briefly, IFN-cured Nneo/3-5B(RG) cells (2 × 106) were washed twice with Dulbecco's phosphate-buffered saline (without Ca2+ and Mg2+) (Invitrogen) and then mixed with 10 μg of replicon RNA in a Gene Pulser cuvette with a 0.2-cm electrode gap (Bio-Rad). Electroporation was immediately performed at 480 V and 25-μF capacitance with two manual pulses. Transfected cells were plated into 96-well plates with 5,000 cells per well. Compounds at various concentrations were added to the cells after 2 h and were cultured for 4 days. Four days was chosen based on the results of time course experiments performed with both wild-type and GDD mutant RNAs in which cells were treated with or without 100 nM BILN-2061 and the luciferase activity was monitored at 2, 4, 6, and 8 h, followed by days 1, 2, 3, 4, and 5. The results showed that at 4 days after transfection, luciferase activity obtained with the wild-type replicon without BILN-2061 was at least 800-fold above the background level as determined with either wild-type replicon cells treated with BILN-2061 or the GDD mutant negative control (data not shown). The cells were lysed with 1× passive lysis buffer, and luciferase activity was measured with the luciferase assay system kit (Promega) and a Wallac 1420 workstation (Perkin-Elmer Life Science) as described by the manufacturers. Luciferase activity was measured 4 h posttransfection without drug to determine the efficiency of transfection. Replication capacity was determined by measuring the luciferase activity of transfected cells after 4 days of culture in the absence of drug. The IC50 was then determined by nonlinear regression analysis with Prism (GraphPad Software, Inc.). Titrations were performed in triplicate, and the values were averaged. All experiments were repeated at least once in their entirety with new transfections to further verify the reproducibility.
RESULTS
A-782759 is a potent inhibitor of the HCV replicon.
A-782759, an N-1 azaquinolone benzothiadiazine, was identified as an inhibitor of HCV NS5B RdRp. This compound had an in vitro 1b HCV replicon (1b) IC50 of 70 nM, as determined by the effect on HCV RNA with no apparent toxicity up to 63 μM, resulting in a therapeutic index of 818-fold (Table 1). BILN-2061 is a highly potent HCV protease inhibitor with an IC50 of 4 nM against the 1b replicon (Table 1).
TABLE 1.
Potency and selectivity of HCV polymerase A-782759 and protease inhibitor BILN-2061
| Compound | Replicon 1b-N
|
|
|---|---|---|
| IC50 (μM)a | TD50 (μM)b | |
| A-782759 HCV NS5B polymerase | 0.070 ± 0.015 | 63 ± 17 |
| BILN-2061 HCV NS3 protease | 0.004 ± 0.002 | 16 ± 26 |
IC50 values are means±standard deviation from three separate experiments determined by the reduction of HCV RNA using a quantitative real-time RT-PCR (Taqman) assay.
TD50 values are means ± standard deviations from three independent experiments determined by MTT assay.
Lower frequency of resistant colonies selected by the combination of A-782759 and BILN-2061 than with either compound alone.
In order to select resistant mutants, 1b-N replicon cells were treated in the presence of neomycin plus either the polymerase inhibitor A-782759 or the protease inhibitor BILN-2061, or both, at concentrations of 5 or 10 times their corresponding IC50s as determined by HCV RNA reduction (Table 1). Since neomycin is included in the culture system but cell splitting is avoided, cells either lacking replicon or containing a drug-susceptible replicon are killed, and any remaining colonies that grow out can be assumed to emerge from a single cell during 3 to 4 weeks of selection. Using the initial cell number, the prevalence (percentage) of resistant mutants preexisting in the replicon quasispecies can be estimated by the following equation: percentage of resistant mutants = number of colonies/number of initial cells used in selection × 100. The results of these experiments with A-782759 or BILN-2061 alone or in combination are provided in Table 2. Using 2 × 104 cells, 123 and 10 colonies were observed with selection with A-782759 or BILN-2061 alone, respectively, while no colonies formed with the combination of these two compounds at both 5 and 10 times their corresponding IC50s. Even with 2 × 105 cells, only two and one colonies were found with combinations at 5 and 10 times their IC50s, respectively. Based on these results, the frequencies of resistance to A-782759 and BILN-2061 alone were estimated to be 0.62% and 0.05%. However, the frequency of replicons resistant to a combination of both compounds was between 0.001% and 0.0005%, which was significantly lower than with either compound alone.
TABLE 2.
Frequency of resistant-colony formation in the presence of HCV inhibitors and neomycin
| No. of cells and frequencya | No. of colonies
|
||
|---|---|---|---|
| A-782759 | BILN-2061 | A-782759 + BILN-2061 | |
| 10b | 10 | 5 + 5 10 + 10 | |
| 2 × 104 cells | 123 | 10 | 0 0 |
| 2 × 105 cells | NDc | ND | 21 |
| Frequency (%) | 0.62 | 0.05 | 0.001 0.0005 |
Frequency was calculated using equation: “number of colonies observed / the number of initial cells used × 100”.
Multiple of IC50.
ND, not done.
Characterization of selected colonies resistant to A-782759 alone or in combination with BILN-2061.
To characterize the phenotypic and genotypic changes, resistant colonies were individually expanded and analyzed. The genotype of each individual colony was determined by population sequencing of the NS3 and/or NS5B gene, and the phenotype was determined by measuring the reduction of SEAP activity following treatment of cells with inhibitors. Each colony selected by A-782759 alone contained one or two mutations in the NS5B polymerase gene and displayed an intermediate to high level of resistance to A-782759 (Table 3). The highest level of resistance was observed with colonies containing M414T (>200-fold).
TABLE 3.
Genotypes and phenotypes of 1b-N replicon colonies selected by A-782759 alone
| Colony no. | Mutation(s) in NS5B gene | A-782759
|
|
|---|---|---|---|
| IC50 (μM) ± SDa | Change in IC50b | ||
| 1b-N | WT 1b-Nc | 0.077 ± 0.031 | 1 |
| 1 | H95Q | 3.447 ± 2.336 | 44 |
| 2 | N411S | 2.217 ± 2.517 | 28 |
| 3 | Y448H | 2.739 ± 2.745 | 35 |
| 4 | M414L | 5.489 ± 2.923 | 70 |
| 5 | M414T | 22.013 ± 0.303 | >200 |
| 6 | V1381, M414T | 16.439 ± 11.115 | >200 |
IC50 values are means±SD from three separate experiments, with duplicates in each experiment determined by the reduction of SEAP reporter.
Change in IC50 (n-fold) = IC50 of mutant tested/IC50 of wild-type replicon.
WT, wild-type.
In addition, one M414L-containing colony showed 70-fold resistance. Other mutations (H95Q, N411S, and Y448H) had IC50 increases ranging from 28- to 44-fold (Table 3). We and others have previously reported that replicons containing mutations in the NS3 protease gene (A156T and D168A or D168V) confer high levels of resistance BILN-2061 (28, 29, 34).
To assess the effect of drug combination on the selection of resistant mutants, the 1b-N replicon was treated with both the polymerase and protease inhibitors at either 5 times the IC50 or 10 times the IC50 each. Using 5 × 105 cells, two and one colonies were selected when 5 and 10 times the IC50 of each compound were used (Table 2). However, no colony was selected when 4 × 104 cells were used for drug treatment (Table 2). Population sequencing revealed that each replicon colony had one mutation in the NS3 protease gene (A156V or D168V), together with one or two mutations in the NS5B polymerase gene (M414T or D55E/M414L) (Table 4). To ensure that the double or triple NS3 plus NS5B mutations (A156V plus M414T, D168V plus M414T, D168V plus D55E/M414L, or V138I/M414T) were derived from the same strand of HCV replicon RNA, the pooled PCR products containing NS3 and NS5B genes from each replicon colony were cloned into a TA cloning vector, and nucleotide sequences were determined from at least three individual clones derived from each replicon colony. The results confirmed the coexistence of the NS3 and NS5B mutations (A156V/M414T, D168V/M414T, or D168V/D55E/M414L) or two NS5B mutations (V138I/M414T) on the same replicon RNA strand. These genetic changes resulted in resistance to A-782759 that ranged from 112- to >187-fold over that of the wild type, while BILN-2061 resistance was also high at 115- to >800-fold over wild type (Table 3).
TABLE 4.
Genotypes and phenotypes of 1b-N replicon colonies selected by a combination of A-782759 and BILN-2061
| Colony no. | Mutation(s) in:
|
IC50 (μM) ± SD (change in IC50)a
|
||
|---|---|---|---|---|
| NS3 | NS5B | BILN-2061 | A-782759 | |
| 1b-N | WTb | WT | 0.004 ± 0.002 (1) | 0.107 ± 0.011 (1) |
| A | A156V | M414T | 0.460 ± 0.071 (115) | >20 (>187) |
| B | D168V | M414T | >3.2 (>800) | >20 (>187) |
| C | D168V | D55E, M414L | >3.2 (>800) | 11.82 ± 1.22 (112) |
IC50 values are means±SD from three separate experiments, with duplicates in each experiment determined by the reduction of SEAP reporter. Change in IC50 (n-fold) = IC50 of mutant tested/IC50 of wild-type replicon.
WT, wild type.
A-782759-resistant mutants remained susceptible to other classes of polymerase inhibitors and IFN.
To address whether the observed mutations in the NS5B gene were responsible for the increased resistance to A-782759, site-directed mutagenesis was performed to introduce these mutations individually into a subgenomic replicon containing a luciferase reporter gene (Table 5). The susceptibilities of these mutant replicons to A-782759 were evaluated using a transient-transfection assay. As shown in Table 5, no significant change in sensitivity to A-782759 was observed with the D55E or V138I mutants compared to the parental replicon. In contrast, the H95Q, N411S, M414L, M414T, and Y448H mutant replicons each displayed significantly reduced susceptibility, with IC50 increases ranging from 20- to >800-fold compared to the wild-type replicon. The changes in IC50 in this assay were even greater than those of colonies, which may be a reflection of the greater potency of A-782759 in this transient-transfection assay (0.004 μM) versus in the stable replicon assay against wild-type replicon (0.077 to 0.107 μM, using a SEAP reporter assay). Nevertheless, these results suggested that the above-mentioned mutations were each responsible for the decreased susceptibility to A-782759.
TABLE 5.
Sensitivities of NS5B mutants and NS3 mutant A156V to polymerase inhibitors and BILN-2061, as well as IFN
| Inhibitor | IC50 (μM) ± SD (IC50 change)a
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| WT-1b-N | D55E | H95Q | V138I | N411S | M414L | M414T | Y448H | A156Vb | |
| A-782759 Abbott, 1 | 0.004 ± 0.003 (1) | 0.013 ± 0.005 (3.3) | 0.81 ± 0.034 (20) | 0.013 ± 0.004 (3) | 1.33 ± 0.08 (333) | 0.41 ± 0.08 (103) | 8.9 ± 1.39 (>800) | 1.22 ± 0.15 (305) | 0.003 ± 0.001 (0.8) |
| Shire, 2 | 0.35 ± 0.08 (1) | 0.29 ± 0.10 (0.7) | 0.143 ± 0.01 (0.4) | 0.26 ± 0.06 (0.7) | 0.24 ± 0.11 (1) | 0.17 ± 0.08 (0.5) | 0.35 ± 0.08 (1) | 0.29 ± 0.11 (1) | 0.44 ± 0.004 (1.3) |
| Merck, 3 | 0.27 ± 0.05 (1) | 0.52 ± 0.14 (1.9) | ND | 0.38 ± 0.13 (1.4) | 0.14 ± 0.7 (0.5) | 0.24 ± 0.08 (1) | 0.17 ± 0.08 (0.5) | 0.35 ± 0.08 (1) | 0.29 ± 0.11 (1) |
| Boehringer Ingelheim, 4 | 1.97 ± 0.23 (1) | 3.77 ± 1.2 (1.9) | 1.24 ± 0.1 (0.7) | 5.35 ± 0.35 (3) | 0.6 ± 0.33 (1) | 1.9 ± 0.23 (1) | 0.64 ± 0.03 (0.3) | 1.32 ± 0.11 (1) | 5 ± 0.6 (2.5) |
| BILN-2061 Boehringer Ingelheim, 5 | 0.001 ± 0.0004 (1) | 0.0020 ± 0.0014 (2) | 0.001 ± 0.0005 (1) | 0.0013 ± 0.0002 (1) | 0.001 ± 0.0001 (1) | 0.001 ± 0.0002 (1) | 0.0019 ± 0.001 (1.9) | 0.002 ± 0.0001 (1) | 0.61 ± 0.16 (610) |
| IFN (μg/ml) | 0.06 ± 0.03 (1) | ND | 0.05 ± 0.03 (0.8) | 0.08 ± 0.001 (1) | 0.15 ± 0.001 (1) | 0.09 ± 0.001 (1.5) | 0.19 ± 0.02 (3) | 0.2 ± 0.02 (3) | 0.04 ± 0.02 (0.7) |
ND, not done due to unavailability of Merck, 3, or the same lot of IFN used for this study. Data are averages of at least two separate experiments done in triplicate. Change in IC50 (n-fold) = IC50 of mutant tested/IC50 of wild-type replicon.
NS3 mutant.
To assess cross-resistance, the A-782759-resistant mutants were tested for sensitivity to other classes of HCV polymerase inhibitors, the protease inhibitor BILN-2061, and IFN using the transient-transfection assay. These polymerase inhibitors included two nonnucleoside (thiophene amide and diamide derivatives discovered by Shire and Boehringer Ingelheim, respectively) and one nucleoside (published by Merck) inhibitors (6, 16, 30). The IC50 values for Shire-thiophene amide, Boehringer Ingelheim-diamide, and Merck-nucleoside were 0.35, 1.97, and 0.27 μM, respectively, which are 68- to 492-fold less potent than A-782759 (0.004 μM) in this transient-transfection assay (Table 5). No significant change in the IC50 to any of these three compounds was observed with the replicon containing the H95Q, N411S, M414L, M414T, or Y448H mutation, despite the fact that these mutations confer high levels of resistance to A-782759. Similarly, each of these mutations had minimal effect on susceptibility to BILN-2061 and IFN, suggesting that this series can be used in combination with any of these three classes of polymerase inhibitors, as well as with protease inhibitors and IFN.
Sensitivity of single NS3 mutation replicons to inhibitors.
Two single NS3 mutations, A156V and D168V, were found in colonies selected by the combination of A-760759 and BILN-2061. Mutations A156T and D168A or D168V in HCV NS3 protease were previously observed in resistance studies using BILN-2061 alone (28, 29, 34). However, A156V is a new mutation first reported in the present study. To ensure that this mutation is responsible for resistance to BILN-2061, we introduced A156V into a replicon construct and determined the susceptibility of the mutant replicon using a transient-transfection assay. As expected, the A156V mutant displayed 610-fold-reduced susceptibility to BILN-2061 compared to the wild type but retained wild-type susceptibility to all polymerase inhibitors tested, and to IFN as well (Table 5).
Replication capacities of mutants.
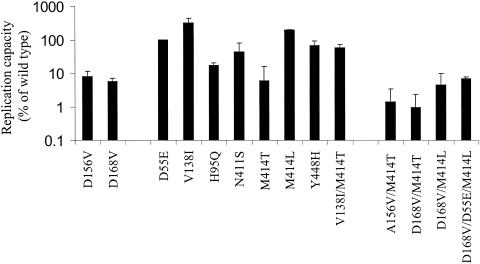
Since the replication capacity may be a significant factor in determining the prevalence of the resistant mutants in a viral population, it is of interest to assess the effects of the resistant mutants on the replication capacity. RNA from mutant replicon constructs was used to transfect cured Nneo/3-5B(RG) cells, which support replication of HCV replicons much more efficiently than parental Huh-7 cells. Since the copy number of the luciferase gene is determined by the replication level of the corresponding replicon RNA, the replication capacity of each mutant can be determined by measuring luciferase activity in a lysate of transfected cells. Each mutant replicon was transfected in the absence or presence of inhibitor, and luciferase activity was measured at 4 h and 4 days posttransfection. Luciferase activities from the mutant replicons were then compared with the luciferase activity from wild-type replicon. All of them had very similar levels of luciferase activity at 4 h after transfection, indicating comparable levels of RNA transfection and translation ability of the RNAs (data not shown). At 4 days after transfection, the NS5B mutants D55E, V138I, and M414L replicated as well as, and in some case better than, the wild type (102 to 320% of the wild-type replication), while N411S and Y448H displayed slightly reduced replication capacity from the wild type (Fig. 2). In contrast, the H95Q and M414T mutants retained only 18% and 6% of wild-type replication capacity, respectively. Similarly, two NS3 mutants, A156V and D168V, exhibited 8% and 6% of wild-type replication capacity, respectively. Furthermore, all double mutants (A156V/M414T, D168V/M414T, and D168V/M414L) were further attenuated compared to the single mutants. For example, the single D168V and M414L mutants had 6% and 200% wild-type replication capacity, respectively; however, the combination of these two mutants (D168V/M414L) resulted in a replicon with only 4% of the replication capacity. In addition, the replication capacities of the A156V/M414T and D168V/M414T mutants were less than 1% that of the wild type.
FIG. 2.
Comparison of replication capacity of wild-type replicon with those of the recombinant mutant replicons. Cells were transfected either with wild-type or with mutant replicon RNA in the absence of BILN-2061, and luciferase activity was measured at 4 hours and 4 days after transfection. The replication capacity of each mutant was calculated by comparing the firefly luciferase activity generated by the mutant to that generated by wild-type replicon at day 4, after adjusting for minor differences in transfection efficiencies (the 4-h luciferase activity). The data are averages of at least two separate experiments with six replicates in each experiment. The error bars represent standard deviations.
As shown in Table 2, one A-782759-resistant replicon colony had both M414T and V138I mutations in NS5B, while another colony selected by both A-782759 and BILN-2061 had D168V in NS3 together with M414L and D55E in NS5B. To understand the effect of V138I and D55E on the replication capacities of M41T and D168V/M414L, the double V138I/M414T and triple D168V/D55E/M414L mutants were created and used to study the replication capacity. Interestingly, the most resistant mutant, M414T, is also very growth impaired (6%), while V138I has a growth advantage over the wild type (300% of wild type, or three times above the wild-type luciferase activity). The combination of both exhibited 56% replication of the wild type, indicating that the impaired growth capacity of M414T is mostly restored by V138I in the transient-replication assay. In contrast, addition of D55E to D168V/M414L did not change the replication capacity of the double mutant (D168V/M414L) significantly.
DISCUSSION
In this study, we selected and characterized in vitro resistance to an HCV NS5B polymerase inhibitor, A-782759, either alone or in combination with the HCV NS3/4A protease inhibitor BILN-2061. The dominant mutations resistant to A-782759 were H95Q, N411S, M414L, M414T, and Y448H in NS5B, as recombinant replicons containing each of these five NS5B mutations displayed intermediate to high levels of reduction in susceptibility to A-782759. Modeling of the mutations into the three-dimensional structure of NS5B polymerase showed that three of the above-mentioned residues (411, 414, and 448) clustered in an alpha-helical region of the thumb domain, a site in close proximity to the interface of the palm and thumb domains of NS5B (Fig. 3). This region has been shown to be the allosteric site bound by several benzothiadiazine analog from GSK (15). In previous studies, Tomei et al. and Nguyen et al. found that the M414T and H95Q mutations led to a reduced sensitivity to benzothiadiazine analog (39, 53). These findings, together with the fact that A-782759 and the GSK benzothiadiazines share the structural feature of ABCD rings (Fig. 1), suggest that the Abbott benzothiadiazine A-782759 may bind to the allosteric site similarly to the GSK benzothiadiazine binding site. Thus, the mutations N411S, M414L/T, and Y448H occur in the inhibitor binding region and may potentially directly affect enzyme-inhibitor interaction. Unlike residues 411, 414, and 448, residue 95 is in the finger domain, distant from the potential inhibitor binding site. This mutation may indirectly influence ligand binding.
In contrast to the mutations H95Q, N411S, M414L, M414T, and Y448H, no significant change in susceptibility to A-782759 was observed with replicons containing the single mutation D55E or V138I. These two mutations were scattered in the finger domain of the NS5B polymerase protein, far away from the inhibitor binding site. Both mutations coexisted with at least one other mutation, suggesting that the two mutations may not be primarily responsible for conferring A-782759 resistance; rather, they may play a compensatory role for replication. This is also supported by the observation that the resistant colonies containing V138I/M414T (NS5B) or D168V (NS3)/D55E/M414L (NS5B) mutations and the recombinant single mutant D55E or V138I replicated well, while the replicons containing mutation M414T or D168V/M414L exhibited greatly reduced HCV RNA replication in the transient assay.
As shown in Table 4, colonies selected by the combination of A-782759 and BILN-2061 containing the single mutation M414T or the double mutation D55E/M414L in NS5B, together with either A156V or D168V in NS3, substantially reduced susceptibility to both A-782759 and BILN-2061. Mutations A156T, D168A, and D168V in HCV NS3 protease have been previously observed in resistance selection studies using BILN-2061 alone (28, 29, 34), while mutation A156S has been reported to confer resistance to VX-950, but not to BILN-2061. However, A156V is a new mutation first reported in the present study. We showed that a molecular clone containing the A156V single mutation confers a high level of resistance to BILN-2061(Table 5). As demonstrated in previous studies, Ala156 is located on the E2 strand in the HCV NS3/4A protease structure. The side chain of Ala156 is within the Van der Waals contact of the P2 group of BILN-2061 (56). Valine substitution would be expected to disrupt the contact between the inhibitor and the protein, leading to decreased affinity of the inhibitor. As expected, this NS3 mutant retained wild-type sensitivity to the polymerase inhibitors and IFN.
In the present study, multiple mutational patterns were observed in response to in vitro selection with A-782759, and single mutations were shown to confer high levels of resistance. Similar observations were demonstrated for the GSK benzothiadiazine polymerase inhibitors and HCV protease inhibitors (28, 29, 34, 39, 53, 54). With all of the inhibitors studied, the levels of resistance caused by some of the single resistant mutants were greater than 100-fold above the wild-type IC50. This level of resistance will not be easily overcome by the drug concentrations in vivo. The characterization of resistances to the HCV polymerase and protease inhibitors is similar to what was found with HIV nonnucleoside reverse transcriptase inhibitors (NNRTIs), in which single mutations in the NNRTI-binding pocket resulted in high levels of resistance to one or more of the NNRTIs (48). In the case of NNRTIs for HIV, resistance usually emerged rapidly when NNRTIs were administered as monotherapy in HIV-infected patients (7). This is likely due to the selection of a preexisting population of mutant viruses within an individual. Given the higher replication rate of HCV in vivo and the longer average time after infection compared to HIV (46), many multiple mutants are likely to preexist in HCV-infected individuals. Therefore, like treatment for HIV infection, monotherapy with these HCV inhibitors will be unlikely to lead to the eradication of HCV due to the selection of preexisting resistant mutants.
In this study, we demonstrated that the major in vitro-resistant mutations selected by A-782759 remain fully susceptible to two other classes of nonnucleoside HCV polymerase inhibitors (diamide derivative from Boehringer Ingelheim and thiophene derivative from Shire). This result suggests that the putative binding site for A-782759 may be distinct from the binding sites for either the diamide derivative or the thiophene derivative. In fact, as shown in Fig. 3, the binding region for the diamide is located in the upper thumb domain, which is distant from the putative benzothiadiazine binding site in the region of residues 411, 414, and 448 (8). Similarly, the phenylalanine amides from Shire have been demonstrated to be bound in the lower thumb domain, away from the region of residues 411, 414, and 448 (Fig. 3) (57). However, the binding mode for Shire-thiophene amide derivatives has not been reported. Nevertheless, unlike the HIV NNRTIs, which all bind to the same hydrophobic pocket in the reverse transcriptase and have overlapping cross-resistances (48), nonnucleoside HCV polymerase inhibitors with distinct structures bind to different allosteric binding sites on the HCV NS5B polymerase enzyme and thus may display nonoverlapping resistances and may be able to be used in combination. Indeed, lack of cross-resistance between A-782759 and the diamide and thiophene derivatives may allow the benzothiadiazine derivative to be used in combination with these two classes of nonnucleoside polymerase inhibitors. Similarly, the lack of cross-resistance of the A-782759-resistant mutants to the HCV protease inhibitor BILN-2061 or IFN observed in this study provides a rationale for combining the benzothiadiazine derivative with the HCV protease inhibitor and IFN.
This is the first report to provide additional evidence that combination therapy with two small-molecule inhibitors may afford an advantage in overcoming the resistance issue. For example, we demonstrated that the frequency of resistance to the combination of A-782759 and BILN-2061 was significantly lower than the frequency of resistance to either single compound alone. In addition, the replicon containing dually resistant mutants had dramatically reduced replication compared to the wild type despite some of the single mutants having replication capacities close to or better than the wild type. Taken together, these findings support the notion that combination therapy will provide an approach to suppress the emergence of resistance and increase the efficacy of HCV therapy.
This study has several limitations. The selection of resistant mutants was conducted using a specific strain of genotype 1b in the subgenomic replicon system. This replicon system includes the replication cycle but lacks the entry, assembly, and release of viral particles, which is different from an in vivo HCV infection. In addition, different patients contain various quasispecies of viral strains. Consequently, whether the same mutations will be selected in patients needs to be determined in the future. Similarly, the relative level of viral replication of mutants in this study may be different in vivo because of the differences between this in vitro replicon system and viral infection in patients.
In conclusion, we have selected HCV replicons in vitro which have increased resistance to either A-782759 alone or in combination with BILN-2061. Single amino acid substitutions at residues 95, 411, 414, and 448 of NS5B polymerase resulted in high levels of resistance to A-782759. Similarly, a novel mutation, A156V, in the HCV NS3 serine protease was found to confer high-level resistance to BILN-2061. Since single mutations confer a high level of resistance, monotherapy will likely lead to the emergence of resistance in vivo. However, combination therapy may provide an advantage in combating resistance due to the low frequency and poor replication capacity of dually resistant mutants observed in this study. Furthermore, lack of cross-resistance may allow A-782759 to be used in combination with other classes of polymerase inhibitors, protease inhibitors, and/or IFN.
Acknowledgments
We thank Stanley Lemon and Minkyung Yi for providing replicon cell lines. We also thank Yupeng He and Tim Middleton for technical assistance and John Randolph, Keith McDaniel, David Betebenner, and Larry Klein for providing compounds, as well as Dale Kempf, Bill Kohlbrenner, and Shing Chang for providing continuous support.
REFERENCES
- 1.Bartenschlager, R. 1999. The NS3/4A proteinase of the hepatitis C virus: unravelling structure and function of an unusual enzyme and a prime target for antiviral therapy. J. Viral Hepat. 6:165-181. [DOI] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., L. Ahlborn-Laake, J. Mous, and H. Jacobsen. 1994. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J. Virol. 68:5045-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaulieu, P. 2004. Allosteric inhibitors of the hepatitis C virus NS5B polymerase: towards novel anti-HCV therapies, abstr. 1. Abstr. 29th Nat. Med. Chem. Symp. American Chemical Society and University of Wisconsin—Madison, Madison, Wis.
- 4.Behrens, S. E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll, S. S., J. E. Tomassini, M. Bosserman, K. Getty, M. W. Stahlhut, A. B. Eldrup, B. Bhat, D. Hall, A. L. Simcoe, R. LaFemina, C. A. Rutkowski, B. Wolanski, Z. Yang, G. Migliaccio, R. De Francesco, L. C. Kuo, M. MacCoss, and D. B. Olsen. 2003. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem. 278:11979-11984. [DOI] [PubMed] [Google Scholar]
- 6.Chan, L., O. Pereira, T. J. Reddy, S. K. Das, C. Poisson, M. Courchesne, M. Proulx, A. Siddiqui, C. G. Yannopoulos, N. Nguyen-Ba, C. Roy, D. Nasturica, C. Moinet, R. Bethell, M. Hamel, L. L'Heureux, M. David, O. Nicolas, P. Courtemanche-Asselin, S. Brunette, D. Bilimoria, and J. Bedard. 2004. Discovery of thiophene-2-carboxylic acids as potent inhibitors of HCV NS5B polymerase and HCV subgenomic RNA replication. Part 2. Tertiary amides. Bioorg. Med. Chem. Lett. 14:797-800. [DOI] [PubMed] [Google Scholar]
- 7.Cheeseman, S. H., D. Havlir, M. M. McLaughlin, T. C. Greenough, J. L. Sullivan, D. Hall, S. E. Hattox, S. A. Spector, D. S. Stein, M. Myers, et al. 1995. Phase I/II evaluation of nevirapine alone and in combination with zidovudine for infection with human immunodeficiency virus. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 8:141-151. [PubMed] [Google Scholar]
- 8.Coulombe, R., P. L. Beaulieu, E. Jolicoeur, G. Kukolj, S. Laplante, and M. A. Poupart. 18 Nov Hepatititis C virus NS5B polymerase inhibitor binding pocket. Int. Patent Appl. WO 2004099241 A1.
- 9.Davis, G. L. 1999. Combination therapy with interferon alfa and ribavirin as retreatment of interferon relapse in chronic hepatitis C. Semin. Liver Dis. 19(Suppl. 1):49-55. [PubMed] [Google Scholar]
- 10.Davis, G. L. 1999. Combination treatment with interferon and ribavirin for chronic hepatitis C. Clin. Liver Dis. 3:811-826. [DOI] [PubMed] [Google Scholar]
- 11.Davis, G. L., R. Esteban-Mur, V. Rustgi, J. Hoefs, S. C. Gordon, C. Trepo, M. L. Shiffman, S. Zeuzem, A. Craxi, M. H. Ling, J. Albrecht, et al. 1998. Interferon alfa-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. N. Engl. J. Med. 339:1493-1499. [DOI] [PubMed] [Google Scholar]
- 12.De Francesco, R., L. Tomei, S. Altamura, V. Summa, and G. Migliaccio. 2003. Approaching a new era for hepatitis C virus therapy: inhibitors of the NS3-4A serine protease and the NS5B RNA-dependent RNA polymerase. Antivir. Res. 58:1-16. [DOI] [PubMed] [Google Scholar]
- 13.Dhanak, D., K. J. Duffy, V. K. Johnston, J. Lin-Goerke, M. Darcy, A. N. Shaw, B. Gu, C. Silverman, A. T. Gates, M. R. Nonnemacher, D. L. Earnshaw, D. J. Casper, A. Kaura, A. Baker, C. Greenwood, L. L. Gutshall, D. Maley, A. DelVecchio, R. Macarron, G. A. Hofmann, Z. Alnoah, H. Y. Cheng, G. Chan, S. Khandekar, R. M. Keenan, and R. T. Sarisky. 2002. Identification and biological characterization of heterocyclic inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 277:38322-38327. [DOI] [PubMed] [Google Scholar]
- 14.Di Bisceglie, A. M., and J. H. Hoofnagle. 2002. Optimal therapy of hepatitis C. Hepatology 36:S121-S127. [DOI] [PubMed] [Google Scholar]
- 15.Duffy, K. J. 2004. The discovery of benzo(1,2,4) thiadiazine as non-nucleoside hepatitis C virus NS5B polymerase inhibitors, abstr. 2. Abstr. 29th Nat. Med. Chem. Symp. American Chemical Society and University of Wisconsin—Madison, Madison, Wis.
- 16.Eldrup, A. B., M. Prhavc, J. Brooks, B. Bhat, T. P. Prakash, Q. Song, S. Bera, N. Bhat, P. Dande, P. D. Cook, C. F. Bennett, S. S. Carroll, R. G. Ball, M. Bosserman, C. Burlein, L. F. Colwell, J. F. Fay, O. A. Flores, K. Getty, R. L. LaFemina, J. Leone, M. MacCoss, D. R. McMasters, J. E. Tomassini, D. Von Langen, B. Wolanski, and D. B. Olsen. 2004. Structure-activity relationship of heterobase-modified 2′-C-methyl ribonucleosides as inhibitors of hepatitis C virus RNA replication. J. Med. Chem. 47:5284-5297. [DOI] [PubMed] [Google Scholar]
- 17.Frecer, V., M. Kabelac, P. De Nardi, S. Pricl, and S. Miertus. 2004. Structure-based design of inhibitors of NS3 serine protease of hepatitis C virus. J. Mol. Graph Model. 22:209-220. [DOI] [PubMed] [Google Scholar]
- 18.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 19.Godofsky, E. 2004. Phase I/II dose escalation trial assessing tolerance, pharmacokinetics, and antiviral activity of NM283, a novel antiviral treatment for hepatitis C, abstr. 407. Abstr. Dig. Dis. Week 2004. Medscape, New Orleans, La.
- 20.Gopalsamy, A., K. Lim, G. Ciszewski, K. Park, J. W. Ellingboe, J. Bloom, S. Insaf, J. Upeslacis, T. S. Mansour, G. Krishnamurthy, M. Damarla, Y. Pyatski, D. Ho, A. Y. Howe, M. Orlowski, B. Feld, and J. O'Connell. 2004. Discovery of pyrano[3,4-b]indoles as potent and selective HCV NS5B polymerase inhibitors. J. Med. Chem. 47:6603-6608. [DOI] [PubMed] [Google Scholar]
- 21.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 67:2832-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu, B., V. K. Johnston, L. L. Gutshall, T. T. Nguyen, R. R. Gontarek, M. G. Darcy, R. Tedesco, D. Dhanak, K. J. Duffy, C. C. Kao, and R. T. Sarisky. 2003. Arresting initiation of hepatitis C virus RNA synthesis using heterocyclic derivatives. J. Biol. Chem. 278:16602-16607. [DOI] [PubMed] [Google Scholar]
- 23.Hinrichsen, H., Y. Benhamou, H. Wedemeyer, M. Reiser, R. E. Sentjens, J. L. Calleja, X. Forns, A. Erhardt, J. Cronlein, R. L. Chaves, C. L. Yong, G. Nehmiz, and G. G. Steinmann. 2004. Short-term antiviral efficacy of BILN 2061, a hepatitis C virus serine protease inhibitor, in hepatitis C genotype 1 patients. Gastroenterology 127:1347-1355. [DOI] [PubMed] [Google Scholar]
- 24.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 25.Johnston, V. K., D. Maley, R. C. Gagnon, C. W. Grassmann, S. E. Behrens, and R. T. Sarisky. 2003. Kinetic profile of a heterocyclic HCV replicon RNA synthesis inhibitor. Biochem. Biophys. Res. Commun. 311:672-677. [DOI] [PubMed] [Google Scholar]
- 26.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol 6:937-943. [DOI] [PubMed] [Google Scholar]
- 28.Lin, C., K. Lin, Y. P. Luong, B. G. Rao, Y. Y. Wei, D. L. Brennan, J. R. Fulghum, H. M. Hsiao, S. Ma, J. P. Maxwell, K. M. Cottrell, R. B. Perni, C. A. Gates, and A. D. Kwong. 2004. In vitro resistance studies of hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061: structural analysis indicates different resistance mechanisms. J. Biol. Chem. 279:17508-17514. [DOI] [PubMed] [Google Scholar]
- 29.Lindenbach, B. D., and C. M. Rice. 2003. Evasive maneuvers by hepatitis C virus. Hepatology 38:769-771. [DOI] [PubMed] [Google Scholar]
- 30.Llinas-Brunet, M., M. D. Bailey, G. Bolger, C. Brochu, A. M. Faucher, J. M. Ferland, M. Garneau, E. Ghiro, V. Gorys, C. Grand-Maitre, T. Halmos, N. Lapeyre-Paquette, F. Liard, M. Poirier, M. Rheaume, Y. S. Tsantrizos, and D. Lamarre. 2004. Structure-activity study on a novel series of macrocyclic inhibitors of the hepatitis C virus NS3 protease leading to the discovery of BILN 2061. J. Med. Chem. 47:1605-1608. [DOI] [PubMed] [Google Scholar]
- 31.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lohmann, V., A. Roos, F. Korner, J. O. Koch, and R. Bartenschlager. 1998. Biochemical and kinetic analyses of NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Virology 249:108-118. [DOI] [PubMed] [Google Scholar]
- 33.Love, R. A., H. E. Parge, X. Yu, M. J. Hickey, W. Diehl, J. Gao, H. Wriggers, A. Ekker, L. Wang, J. A. Thomson, P. S. Dragovich, and S. A. Fuhrman. 2003. Crystallographic identification of a noncompetitive inhibitor binding site on the hepatitis C virus NS5B RNA polymerase enzyme. J. Virol. 77:7575-7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu, L., T. J. Pilot-Matias, K. D. Stewart, J. T. Randolph, R. Pithawalla, W. He, P. P. Huang, L. L. Klein, H. Mo, and A. Molla. 2004. Mutations conferring resistance to a potent hepatitis C virus serine protease inhibitor in vitro. Antimicrob. Agents Chemother. 48:2260-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, J. K. Albrecht, et al. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 36.Migliaccio, G., J. E. Tomassini, S. S. Carroll, L. Tomei, S. Altamura, B. Bhat, L. Bartholomew, M. R. Bosserman, A. Ceccacci, L. F. Colwell, R. Cortese, R. De Francesco, A. B. Eldrup, K. L. Getty, X. S. Hou, R. L. LaFemina, S. W. Ludmerer, M. MacCoss, D. R. McMasters, M. W. Stahlhut, D. B. Olsen, D. J. Hazuda, and O. A. Flores. 2003. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J. Biol. Chem. 278:49164-49170. [DOI] [PubMed] [Google Scholar]
- 37.Mo, H., L. Lu, T. Dekhtyar, K. D. Stewart, E. Sun, D. J. Kempf, and A. Molla. 2003. Characterization of resistant HIV variants generated by in vitro passage with lopinavir/ritonavir. Antivir. Res. 59:173-180. [DOI] [PubMed] [Google Scholar]
- 38.Molla, A., M. Korneyeva, Q. Gao, S. Vasavanonda, P. J. Schipper, H. M. Mo, M. Markowitz, T. Chernyavskiy, P. Niu, N. Lyons, A. Hsu, G. R. Granneman, D. D. Ho, C. A. Boucher, J. M. Leonard, D. W. Norbeck, and D. J. Kempf. 1996. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat. Med. 2:760-766. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen, T. T., A. T. Gates, L. L. Gutshall, V. K. Johnston, B. Gu, K. J. Duffy, and R. T. Sarisky. 2003. Resistance profile of a hepatitis C virus RNA-dependent RNA polymerase benzothiadiazine inhibitor. Antimicrob. Agents Chemother. 47:3525-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ni, Z. J., and A. S. Wagman. 2004. Progress and development of small molecule HCV antivirals. Curr. Opin. Drug Discov. Devel. 7:446-459. [PubMed] [Google Scholar]
- 41.Olsen, D. B., A. B. Eldrup, L. Bartholomew, B. Bhat, M. R. Bosserman, A. Ceccacci, L. F. Colwell, J. F. Fay, O. A. Flores, K. L. Getty, J. A. Grobler, R. L. LaFemina, E. J. Markel, G. Migliaccio, M. Prhavc, M. W. Stahlhut, J. E. Tomassini, M. MacCoss, D. J. Hazuda, and S. S. Carroll. 2004. A 7-deaza-adenosine analog is a potent and selective inhibitor of hepatitis C virus replication with excellent pharmacokinetic properties. Antimicrob. Agents Chemother. 48:3944-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pace, P., E. Nizi, B. Pacini, S. Pesci, V. Matassa, R. De Francesco, S. Altamura, and V. Summa. 2004. The monoethyl ester of meconic acid is an active site inhibitor of HCV NS5B RNA-dependent RNA polymerase. Bioorg. Med. Chem. Lett. 14:3257-3261. [DOI] [PubMed] [Google Scholar]
- 43.Pause, A., G. Kukolj, M. Bailey, M. Brault, F. Do, T. Halmos, L. Lagace, R. Maurice, M. Marquis, G. McKercher, C. Pellerin, L. Pilote, D. Thibeault, and D. Lamarre. 2003. An NS3 serine protease inhibitor abrogates replication of subgenomic hepatitis C virus RNA. J. Biol. Chem. 278:20374-20380. [DOI] [PubMed] [Google Scholar]
- 44.Pauwels, R., J. Balzarini, M. Baba, R. Snoeck, D. Schols, P. Herdewijn, J. Desmyter, and E. De Clercq. 1988. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J. Virol. Methods 20:309-321. [DOI] [PubMed] [Google Scholar]
- 45.Pratt, J. K., D. Beno, P. Donner, W. Jiang, W. Kati, D. J. Kempf, G. Koev, D. Liu, Y. Liu, C. Maring, S. Masse, T. Middleton, H. Mo, A. Molla, D. Montgomery, and Q. Xie. 2004. N-1-AZA-4-hydroxyquinolone benzothiadianes: inhibitors of HCV genotype 1 NS5B RNA-dependent RNA polymerase exhibiting replication potency and favorable rat pharmacokinetics, abstr. 1224a. Abstr. 55th Ann. Meet. Am. Assoc. Study Liver Dis.
- 46.Ramratnam, B., S. Bonhoeffer, J. Binley, A. Hurley, L. Zhang, J. E. Mittler, M. Markowitz, J. P. Moore, A. S. Perelson, and D. D. Ho. 1999. Rapid production and clearance of HIV-1 and hepatitis C virus assessed by large volume plasma apheresis. Lancet 354:1782-1785. [DOI] [PubMed] [Google Scholar]
- 47.Sarisky, R. T. 2004. Non-nucleoside inhibitors of the HCV polymerase. J. Antimicrob Chemother. 54:14-16. [DOI] [PubMed] [Google Scholar]
- 48.Shulman, N., and M. Winters. 2003. A review of HIV-1 resistance to the nucleoside and nucleotide inhibitors. Curr. Drug Targets Infect. Disord. 3:273-281. [DOI] [PubMed] [Google Scholar]
- 49.Sommadossi, J. P., and P. Lacolla. 29November2001. Methods and compositions for treating hepatitis C virus. Int. patent appl. WO 0190121 A2.
- 50.Stansfield, I., S. Avolio, S. Colarusso, N. Gennari, F. Narjes, B. Pacini, S. Ponzi, and S. Harper. 2004. Active site inhibitors of HCV NS5B polymerase. The development and pharmacophore of 2-thienyl-5,6-dihydroxypyrimidine-4-carboxylic acid. Bioorg. Med. Chem. Lett. 14:5085-5088. [DOI] [PubMed] [Google Scholar]
- 51.Summa, V., A. Petrocchi, P. Pace, V. G. Matassa, R. De Francesco, S. Altamura, L. Tomei, U. Koch, and P. Neuner. 2004. Discovery of α,γ-diketo acids as potent selective and reversible inhibitors of hepatitis C virus NS5b RNA-dependent RNA polymerase. J. Med. Chem. 47:14-17. [DOI] [PubMed] [Google Scholar]
- 52.Tomei, L., S. Altamura, L. Bartholomew, A. Biroccio, A. Ceccacci, L. Pacini, F. Narjes, N. Gennari, M. Bisbocci, I. Incitti, L. Orsatti, S. Harper, I. Stansfield, M. Rowley, R. De Francesco, and G. Migliaccio. 2003. Mechanism of action and antiviral activity of benzimidazole-based allosteric inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 77:13225-13231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomei, L., S. Altamura, L. Bartholomew, M. Bisbocci, C. Bailey, M. Bosserman, A. Cellucci, E. Forte, I. Incitti, L. Orsatti, U. Koch, R. De Francesco, D. B. Olsen, S. S. Carroll, and G. Migliaccio. 2004. Characterization of the inhibition of hepatitis C virus RNA replication by nonnucleosides. J. Virol. 78:938-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trozzi, C., L. Bartholomew, A. Ceccacci, G. Biasiol, L. Pacini, S. Altamura, F. Narjes, E. Muraglia, G. Paonessa, U. Koch, R. De Francesco, C. Steinkuhler, and G. Migliaccio. 2003. In vitro selection and characterization of hepatitis C virus serine protease variants resistant to an active-site peptide inhibitor. J. Virol. 77:3669-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsantrizos, Y. S. 2004. The design of a potent inhibitor of the hepatitis C virus NS3 protease: BILN 2061—from the NMR tube to the clinic. Biopolymers 76:309-323. [DOI] [PubMed] [Google Scholar]
- 56.Tsantrizos, Y. S., G. Bolger, P. Bonneau, D. R. Cameron, N. Goudreau, G. Kukolj, S. R. LaPlante, M. Llinas-Brunet, H. Nar, and D. Lamarre. 2003. Macrocyclic inhibitors of the NS3 protease as potential therapeutic agents of hepatitis C virus infection. Angew. Chem. Int. Ed. Engl. 42:1356-1360. [DOI] [PubMed] [Google Scholar]
- 57.Wang, M., K. K. Ng, M. M. Cherney, L. Chan, C. G. Yannopoulos, J. Bedard, N. Morin, N. Nguyen-Ba, M. H. Alaoui-Ismaili, R. C. Bethell, and M. N. James. 2003. Non-nucleoside analogue inhibitors bind to an allosteric site on HCV NS5B polymerase. Crystal structures and mechanism of inhibition. J. Biol. Chem. 278:9489-9495. [DOI] [PubMed] [Google Scholar]
- 58.Yi, M., F. Bodola, and S. M. Lemon. 2002. Subgenomic hepatitis C virus replicons inducing expression of a secreted enzymatic reporter protein. Virology 304:197-210. [DOI] [PubMed] [Google Scholar]
- 59.Yip, Y., F. Victor, J. Lamar, R. Johnson, Q. M. Wang, D. Barket, J. Glass, L. Jin, L. Liu, D. Venable, M. Wakulchik, C. Xie, B. Heinz, E. Villarreal, J. Colacino, N. Yumibe, M. Tebbe, J. Munroe, and S. H. Chen. 2004. Discovery of a novel bicycloproline P2 bearing peptidyl alpha-ketoamide LY514962 as HCV protease inhibitor. Bioorg. Med. Chem. Lett. 14:251-256. [DOI] [PubMed] [Google Scholar]