Abstract
Proteus mirabilis is a urinary tract pathogen that differentiates from a short swimmer cell to an elongated, highly flagellated swarmer cell. Swarmer cell differentiation parallels an increased expression of several virulence factors, suggesting that both processes are controlled by the same signal. The molecular nature of this signal is not known but is hypothesized to involve the inhibition of flagellar rotation. In this study, data are presented supporting the idea that conditions inhibiting flagellar rotation induce swarmer cell differentiation and implicating a rotating flagellar filament as critical to the sensing mechanism. Mutations in three genes, fliL, fliF, and fliG, encoding components of the flagellar basal body, result in the inappropriate development of swarmer cells in noninducing liquid media or hyperelongated swarmer cells on agar media. The fliL mutation was studied in detail. FliL− mutants are nonmotile and fail to synthesize flagellin, while complementation of fliL restores wild-type cell elongation but not motility. Overexpression of fliL+ in wild-type cells prevents swarmer cell differentiation and motility, a result also observed when P. mirabilis fliL+ was expressed in Escherichia coli. These results suggest that FliL plays a role in swarmer cell differentiation and implicate FliL as critical to transduction of the signal inducing swarmer cell differentiation and virulence gene expression. In concert with this idea, defects in fliL up-regulate the expression of two virulence genes, zapA and hpmB. These results support the hypothesis that P. mirabilis ascertains its location in the environment or host by assessing the status of its flagellar motors, which in turn control swarmer cell gene expression.
Epithelial surfaces of eukaryotic organisms are continuously exposed to bacteria. These surfaces are protected by numerous defense mechanisms, such as the antibacterial peptides that are part of the innate defense system. Pathogens, however, appear to overcome the innate immune defense systems, giving rise to various infections. In some conditions, such as chronic ulcers of the skin or urinary tract infections, bacteria such as Proteus mirabilis, an opportunistic pathogen, manage to infect and persist for long periods of time (22, 48, 62).
Infections of P. mirabilis persist in part due to the ability of these bacteria to differentiate from short vegetative swimmer cells into elongated, multinucleated, and hyperflagellated swarmer cells (5, 10, 76). This differentiation takes place in response to growth on surfaces or viscous liquids (1, 35, 48). Swarming is a demanding process, with a substantial proportion of metabolism given over to the assembly and operation of flagella and other swarmer cell-dependent proteins. At the genetic level, differentiation involves the coordinate expression of a global regulon of >50 genes (12). Included in this group of swarmer cell-dependent proteins is a set of virulence factors, including flagellin, urease, hemolysin, and the ZapA metalloprotease, which is capable of degrading antibacterial peptides, such as β-defensin 1 and LL-37 (7, 15). It has been postulated, based in part on the evidence of coordinate expression of virulence factors during cellular differentiation, that the swarmer cell and swarming behavior are important in pathogenesis (6, 7, 19, 48).
Wild-type P. mirabilis strains are unable to differentiate in liquid cultures. Swarmer cell differentiation is initiated upon contact with a solid surface, hypothesized to inhibit flagellar rotation, which serves as the signal from the surface inducing swarmer cell genes (1, 10, 30, 60). Cell contact with the surface is absolute, and dedifferentiation of swarmer cells occurs rapidly once the cells are removed from the surface and placed into a liquid medium. It has been proposed that the inhibition of flagellar rotation is one signal in the differentiation process that leads to the P. mirabilis swarmer cell (1). In Vibrio parahaemolyticus, a marine swarming bacterial species, the inhibition of polar flagellum rotation can trigger the expression of a differentiated swarmer cell (17, 45). Similarly, in P. mirabilis, the correct flagellar assembly is required for differentiation, and certain flagellar mutations result in a non-wild-type response to the surface signal (14, 35). Cell-cell signaling via putrescine is also critical in P. mirabilis swarming motility (68), although how this signal is integrated into the regulatory hierarchy to control cell differentiation is not currently understood.
It is attractive to propose that the P. mirabilis flagellum itself acts as a surface sensor and has a role in swarmer cell differentiation; however, the mechanism involved in surface sensing remains to be identified. Further, since many genera of bacteria, including Aeromonas, Bacillus, Escherichia, Salmonella, Proteus, Pseudomonas, Vibrio, and Yersinia (2, 37, 38, 40, 58, 59, 80), are now known to swarm and undergo swarmer cell differentiation, a fundamental understanding of the surface-sensing mechanism inherent in this process is certain to lead to better, more effective treatment for the diseases caused by these pathogens. In this report, we test a hypothesis that P. mirabilis uses flagellar rotation to sense surfaces and control both swarmer cell differentiation and the expression of virulence genes.
MATERIALS AND METHODS
Strains, plasmids, oligonucleotides, and media.
The strains, plasmids, and oligonucleotides used in this study are listed in Table 1. P. mirabilis BB2000 is wild type for swimming and swarming behaviors. P. mirabilis strains were maintained as previously described (12, 13) in Luria-Bertani (LB) broth (61) or, when isolated colonies were required, on LSW− agar (10 g tryptone, 5 g yeast extract, 0.4 g NaCl, 5 ml glycerol, 20 g agar/liter distilled H2O) to phenotypically inhibit swarming. Escherichia coli strains were maintained either in LB broth or on LB agar. All bacterial cultures are incubated overnight at 37°C, unless otherwise noted. Antibiotics were added to the media at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 40 μg/ml; rifampin, 100 μg/ml; and tetracycline, 15 μg/ml.
TABLE 1.
Strains, plasmids, and oligonucleotides used
| Strain, plasmid, or oligonucleotide | Relevant characteristic(s)aor sequence | Derivation or description | Source and/or reference |
|---|---|---|---|
| E. coli K-12 | |||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17(rK− mK+) deoR thi-1 supE44 λ−gyrA96 relA1 | Laboratory stock (61) | |
| SM10 (λpir) | Rec− RP4-2Tc::Mu λpir | C600 | 24, 65 |
| INVαF′ | F′ endA1 recA1 hsdR17(rK− mK+) supE44 gyrA96 relA1 φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR thi-1 λ− | Invitrogen | |
| P. mirabilis | |||
| BB2000 | Wild type, Rfr Tcr | Spontaneous from PRM1 | 13 |
| BB2021 | flgK2021::Tn5-Cm, Cmr Rfr Tcr, non-wild-type elongation | Mini-Tn5-CM insertion in BB2000 | This study |
| BB2030 | flgL2030::Tn5-Cm, Cmr Rfr Tcr, non-wild type elongation | Mini-Tn5-CM insertion in BB2000 | This study |
| BB2043 | fliF2234::Tn5-Cm, Cmr Rfr Tcr, non-wild type elongation | Mini-Tn5-CM insertion in BB2000 | This study |
| BB2107 | fliM2107::Tn5-Cm, Cmr, Rfr Tcr, non-wild type elongation | Mini-Tn5-CM insertion in BB2000 | This study |
| BB2109 | flgK2109::Tn5-Cm, Cmr, Rfr, Tcr, non-wild type elongation | Mini-Tn5-CM insertion in BB2000 | This study |
| BB2122 | fliQ2122::Tn5-Cm, Cmr Rfr Tcr, non-wild type elongation | Mini-Tn5-CM insertion in BB2000 | This study |
| BB2148 | fliG2148::Tn5-Cm, Cmr Rfr Tcr, non-wild type elongation | Mini-Tn5-CM insertion in BB2000 | This study |
| BB2156 | flhA2156::Tn5-Cm, Cmr Rfr Tcr, non-wild type elongation | Mini-Tn5-CM insertion in BB2000 | This study |
| BB2189 | flgK2189::Tn5-Cm, Cmr Rfr Tcr, non-wild-type elongation | Mini-Tn5-CM insertion in BB2000 | This study |
| BB2193 | flgB2193::Tn5-Cm, Cmr Rfr Tcr, non-wild-type elongation | Mini-Tn5-CM insertion in BB2000 | This study |
| BB2196 | flgK2196::Tn5-Cm, Cmr Rfr Tcr, non-wild-type elongation | Mini-Tn5-CM insertion in BB2000 | This study |
| BB2199 | fliQ2199::Tn5-Cm, Cmr Rfr Tcr, non-wild-type elongation | Mini-Tn5-CM insertion in BB2000 | This study |
| BB2201 | fliP2201::Tn5-Cm, Cmr Rfr Tcr, non-wild-type elongation | Mini-Tn5-CM insertion in BB2000 | This study |
| BB2202 | flhA2202::Tn5-Cm, Cmr Rfr Tcr, non-wild-type elongation | Mini-Tn5-CM insertion in BB2000 | This study |
| BB2203 | flhD2203::Tn5-Cm, Cmr Rfr Tcr, non-wild-type elongation | Mini-Tn5-CM insertion in BB2000 | This study |
| BB2204 | fliL2204::Tn5-Cm, Cmr Rfr Tcr, non-wild-type elongation | Mini-Tn5-CM insertion in BB2000 | This study |
| Plasmids | |||
| Vectors | |||
| pACYC177 | Apr Kmr | p15A ori low-copy-number plasmid vector | Laboratory stock |
| pBluescriptII | Apr | pBluescriptII SK(+) and KS (−) | Stratagene |
| pBB402 | Tcr | pSB401 with deletion of EcoRI fragment containing Plux1 | This study |
| pBB403 | Apr Kmr | pCR2.1 containing promoterless luxCDABE cassette amplified by PCR using 401FOR and 401REV with pBB402 template | This study |
| pBB404 | Kmr | pACYC177 with PstI fragment from pBB403 containing a promoterless luxCDABE cassette with an EcoRI cloning site 5′ to luxC | This study |
| pBB413 | Kmr | 555-bp fragment bearing PzapA inserted upstream of luxCDABE | This study |
| pBB415 | Kmr | 721-bp fragment bearing PhpmB inserted upstream of luxCDABE | This study |
| pCR2.1 | Apr Kmr | PCR TOPO TA cloning vector | Invitrogen |
| pRSI100 | Apr Kmr | pCR2.1 containing a 770-bp DNA containing 211 bp 5′ to the start of fliL and 76 bp 3′ to its stop codon | Invitrogen |
| pSB401 | Tcr | Promoterless luxCDABE cassette on pACYC184 | 77 |
| Oligodeoxyribonucleotides | |||
| 401FOR | 5′-CTGCAGGAATTCAGGCTTGGAGGATACGTATGACTAAAAAAATTTC-3′ | PCR primer for cloning the promoterless luxCDABE cassette from pBB402; contains PstI (underlined) and EcoRI (double underlined) sites at the 5′ end | This study |
| 401REV | 5′-CTGCAGGCGCTGATGTCCGGCGG-3′ | PCR primer for cloning the promoterless luxCDABE cassette from pBB402; contains a PstI site at the 5′ end (underlined) | This study |
| fliL-F | 5′-CTCTGCTCGTGGTGGTGTCG-3′ | PCR primer for cloning P. mirabilis fliL | This study |
| fliL-R | 5′-GCGTCGTCACCTGATGTGTC-3′ | PCR primer for cloning P. mirabilis fliL | This study |
| Tn5-I | 5′-AGATCTGATCAAGAGACAG-3′ | Primer for sequencing DNA flanking mini Tn5-CM insertions | 34, 51 |
| Tn5-O | 5′-ACTTGTGTATAAGAGTCAG-3′ | Primer for sequencing DNA flanking mini-Tn5-CM insertions | 34, 1 |
| zapApF | 5′-GGATCCAGATATTATTTTTATTAATAATAAGGATTAC-3′ | PCR primer for cloning the upstream regulatory region of zapA (PzapA) and containing a BamHI site at the 5′ end (underlined) | This study |
| zapApR | 5′-AATATCCTCCAGATATAAATTTAGGTTAAGTTTATT-3′ | PCR primer for cloning PzapA | This study |
| hpmBpF | 5′-GGATCCTATGTAGTTTTTTTTTATCTATCATAAAC-3′ | PCR primer for cloning the upstream regulatory region of hpmB (PhpmB) and containing a BamHI site at the 5′ end (underlined) | This study |
| hpmBpR | 5′-TTTACCTCGAGATAAAGGTATTAATTCAA-3′ | PCR primer for cloning PhpmB | This study |
| This study |
Apr, Ampicillin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; Rfr, rifampin resistance; Tor, tetracycline resistance.
A high-copy-number ColE1 plasmid bearing P. mirabilis fliL+ was constructed by PCR amplification using two oligonucleotide primers (Table 1), fliL-F and fliL-R, which bind 211 bp 5′ to the start of fliL and 76 bp 3′ to the fliL stop codon, respectively. The 770-bp fliL DNA was cloned into pCR2.1 via TOPO TA cloning, resulting in pRSI100. This plasmid was then electroporated into E. coli DH5α and P. mirabilis as indicated.
Tn5 mutagenesis.
P. mirabilis mutants with defects in swarmer cell elongation were identified from a bank composed of 212 swarming mutants previously produced (12, 13) through mini-Tn5-CM (24) mutagenesis.
DNA manipulation.
Electroporation using standard methods was employed for transformation of E. coli and P. mirabilis (8). Chromosomal DNA was isolated using a modification of the cetyltrimethylammonium bromide procedure (8). Plasmid DNA was purified using the QIAGEN Plasmid Mini kit as described by the manufacturer. Oligonucleotide primers were obtained commercially (Sigma-Genosys).
PCR DNA amplification.
PCR was performed with either 2.5 U of recombinant Taq polymerase (AmpliTaq; Perkin-Elmer Cetus, Norwalk, Conn.), Vent (New England BioLabs), or Pfu Turbo (Stratagene) polymerase, and an MJ Research DNA engine thermocycler. PCR DNA amplicons were cloned using plasmid pCR2.1 and the TA TOPO cloning kit (Invitrogen Corporation, Carlsbad, CA) according to the recommendations of the manufacturer. The primers used are listed in Table 1.
Nucleotide sequencing and analysis.
Double-stranded DNA was used as a template for nucleotide sequencing using the recommended procedures of the Prism Ready Reaction Dye Deoxy termination kit (Applied Biosystems) in conjunction with Taq polymerase and a model 310 DNA sequencer (Applied Biosystems). The gene disrupted by Tn5 insertion was identified by searching for homologs in the P. mirabilis HI4320 genome database. The P. mirabilis genomic sequence data were produced by the Proteus mirabilis Sequencing Group at the Sanger Institute and can be obtained from ftp://ftp.sanger.ac.uk/pub/pathogens/pm/.
Protein analyses.
The protein concentration was determined using the microbicinchoninic acid protein assay reagent as recommended by the manufacturer (Pierce Chemical Co.). sodium dodecyl sulfate-polyacrylamide gel electrophoresis was done using the system established by Laemmli (43). Western blot analyses of proteins were performed using the appropriate antiserum as described previously (13).
Swarmer cell differentiation and swarming motility.
Swarming motility was observed every 15 to 30 min using a stereo dissection microscope and microcalipers to determine distances the cells moved over each time point. Swarmer cell differentiation, i.e., the overproduction of flagella, cellular elongation, and polyploidy, was also examined microscopically as described by Belas et al. (12). Cells obtained from an agar surface are not of uniform length; rather, the population is a mixture of cell types that include elongated swarmer cells, whose mean length is 45.3 ± 9.1 μm, as well as swimmer cells, with a mean length of 1.9 ± 0.3 μm. In the current study, any cell with a length of ≥35 μm or greater was defined as a swarmer cell. Using this definition, the percentage of swarmer cells in samples acquired from the periphery of wild-type swarming colonies typically comprises between 40 and 90% (averaging ca. 65%) of the total population depending on the sample measured. Therefore, in measuring swarmer cell lengths, a minimum size of 35 μm was applied, and only cells falling into this criterion were measured and called “swarmer cells.”
Swarmer cell differentiation during conditions of flagellar filament tethering was measured using LB broth supplemented with either 15% (wt/vol) polyvinylpyrrolidone 360 (PVP) (Sigma) or a 1:100 dilution of rabbit polyvalent antiserum to purified FlaA (α-Fla), which has been previously shown to tether the flagella of P. mirabilis (9). Vegetative swimmer cells were harvested by centrifugation from an overnight LB broth culture and resuspended in fresh broth to an optical density at 600 nm (OD600) of 0.6. The washed swimmer cells were inoculated at a 1:100 dilution in fresh LB broth supplemented with PVP or α-Fla. Swimming motility and cell elongation were then measured over the course of 6 h at 37°C with shaking.
Swimming motility.
Swimming motility was assessed macroscopically on Mot agar and microscopically in Mot broth as previously described (12).
Isolation of flagella and preparation of flagellin.
Flagella were isolated by mechanical shearing and the flagellin purified using the procedures described by Belas et al. (13).
Lux transcriptional fusions and measurement of gene expression.
Transcriptional fusions were constructed between the respective upstream regulatory regions of two known virulence genes, zapA and hpmB, and a promoterless bacterial luciferase (luxCDABE) cassette from Photorhabdus luminescens (78). Briefly, an EcoRI fragment containing PluxI was excised from pSB401 to yield pBB402 (Table 1). The promoterless luxCDABE genes were then amplified by PCR with primers 401FOR and 401REV and TOPO cloned into pCR2.1 to yield pBB403. Plasmid pBB403 was then cleaved with PstI, and the DNA fragment bearing luxCDABE was ligated into PstI-cut pACYC177. The resulting plasmid, pBB404, is a low-copy-number p15A ori plasmid with a unique EcoRI site immediately 5′ to the start codon of luxC and confers kanamycin resistance.
The upstream regulatory region of zapA was amplified from a genomic DNA template using primers zapApF and zapApR (Table 1), which introduce a BamHI site at the 5′ end of the fragment when in the correct orientation to permit PzapA transcription of luxCDABE. This PCR product was TOPO cloned in pCR2.1, and a 555-bp EcoRI fragment containing PzapA was isolated. The 555-bp fragment was then ligated into the EcoRI site of pBB404 and the orientation of the region confirmed by BamHI digestion, resulting in pBB413 (zapA::lux). A similar strategy was used to amplify the 721-bp upstream regulatory region of hpmB using primers hpmBpF and hpmBpR (Table 1) and clone the resulting DNA in pBB404, resulting in pBB415 (hpmB::lux).
The level of zapA and hpmB expression as the swarming colony migrated and matured on LB agar, i.e., spatial expression, was measured using the method of Belas et al. (16) with slight modifications. Luminescence from the expression of the lux cassette was recorded using a low-light charge-coupled device digital camera detection system (Photometrics Quantix 1400 camera) and IPLab (Scanalytics, Fairfax, VA) optical measurement software. To maintain a constant reference, all images were recorded at a 10-s exposure, determined empirically to yield nonsaturated pixels at all incubation times. The 256-level gray-scale images of colony luminescence were converted into false color representations using ImageJ (Rasbad, NIH) and Scion Image (Scion Corp.) software; no other image enhancement was done. The false color images were then arranged using Adobe Photoshop V7.01 software. A similar method was used to obtain natural images of the swarming colony without postimaging false color representation.
Using a slight modification of the method described by Wang et al. (74), temporal expression of zapA and hpmB transcription was measured by inoculating a 1:100 dilution of washed swimmer cells, obtained from overnight LB broth culture and resuspended to an OD600 of 0.6, in fresh, prewarmed LB broth. For LB agar lux expression the same inoculum was used, which was spread over the agar surface with a sterile glass rod. The cells were then incubated at 37°C and, in the case of the broth cultures, with vigorous shaking. LB broth cultures were processed at each time point by measuring the OD600 of the culture directly, while LB agar cultures were resuspended in 5 ml of LB broth using a sterile glass spreader to disperse the cells off the surface. The OD600 of the cell suspension was then recorded. The bioluminescence (as relative light units [RLU]) of triplicate 100-μl samples of the LB broth and LB agar cultures was measured using an EG&G Berthold Microlumat LB 96P luminometer. Luminescence was recorded as the mean number of RLU per OD600 plotted against time.
Materials and reagents.
All reagents were of the highest purity available. Components of bacteriological media were purchased from BBL (Difco). Restriction endonucleases and DNA-modifying enzymes were obtained from New England Biolabs, Stratagene, Invitrogen, Perkin-Elmer, or Promega and were used according to the supplier's recommendations.
RESULTS
Conditions that inhibit flagellar rotation induce swarmer cell differentiation.
In another swarming bacterium, V. parahaemolyticus, conditions that restrict polar flagellar rotation lead to swarmer cell differentiation. These include increasing viscosity, using antibodies to inhibit flagellar function, and decreasing motor speed by controlled use of a sodium channel-blocking drug (17, 45, 46). Certain mutations in flagellar genes have also been shown to prevent V. parahaemolyticus swarmer cell development (45).
It is not known what signal or signals are required by P. mirabilis to induce swarmer cell differentiation and expression of virulence genes; however, there is anecdotal evidence that forces that prevent rotation of the flagella of P. mirabilis induce swarmer cell differentiation in this species. Thus, inhibition of the rotation of the flagellar filaments is thought to be the signal sensed by P. mirabilis when it nears or is on a surface.
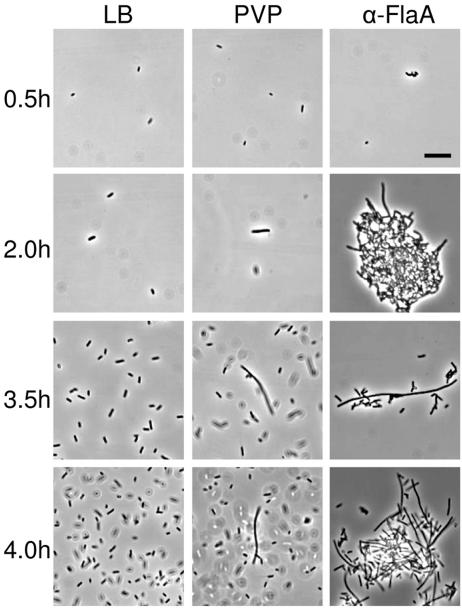
Vegetative swimmer cells were incubated in LB broth, LB broth containing 15% PVP (molecular weight, 360,000), or LB broth with a 1:100 dilution of rabbit antiflagellin polyvalent antiserum (α-FlaA), and the swimming behavior and cell morphology were examined by light microscopy. The results are shown in Fig. 1. Vegetative cells placed in PVP (Fig. 1, middle column) or α-FlaA (Fig. 1, right column) rapidly (<15 min) ceased swimming due to the high viscosity (∼16 centipoise) of 15% PVP and the tethering of flagella by the antiserum, respectively. As a comparison, cells in LB broth (left column in Fig. 1) continued to swim throughout the course of this experiment. At 2 h postinoculation, elongation of cells in PVP and α-FlaA became apparent, reaching a maximum at 4 h. However, after 4.5 h, cell elongation decreased, while at the same time, motility increased in both PVP and α-FlaA. The length of cells in PVP and α-FlaA equaled and often exceeded the mean length of wild-type swarmer cells obtained from an LB agar surface. The simplest explanation of these results is that conditions such as high viscosity and antibody tethering of flagella, which inhibit the rotation of the flagella, induce swarmer cell differentiation. These data support the hypothesis that P. mirabilis ascertains its location in the environment or host by assessing the “health” of its flagellar motors.
FIG. 1.
Conditions that inhibit flagellar rotation induce swarmer cell differentiation. Wild-type cells were incubated in LB broth (left column), LB broth containing 15% PVP (wt/vol) (middle column, labeled PVP), and LB broth with a 1:100 dilution of polyvalent rabbit antiflagellin antiserum (right column, labeled α-FlaA), and the swimming behavior and cell morphology were analyzed. Within less than 0.5 h, cells placed in PVP or α-FlaA became immobilized, presumably as a consequence of either the increased viscosity due to addition of PVP or the tethering of flagellar filaments by α-FlaA. Tethering of cells was manifested by the formation of clumps, as can be seen in the α-FlaA micrographs (right column). Within 2.0 h, cells in the presence of either PVP or α-FlaA began to elongate, with the population obtaining a maximum percentage of elongated cells at 4.0 h. At this time, cells of 20 to >40 μm in length were common, as can be observed in at times 3.5 and 4.0 h for PVP- or α-FlaA-treated cells. After 4.0 h, cell elongation decreased with a concomitant increase in a swimming motility that was predominantly smooth. In contrast, cells incubated in LB were motile and did not clump at any time (left column). Bar = 5 μm.
Mutations affecting flagellar genes result in a non-wild-type response to surface signals.
The results suggest that a functioning flagellum is critical for sensing and/or responding to the surface signal. Since the elongated swarmer cell is the most easily observed swarmer cell phenotype (as demonstrated in Fig. 1), a bank of 212 Tn5-CM transposon-insertion mutants (12) with defects in swarming motility was screened for defects in the swarmer cell elongation phenotype. Identification of the mutated gene was accomplished by cloning the antibiotic resistance gene of the transposon along with a portion of the flanking P. mirabilis genome using either ClaI, PstI, or XbaI restriction digestion and ligation, followed by DNA sequence analysis with oligonucleotide primers Tn5-I and Tn5-O (Table 1).
In this bank, 19 mutants, with mutations in a group of 11 genes, were found to have aberrant swarmer cell elongation (Table 2). These mutants could be separated into two categories: those that failed to produce an elongated swarmer cell and a second group that produced an elongated swarmer cell under normally noninducing conditions and/or produced an unusually long swarmer cell. These results are shown in Tables 1 and 2. The group of mutants that failed to produce an elongated swarmer cell included mutations in flhD, the first gene in the flhDC operon and referred to as the “master regulator” of the flagellar gene hierarchy and other cellular functions (56), fliM, encoding the switch protein of the flagellar C ring (66), components of the flagellar type three secretion system (fliP, fliQ, and flhA), as well as proteins composing the hook (flgK and flgL) and basal body rod (flgB). The second group of mutants produced abnormally long swarmer cells or swarmer cells under noninducing conditions and resulted from mutations in three genes encoding proteins associated with the basal body, including FliF (MS ring), FliG (part of the motor switch complex), and FliL (a protein of unknown function associated with the basal body) (27, 63, 71). Mutation of either fliG or fliL resulted in the formation of elongated swarmer cells during growth in broth and the production of unusually long swarmer cells when these mutants were grown on nutrient agar (Table 2). Unlike fliG or fliL mutants, the fliF mutation, while producing hyperelongated cells when grown on agar, did not result in the formation of swarmer cells in liquid medium. Thus, all three mutants share a common phenotype of unusually elongated cells when grown on nutrient agar but differ in expression of the elongated swarmer cell in liquid medium. As expected, all 11 mutant classes were defective in swarming motility, and all but two were also defective in swimming motility (Table 2).
TABLE 2.
Phenotypic analysis of mutations giving rise to defects in biofilm formation
| Class | Mutated genea | Function and/or cellular location | Motilityb
|
Mean cell length (μm)c
|
||
|---|---|---|---|---|---|---|
| Swimming | Swarming | Broth | Agar | |||
| I | flhD | Master regulator | − | − | 1.7 ± 0.1 | 2.1 ± 0.1 |
| II | fliF | MS ring | − | − | 2.2 ± 0.2 | 58.9 ± 11.8 |
| fliG | Motor switch | − | − | 33.8 ± 10.6 | 45.0 ± 9.4 | |
| fliL | Hook basal body | − | − | 35.4 ± 7.7 | 69.3 ± 22.9 | |
| fliM | C ring | +/− | − | 1.9 ± 0.3 | 2.0 ± 0.2 | |
| fliP | Export pathway | ++ | − | 1.9 ± 0.1 | 2.2 ± 0.3 | |
| fliQ | Export pathway | − | − | 1.9 ± 0.2 | 2.0 ± 0.2 | |
| flhA | Export pathway | − | − | 1.7 ± 0.1 | 1.9 ± 0.2 | |
| III | flgK | Hook junction | − | − | 1.8 ± 0.2 | 1.9 ± 0.3 |
| flgL | Hook junction | − | − | 1.9 ± 0.1 | 1.9 ± 0.2 | |
| flgB | Basal body rod | − | − | 2.0 ± 0.2 | 2.3 ± 0.2 | |
Based on BLAST protein:protein homology to sequences in the P. mirabilis HI4320 genome database (ftp://ftp.sanger.ac.uk/pub/pathogens/pm/).
Motility is scored on a relative scale from − to ++++, where − indicates no motility and ++++, indicates wild-type motility.
Mean cell length (± standard error of the mean) as determined using light-microscopic measurements of 50 cells for each determination. The mean cell length of wild-type swarmer cells obtained from the periphery of a swarming colony is 45.3 ± 9.1 μm, while the mean swimmer-cell length is 1.9 ± 0.3 μm.
Taken together, these data underscore the essential role of the flagellum in sensing a surface and show that defects in flagellar genes lead to one of two phenotypes, both of which result in the inappropriate processing of the surface signal. To gain greater insight into the nature of these defects and their role in surface signal transduction, the fliL mutation, chosen because it resulted in the most exaggerated swarmer cell elongation phenotype, was examined in greater detail.
Mutation of fliL results in cells that are nonmotile, do not synthesize flagellin, and have elongated cell morphology.
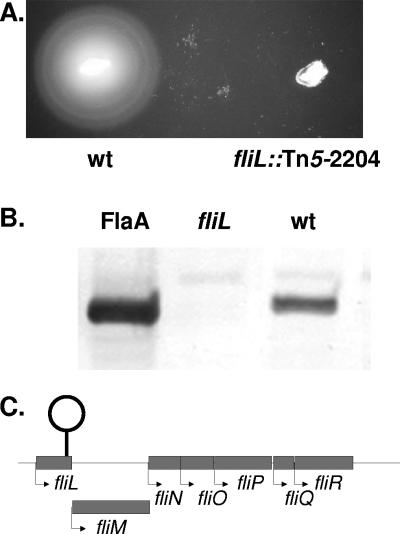
The phenotype of a representative fliL mutant, strain BB2204 (fliL::Tn5-CM), was examined. The motility of FliL− cells was determined using a combination of light-microscopic examination of cells grown in Mot broth and semisolid Mot agar to detect swimming motility (Fig. 2A), while swarming was measured on LB agar (12). The results of these experiments confirm that mutants with defects in fliL do not swim or swarm (Table 2 and Fig. 2A).
FIG. 2.
FliL− bacteria are nonmotile and defective in flagellar synthesis. (A) The swimming motility of a FliL− mutant (fliL2204::Tn5-Cm) was measured using semisolid Mot agar and compared to that of the wild-type strain. FliL− defects result in a loss of swimming and swarming (see Table 2). (B) Detection of FlaA flagellin in fliL and wild-type whole-cell homogenates. Western blots with polyvalent antiserum to FlaA, the principal flagellin of P. mirabilis, were used to detect the presence of FlaA in whole-cell homogenates. The antiserum hybridized to a single prominent band of ca. 40 kDa both in the control lane (FlaA; left lane) containing purified FlaA protein and in homogenates from wild-type cells (wt; right lane) but failed to detect FlaA in the fliL strain (fliL; middle lane). (C.) Map of fliL locus. Nucleotide sequence analysis indicates that the transposon insertion giving rise to fliL2204::Tn5-Cm is located at nucleotide 430 in the 483-bp fliL gene and may have resulted in a polar effect on the genes located downstream (3′) in the putative operon.
Western blots using α-FlaA were used to detect flagellin in preparations of sheared flagellar filaments from broth-grown wild-type and fliL cells (Fig. 2B). The fliL mutant lacked external FlaA, which was confirmed by light microscopy of stained cells. The insertion site of Tn5-CM in fliL was determined to be at base 432 of the fliL gene, 50 bases from the 3′ end of the coding sequence (Fig. 2C); thus, the transposon inserted in the first gene of a seven-gene operon (fliLMNOPQR). Insertion of Tn5 in fliL could result in polar effects on the downstream genes responsible for flagellin export, i.e., fliOPQR, which may help to explain the failure of fliL cells to produce flagella. Polar effects of Tn5 insertion in fliL may also affect the hyperelongated phenotype of this mutant; however, this is not likely to be the case, since transposon insertions in fliM, fliP, and fliQ were also analyzed in this study, and none of these mutations resulted in the hyperelongated swarmer cell phenotype observed in the fliL mutant. In fact, all three mutants failed to form elongated swarmer cells under all conditions (Table 2).
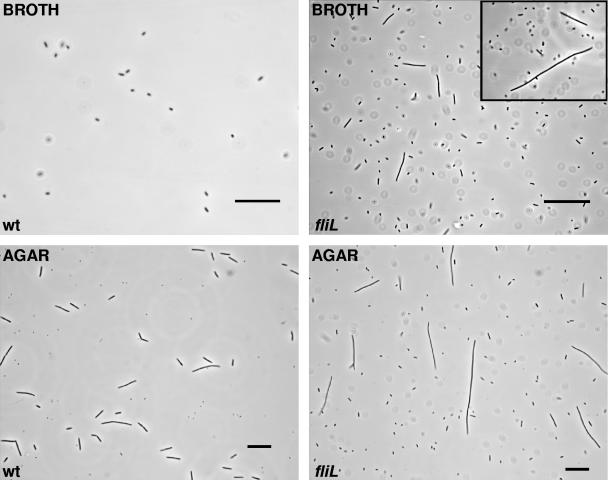
Mutations in fliL resulted in cells that were abnormally elongated when grown in either broth or agar (Fig. 3) compared to wild-type cells. For example, when BB2204 was grown in LB broth, a non-swarmer cell-inducing environment, swarmer cells comprised ca. 25 to 40% of the population, while the wild-type strain produces <0.1% swarmer cells in liquid media. The mean cell length of liquid-grown swarmer cells with a fliL mutation was 35.4 μm, compared to a wild-type swimmer cell length of 1.9 μm (Table 2 and Fig. 3). In liquid media, FliL− cells exceeding the mean length were frequently observed, as shown in the inset in Fig. 3. Up-regulation of swarmer cell elongation (mean cell length = 69.3 μm) was also apparent in FliL− cells grown on LB agar (Fig. 3). This is a significant difference from the mean length of wild-type swarmer cells (45.3 μm). Many fliL cells exceeded 80 μm length (Fig. 3). The percentage of swarmer cells in samples of bacteria obtained from the periphery of swarming colonies was essentially the same for the fliL mutation and wild-type, i.e., elongated swarmer cells comprised between 40 and 90% of the population. One interpretation of these data is that fliL defects result in an up-regulation of the genes, resulting in the differentiated swarmer cell.
FIG. 3.
Mutation in fliL results in cells that are locked into an elongated morphotype. The morphology of the wild-type (wt) and fliL (fliL) strains was examined by phase-contrast light microscopy after incubation (8 h at 37°C) in LB broth and on L agar. Wild-type cells differentiated to the swarmer cell morphotype when grown on L agar but not in LB broth (left column). In contrast, the FliL-defective strain differentiated into elongated cells, reminiscent of wild-type swarmer cells but lacking flagella, in liquid medium (LB broth; right top micrograph), a condition that does not induce differentiation in the wild type. The FliL− strain also produced elongated cells that were significantly longer than wild-type swarmer cells (mean FliL− cell length = 69.3 ± 10 μm versus 45.3 ± 9.1 μm for wild-type cells) when grown on agar-gelled LB. Bars = 25 μm.
Complementation of defects in fliL restores cell elongation but not motility.
A ColE1 plasmid (pRSI100) bearing an intact copy of fliL+ under the control of the lac promoter was constructed and transformed into BB2204 (fliL2204::Tn5-Cm). The lac promoter is constitutively expressed in P. mirabilis, resulting in overexpression of FliL. When the FliL− strain was complemented in trans by the plasmid-borne copy of fliL+, the cells lost the elongated phenotype, i.e., their mean length (2.1 μm) was equivalent to that of wild-type swimmer cells regardless of whether they were grown in liquid or on agar medium (data not shown). Therefore, fliL+ in trans can complement the fliL defect in BB2204; however, fliL+ did not complement the motility defects, and these cells remained nonswimming and nonswarming. These data support an involvement of FliL in surface signal reception and swarmer cell differentiation.
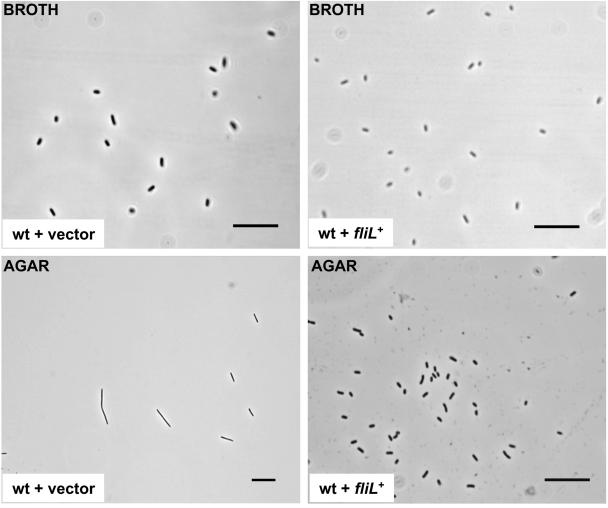
Overexpression of fliL+ in wild-type cells results in constitutive swimmer cell morphology and loss of swimming and swarming motility.
Plasmid pRSI100 was also transferred to wild-type P. mirabilis and E. coli. Overexpression of fliL+ in a wild-type background results in an inability of the cells to differentiate to swarmer cells on LB agar surfaces (Fig. 4), a severe reduction in swimming motility, a loss of swarming motility, and a marked decrease in flagellar filament synthesis, as determined by Western blots using anti-FlaA antisera. The plasmid bearing fliL+ from P. mirabilis was also transformed into E. coli DH5α. When the expression of P. mirabilis fliL+ was induced with isopropyl-β-d-thiogalactopyranoside, the swimming motility and flagellar synthesis of E. coli both decreased. These results may be due to failure of the cell to assemble a flagellum if too much FliL is made or may indicate a more direct involvement of FliL in surface sensing. The data do not permit a resolution of these two possibilities; however, they do suggest that FliL plays a role in flagellum function, both in P. mirabilis and in E. coli.
FIG. 4.
Overexpression of FliL results in wild-type cells unable to undergo swarmer cell differentiation. A multicopy plasmid bearing fliL was transformed into wild-type P. mirabilis, and the cellular morphology was measured by phase-contrast light microscopy. When incubated in LB broth (top row), the length of wild-type cells harboring either the vector alone (left top) or the fliL-containing plasmid (right top) was indicative of the vegetative, swimmer cell (averaging 2 μm). Wild-type cells bearing the vector control grown on L agar differentiated to swarmer cell length (bottom left; mean, 45.3 ± 9.1 mm), while those overexpressing FliL (wt + fliL+; lower right) did not and instead remained as vegetative, swimmer cells. Bars = 25 μm.
Defects in fliL up-regulate virulence gene expression.
A low-copy-number plasmid with a promoterless P. luminescens luxCDABE cassette downstream of an EcoRI cloning site, pBB404, was used to measure gene expression of two known virulence genes, zapA and hpmB, encoding the major extracellular zinc metalloprotease and hemolysin, respectively. The upstream regulatory region of each respective gene was inserted 5′ to the lux genes, and light (Lux+) was measured as a function of transcriptional activity in wild-type and fliL strains grown in LB broth or on LB agar.
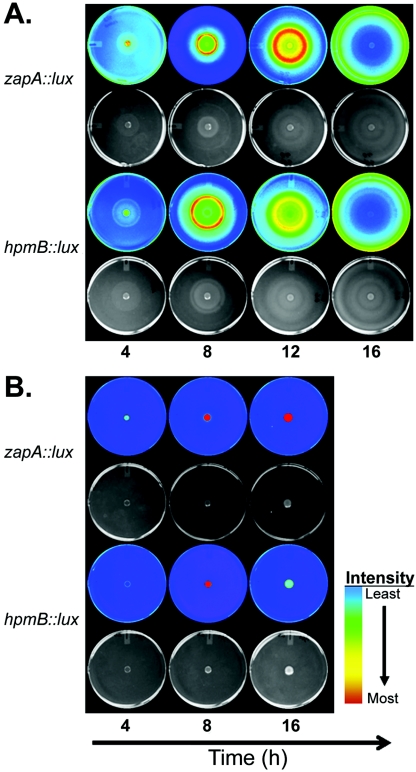
The expression of zapA::lux and hpmB::lux was measured over time after broth-grown cells were inoculated on the center of LB agar and incubated at 37°C. The results are shown in Fig. 5, where the Lux imagery is paired with a visible light photograph of the same culture (shown below the Lux image for each strain), and the Lux intensity is depicted in false color, where blue is low or background Lux levels while red represents the most intense Lux values. The data indicate that both zapA and hpmB are up-regulated after the wild-type cells are exposed to the agar surface (Fig. 5A). Not only are zapA and hpmB temporally regulated during swarmer cell differentiation, but their expression is also spatially regulated, with maximal expression occurring at or near the periphery of the swarming colony. These results are in good agreement with previously published results (7, 73).
FIG. 5.
The spatial expression of the virulence genes zapA and hpmB is coordinately regulated with swarming and is affected by a mutation in fliL. The upstream promoter regions of zapA and hpmB, respectively, were fused to a promoterless luxCDABE cassette harbored on a low-copy-number plasmid and transformed into wild-type and fliL strains. Each of the four resulting strains was inoculated as a 5-μl spot in the center of an L agar plate and incubated at 37°C. Gene expression was measured as luminescence (Lux), which is displayed in false color corresponding to relative Lux intensity, where blue is lowest and red is most intense (Materials and Methods). An image of the colony growth photographed in natural light is shown below its corresponding Lux image. (A) Expression of zapA (first two rows) and hpmB (third and fourth row) in the wild type at 4, 8, 12, and 16 h. (B.) Expression of zapA and hpmB in the fliL strain at 4, 8, and 16 h.
The zapA::lux and hpmB::lux plasmids were also transformed into BB2204 (fliL2204::Tn5-Cm), and the expression of the two virulence genes was measured (Fig. 5B). Despite the absence of swarming motility, both zapA and hpmA are expressed in a fliL background. The expression of zapA is significantly greater in a fliL background than in the wild type. This may be seen when the Lux intensities of wild-type and fliL strains harboring zapA::lux are compared at 16 h (Fig. 5B). Similarly, expression of the hpmB::lux fusion, although not as dramatic as that of zapA::lux, was also up-regulated in the fliL background. Because the fliL mutation rendered the cells nonswarming, it was not possible to determine if the spatial expression of either virulence gene was altered during swarming. These semiquantitative measurements indicate that the expression of both zapA and hpmB is up-regulated when fliL is mutated.
The up-regulation of zapA and hpmB in a fliL background was verified by a quantitative assay of temporal gene expression (Fig. 6A and B) using the method of Wang et al. (74) with slight modification (Materials and Methods). The difference between this method and the one used in Fig. 5 is that the cells do not establish a migrating colony, which minimized heterogeneity in the cells assayed (74). Using this approach, washed swimmer cells were diluted to a fixed concentration and inoculated into prewarmed LB broth (plus antibiotic) or an inoculum was spread on prewarmed LB agar (with antibiotics). Cells in broth were grown in flasks on rotary shakers, while those on plates were evenly spread over the entire plate and placed in an incubator. Multiple agar plates were inoculated, allowing individual agar cultures to be sacrificed at each sampling. Cells growing on agar were removed by scraping them off the surface, while aliquots of the broth-grown cells were taken directly from the flask culture. These samples were diluted to a fixed cell density (OD600), and the luminescence of each sample was measured in triplicate.
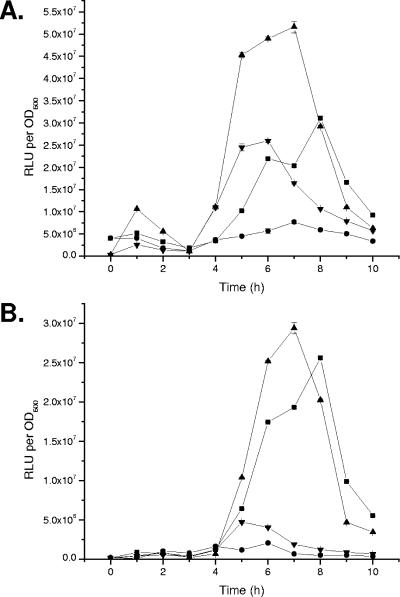
FIG. 6.
Mutation of fliL results in overexpression of zapA and hpmB in both vegetative and swarmer cells. Wild-type and fliL cells were inoculated into LB broth or spread onto the surface of L agar, and the expression of zapA::lux (A) and hpmB::lux (B) was measured over time. The mean luminescence at each point is expressed as relative light units per OD600. Symbols: ▪, wild-type cells grown on agar; •, wild-type cells grown in LB broth; ▴, fliL cells grown on agar; ▾, fliL cells grown in LB broth. Error bars show standard errors of the mean; n = 3.
Both zapA and hpmB are up-regulated during swarming of the wild-type cells (compare the squares in panels A and B of Fig. 6); however, their expression is significantly increased in the fliL background. In broth, the expression of zapA in the fliL strain is ca. five times greater than that in a wild-type background, while when the cells are grown on LB agar, zapA expression is >1.5× in the fliL background compared to wild-type expression. Expression of hpmB is also up-regulated in a fliL background compared to that in wild-type cells (an increase of ca. 2.5× to 3× for swimmer cells and a 1.5× increase for swarmer cells). Taken together, these data support the idea that FliL is critical not only to swarmer cell differentiation but also to the regulation of zapA and hpmB.
FlaA antiserum tethering of flagellar filaments causes an increase in the expression of hpmB but not zapA.
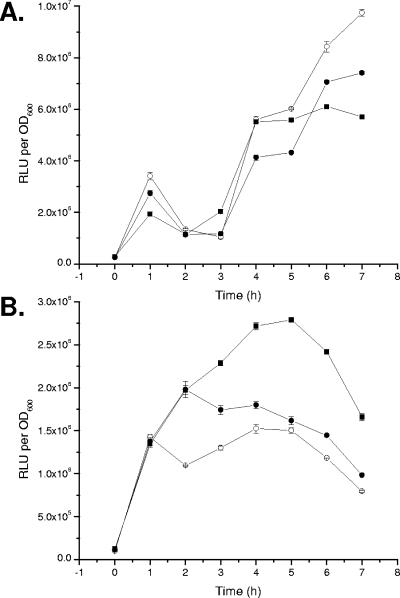
The expression of zapA and hpmB was measured in LB broth containing α-FlaA antiserum at a 1:100 dilution. The expression of zapA (Fig. 7A) is not affected by the addition of flagellin antiserum, while the expression of hpmB (Fig. 7B) is up-regulated (ca. 1.5×) compared to that for the controls (LB alone or LB plus preimmunization serum). These results conflict with the data presented earlier in this study and suggest either that the antiserum did not produce sufficient tethering to induce zapA expression or that multiple regulatory pathways may control the expression of zapA. These possibilities are addressed in the Discussion.
FIG. 7.
Conditions that inhibit flagellar rotation induce the expression of hpmB but not zapA. Wild-type cells harboring either a (A) zapA::lux or (B) hpmB::lux transcriptional fusion were inoculated into LB broth (○), LB broth with a 1:100 dilution of prebleed rabbit serum (•), or LB broth with a 1:100 dilution of rabbit antiflagellin serum (▪), and the luminescence as relative light units per OD600 was recorded over time. The data are expressed as the mean RLU per OD600. Error bars show standard errors of the mean; n = 3.
The expression of zapA and hpmB is not induced by addition of putrescine.
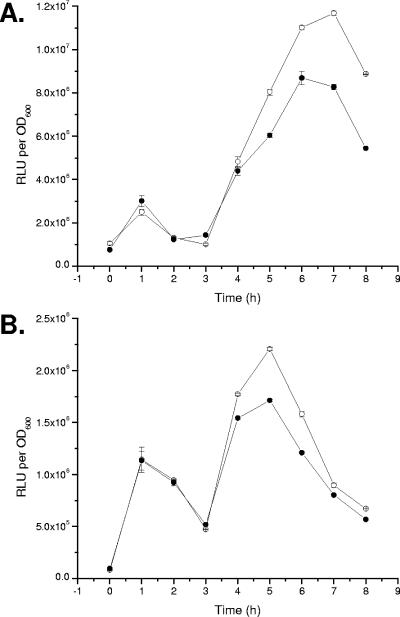
Putrescine has been shown to have a stimulatory effect on swarming behavior. Mutants defective in putrescine metabolism are delayed (but not defective) in swarming, and addition of putrescine restores swarming and cellular differentiation (68). How putrescine functions in this process is currently not known; however, one possible role for putrescine is as an inducer of gene expression, specifically acting in concert with the flagellum-mediated surface signal to up-regulate swarmer cell-dependent genes during the differentiation process.
Putrescine (final concentration, 1 mM) was added to washed suspensions of wild-type cells harboring the zapA::lux or hpmB::lux transcriptional fusion, and the bioluminescence and culture density (OD600) were measured. The results, shown in Fig. 8, indicated that neither the expression of zapA (Fig. 8A) nor that of hpmB (Fig. 8B) is affected by addition of putrescine. Adjusting the concentration of putrescine from 0.1 mM to 500 mM did not affect the expression of either gene. These results indicate that putrescine is not directly involved in regulating the expression of zapA and hpmB.
FIG. 8.
The expression of zapA and hpmB is not affected by putrescine. Wild-type cells harboring either a (A) zapA::lux or (B) hpmB::lux transcriptional fusion were inoculated into LB broth (○) and LB broth with 1 mM putrescine (•). Luminescence was measured as relative light units per OD600. The data are expressed as the mean RLU per OD600. Error bars show standard errors of the mean; n = 3.
DISCUSSION
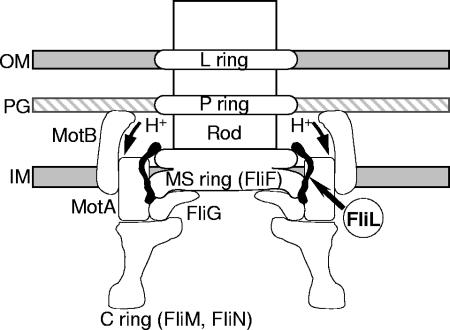
Bacteria swim and swarm by rotating their helical flagella (64). The flagellum may be subdivided into two substructures: a hook-basal body (HBB) that spans the bacterial membranes and the external filament. Within the HBB (Fig. 9), the LP-ring assembly is at the level of the outer (lipopolysaccharide) membrane and peptidoglycan layer and is thought to function as a bushing for the central rod. The MS ring is within and above the cytoplasmic membrane (M, membrane; S, supramembrane). The MS ring and the cell-proximal part of the rod are formed from a single protein, FliF (70). Mounted on the cytoplasmic face of the MS ring is a drum-shaped structure termed the C ring (28, 41). FliG, FliM, and FliN form a rotor-mounted assembly termed the switch complex, which is essential for flagellar assembly and clockwise-counterclockwise switching of flagellar rotation in chemotaxis, as well as for rotation (79). MotA and MotB are membrane proteins (23) that form the stator (20) and function together to conduct H+ across the membrane (18). Rotation of the HBB structure is achieved via the motor force generators, which are proton pumps that act as stators against the C ring.
FIG. 9.
A model of the possible location and function of FliL in P. mirabilis swarmer cell differentiation. An analysis of the deduced amino acid sequence of FliL indicates that the protein contains an amphipathic helical region within the N-terminal section of the polypeptide that may function as a membrane-spanning domain. P. mirabilis FliL is homologous to the FliL proteins of E. coli and S. enterica serovar Typhimurium, and the genetic organization of P. mirabilis fliL as the first gene in a putative operon containing six other genes encoding proteins that are homologous to known flagellar proteins strengthens the idea that P. mirabilis FliL is physically associated with the flagellar hook-basal body structure, as has been demonstrated for FliL from both S. enterica and C. crescentus (39, 63). Mutations in fliL, fliF, and fliG each result in swarmer cell differentiation in noninducing conditions, which suggests that FliL, FliF, and FliG constitute critical components of the P. mirabilis surface-sensing mechanism.
FliM and FliN are encoded by a large flagellar class II operon. This operon, fliLMNOPQR (Fig. 2C), whose first gene is fliL, includes the motor/switch (fliMN) and export apparatus genes (fliOPQR) (47, 69). FliL has no apparent mutant phenotype in E. coli or Salmonella, even when virtually all of its sequence is deleted (57), while mutations in any of the other genes in the fliL operon have deleterious effects on either flagellar switching, assembly, or rotation (47, 69).
FliL is a small protein with deduced and apparent molecular masses of ca. 17 kDa in Salmonella (63) and ca. 18 kDa in P. mirabilis. Its deduced amino acid sequence of 160 amino acids has a basic N terminus followed by a hydrophobic sequence that suggests it is likely a transmembrane domain. Schoenhals and Macnab (63) have shown that Salmonella FliL is an inner membrane protein that associates with the basal body. Based on their observations, Schoenhals and Macnab suggest that FliL is located in the vicinity of the Mot proteins and interacts with the MS ring (63), where it functions to stabilize the Mot complexes, perhaps through an interaction between FliL and MotB. Alternatively, FliL might facilitate the interaction between the stationary Mot complex and the rotating MS ring (63).
In contrast to those of E. coli and Salmonella, the FliL protein of Caulobacter crescentus is required for flagellar rotation and normal cell division (81). In-frame deletions of fliL result in cells that are nonmotile, and FliL is required for the efficient turnover of FliF (3, 39). The interaction between C. crescentus FliL and FliF suggests that FliL may be involved in MS-ring function and flagellar rotation in this bacterium (3).
The results of the current report demonstrate that inhibition of flagellar rotation is critical for P. mirabilis swarmer cell elongation and differentiation. Conditions, such as increased medium viscosity or tethering of flagellar filaments by antisera, that reduce the rotation of the filaments (even in normally noninducing environments, such as liquid medium) induce the formation of the elongated swarmer cell, one of the most conspicuous features of P. mirabilis swarmer cell differentiation (1). Conditions that reduce the ability of the polar flagellum to rotate also induce swarmer cell differentiation in another swarming bacterium, V. parahaemolyticus (45). These data show that P. mirabilis and V. parahaemolyticus monitor the rotation of the flagellar filament to sense and respond to surfaces. On the other hand, a recent study by Wang et al. (75) demonstrates that the flagella of Salmonella enterica serovar Typhimurium act as sensors of surface wetness, providing an alternative mechanism for controlling swarming. Apparently, the various species of swarming bacteria have developed multiple ways to sense a surface.
In a search to understand the mechanism underlying P. mirabilis surface signal transduction, transposon insertion mutagenesis was used to locate mutants with defects that affect swarmer cell elongation. Most (193 out of 212) of the mutations affecting swarming motility do not affect swarmer cell elongation. The remaining 19 mutants with abnormal swarmer cell elongation had defects in 11 flagellar genes associated with the regulation, biosynthesis, or export of the bacterial flagellum. Eight of the eleven mutations, including defects in flhD, fliM, fliP, fliQ, flhA, flgK, flgL, and flgB, result in cells that do not elongate under any condition and may be considered constitutively null for the swarmer cell elongation phenotype. Although not identified in this study, defects in flaA, the major flagellin-encoding gene, have also been shown to result in a swarmer cell elongation null phenotype (9). Together these data underscore the importance of the flagellar filament in P. mirabilis surface sensing.
The defects in fliF, fliG, and fliL may be divided into two classes based on the resulting swarmer cell elongation phenotype. The first class, represented by the fliF mutation, forms hyperelongated swarmer cells (mean cell length of 130% of that of the wild type) when grown on agar but does not produce a swarmer cell in liquid medium. The second class, represented by mutations in fliG and fliL, abnormally produces swarmer cells when grown in broth and hyperelongated swarmer cells when grown on agar medium (Table 2). The data do not provide clues to the reason why the two classes differ in the formation of swarmer cells in liquid medium, but this phenotypic difference undoubtedly reflects molecular differences in how these proteins interact to transduce the signal from the external surface.
Mutations in fliL, fliG, and fliF are unusual, because defects in all other flagellar genes thus far studied (Table 2) result in cells that do not differentiate and have low virulence gene expression (9, 29, 31, 35, 36, 49). Mutations in fliL, fliG, and fliF, in contrast, produce the opposite phenotype. For example, defects in FliL inhibit cell division, giving rise to an elongated cell, and up-regulate the expression of virulence genes, specifically zapA and hpmB (Fig. 5 and 6).
When these mutants formed hyperelongated swarmer cells, the elongated cells comprised ca. 40 to 90% of the population, while the remainder of the cells had lengths that ranged from swimmer cell size (ca. 2.0 μm) up to 35 μm, the lower cutoff length for swarmer cells in this study (Materials and Methods). This variability in cell length is a common feature of swarmer cell populations and can be observed in wild-type P. mirabilis during swarmer cell differentiation (see Fig. 1, 3.5-h exposure in PVP or α-FlaA, as an example).
Defects in fliL result in nonmotile cells that do not synthesize flagella (Fig. 2), abnormal formation of swarmer cells in broth, and formation of hyperelongated (153% longer than wild-type) swarmer cells on agar medium. Complementation of the fliL defect restores cell elongation but not motility, while overexpression of FliL in a wild-type background prevents swarmer cell elongation, both swimming and swarming motility, and flagellum synthesis. Overexpression of P. mirabilis fliL+ in an E. coli background also causes a loss of swimming motility and flagellum synthesis. This is interesting, because it implies that FliL has a role in flagellum function in both P. mirabilis and E. coli and is not cryptic, as originally suggested (63). The expression of two virulence genes, zapA and hpmB, is also up-regulated in a fliL background. Taken as a whole, these results link P. mirabilis surface sensing with flagellum function and virulence gene expression. The data also suggest that FliL may play a role in the transmission of the signal from the exterior flagellar filament into the cell, although much more detailed studies are needed to confirm the role of this protein in swarmer cell differentiation and virulence gene regulation.
Expression of the genes encoding the major extracellular metalloprotease, zapA (15, 73), and hemolysin, hpmB (and hpmA) (50, 72), was used as a general measure of virulence gene expression in this study. The gene expression data were obtained using plasmid-borne transcriptional fusions between the upstream regulatory regions from either zapA or hpmB and a promoterless luxCDABE. The data presented are in good agreement with those reported by Fraser et al. (29), who also used lux transcriptional fusions and Northern blots to measure the expression of hpmB and zapA (29). In the earlier report, both zapA and hpmB were found to be expressed concomitantly with swarmer cell differentiation and behavior, as has been observed here (Fig. 5 and 6), but were also shown to be under separate control circuits (7, 29). The difference in regulatory control between zapA and hpmB may help to explain why adding α-FlaA antisera to LB broth induced the expression of hpmB but not zapA (Fig. 7). However, there may be a simpler explanation resulting from the observation that α-FlaA tethering dramatically decreases beyond 4 h after addition (Fig. 1). Since zapA expression peaks near 8 h, it is reasonable to question whether tethering the flagella for a 4-h period is sufficient to induce expression of zapA. Increasing the concentration of α-FlaA results in an increase in the duration of tethering but also causes cell death (11), so this anomaly remains unanswered, and this possibility is currently under investigation.
Putrescine metabolism has been demonstrated to play a role in swarming motility (68). Mutation of speB, required for putrescine metabolism, delayed swarmer cell differentiation by 2 h relative to results with wild-type cells (68). These data suggest that the addition of exogenous putrescine to swimmer cells in LB broth should induce the expression of swarmer cell-dependent genes, such as zapA and hpmB. The results in Fig. 8 indicate that this is not the case. Swarmer cell differentiation and virulence gene expression are not affected by exogenously applied putrescine. One interpretation of these results is that putrescine acts independently from the status of flagellar health to control gene expression. This is not surprising, since the body of evidence thus far assembled suggests that regulation of P. mirabilis swarmer cell differentiation and ensuing swarming motility is complex and requires the input of multiple signals.
When interpreting the fliL data, one should be aware of the possibility that the Tn5 insertion may have produced polar effects on the genes downstream in the fliL operon. Polar effects would explain the loss of flagellar synthesis in the fliL mutant through the loss of export functions and may be responsible for some of the other phenotypes observed in this study. This possibility cannot be discounted; however, it is important to emphasize that Tn5 insertions in fliM, fliP, and fliQ were also analyzed in this work (Table 2), and none of these mutants has a phenotype similar to that of the fliL mutant. Indeed, it would be predicted that the phenotype of a fliM mutant should be identical to that of a fliL mutant, assuming the cryptic nature of fliL mutants from the earlier E. coli studies (63), but instead, the phenotypes of the fliL and fliM mutants are diametrically opposite one another (Table 2). So, the fliL phenotype is unlikely to be due solely to polar effects on downstream genes in the fliL operon and more likely reflects the function of fliL itself. In this context it is relevant to note that the mutation in BB2204 (fliL2204::Tn5-Cm) results in a truncated FliL protein lacking the last 10 amino acid residues, DVLFTTFILR, of the native protein. A truncated FliL protein may have altered function, leading to the phenotype observed in this study. Attempts to construct a nonpolar mutation in fliL have been unsuccessful, despite repeated tries; thus, polarity of the mutation remains an open issue.
The results show that overexpression of FliL leads to increased cell division and reduced flagellum synthesis, which in turn results in defective swimming and swarming motility, while fliL defects result in decreased cell division and reduced motility. This implicates FliL as a protein that is involved in relaying information about the nature of the environment, in this case the presence of a surface, from the flagellar filament into the cell. A model of P. mirabilis FliL function (Fig. 9) may be developed which is based on the current results and the data gathered on FliL from E. coli, S. enterica serovar Typhimurium, and C. crescentus that show a physical association between FliL and the flagellar basal body (39, 63, 81). In this model, FliL is hypothesized to sense the torque applied to the basal body and motor components when the flagellar motor stalls when faced with high-viscosity environments, or FliL may be involved in counting the rate of protons flowing through the motor. In support of a proton counting function, the addition of carbonylcyanide-p-trifluoromethoxyphenylhydrazone to LB broth prevents swimming of P. mirabilis and induces the elongated swarmer cell (11). It is also possible that overexpression of FliL may have an indirect consequence on the assembly of the flagellum. Although the current study does not distinguish between these possibilities, all three provide a function for FliL that is likely to extend beyond its role in P. mirabilis to other motile bacterial species.
It is doubtful that FliL directly interacts with DNA to control transcription; rather, FliL is more likely to be an intermediate in the surface signal transduction pathway, passing information to the flagellar master operon, FlhDC (44). FlhDC is at the apex of the flagellar hierarchy, controlling flagellar gene expression by assimilation of environmental and physiological signals, and in E. coli and related bacteria it is tightly regulated at the transcriptional (25) and translational (21) levels. Mutations in flhDC result in the loss of swimming and swarming motility in P. mirabilis (25, 31) and other swarming bacteria, such as Serratia liquefaciens (26, 33), Xenorhabdus nematophila (32, 42), and Yersinia enterocolitica (80). More significantly, the flhDC operon is strongly upregulated during P. mirabilis swarming (29) and contributes to virulence factor expression in several pathogens (4, 25, 42, 80). In addition, microarray comparisons of mRNA levels from E. coli wild-type and flhDC mutant strains have suggested that the flagellar master operon regulates several nonflagellar genes, particularly those involved in cell division (52-56, 67). Thus, the phenotype of the fliL mutation found in this study is likely a consequence of FliL directly or indirectly affecting flhDC expression. FlhDC would then modulate the expression of flagellar export and biosynthesis, as well as controlling cell division. These actions are predicted to result in the hyperelongated swarmer cell phenotype observed when fliG, fliF, or fliL is mutated. This hypothesis is currently being tested in our laboratory.
Acknowledgments
We thank the members of the Belas laboratory for thoughtful comments and suggestions regarding this work.
An award from the National Institutes of Health (DK48720) supported this research.
REFERENCES
- 1.Alavi, M., and R. Belas. 2001. Surface sensing, swarmer cell differentiation and biofilm development. Methods Enzymol. 336:29-40. [DOI] [PubMed] [Google Scholar]
- 2.Alberti, L., and R. M. Harshey. 1990. Differentiation of Serratia marcescens 274 into swimmer and swarmer cells. J. Bacteriol. 172:4322-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aldridge, P., and U. Jenal. 1999. Cell cycle-dependent degradation of a flagellar motor component requires a novel-type response regulator. Mol. Microbiol. 32:379-391. [DOI] [PubMed] [Google Scholar]
- 4.Allison, C., L. Emody, N. Coleman, and C. Hughes. 1994. The role of swarm cell differentiation and multicellular migration in the uropathogenicity of Proteus mirabilis. J. Infect. Dis. 169:1155-1158. [DOI] [PubMed] [Google Scholar]
- 5.Allison, C., and C. Hughes. 1991. Bacterial swarming: an example of prokaryotic differentiation and multicellular behaviour. Science Prog. 75:403-422. [PubMed] [Google Scholar]
- 6.Allison, C., H. C. Lai, D. Gygi, and C. Hughes. 1993. Cell differentiation of Proteus mirabilis is initiated by glutamine, a specific chemoattractant for swarming cells. Mol. Microbiol. 8:53-60. [DOI] [PubMed] [Google Scholar]
- 7.Allison, C., H. C. Lai, and C. Hughes. 1992. Co-ordinate expression of virulence genes during swarm-cell differentiation and population migration of Proteus mirabilis. Mol. Microbiol. 6:1583-1591. [DOI] [PubMed] [Google Scholar]
- 8.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. A. Smith, J. G. Seidman, and K. Struhl. 1987. Current protocols in molecular biology. Greene Publishing Associates and John Wiley & Sons, Inc., New York, N.Y.
- 9.Belas, R. 1994. Expression of multiple flagellin-encoding genes of Proteus mirabilis. J. Bacteriol. 176:7169-7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belas, R. 1996. Proteus mirabilis swarmer cell differentiation and urinary tract infection, p. 271-298. In H. Mobley and J. Warren (ed.), Urinary tract infections: molecular pathogenesis and clinical management. American Society for Microbiology, Washington, D.C.
- 11.Belas, R. 2005. Unpublished data.
- 12.Belas, R., D. Erskine, and D. Flaherty. 1991. Proteus mirabilis mutants defective in swarmer cell differentiation and multicellular behavior. J. Bacteriol. 173:6279-6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belas, R., D. Erskine, and D. Flaherty. 1991. Transposon mutagenesis in Proteus mirabilis. J. Bacteriol. 173:6289-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belas, R., M. Goldman, and K. Ashliman. 1995. Genetic analysis of Proteus mirabilis mutants defective in swarmer cell elongation. J. Bacteriol. 177:823-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belas, R., J. Manos, and R. Suvanasuthi. 2004. Proteus mirabilis ZapA metalloprotease degrades a broad spectrum of substrates, including antimicrobial peptides. Infect. Immun. 72:5159-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belas, R., R. Schneider, and M. Melch. 1998. Characterization of Proteus mirabilis precocious swarming mutants: identification of rsbA, encoding a regulator of swarming behavior. J. Bacteriol. 180:6126-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belas, R., M. Simon, and M. Silverman. 1986. Regulation of lateral flagella gene transcription in Vibrio parahaemolyticus. J. Bacteriol. 167:210-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blair, D. F., and H. C. Berg. 1990. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell 60:439-449. [DOI] [PubMed] [Google Scholar]
- 19.Chippendale, G. R., J. W. Warren, A. L. Trifillis, and H. L. Mobley. 1994. Internalization of Proteus mirabilis by human renal epithelial cells. Infect. Immun. 62:3115-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chun, S. Y., and J. S. Parkinson. 1988. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science 239:276-278. [DOI] [PubMed] [Google Scholar]
- 21.Claret, L., and C. Hughes. 2000. Rapid turnover of FlhD and FlhC, the flagellar regulon transcriptional activator proteins, during Proteus swarming. J. Bacteriol. 182:833-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coker, C., O. O. Bakare, and H. L. Mobley. 2000. H-NS is a repressor of the Proteus mirabilis urease transcriptional activator gene ureR. J. Bacteriol. 182:2649-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dean, G. E., R. M. Macnab, J. Stader, P. Matsumura, and C. Burks. 1984. Gene sequence and predicted amino acid sequence of the motA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J. Bacteriol. 159:991-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Lorenzo, M., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dufour, A., R. B. Furness, and C. Hughes. 1998. Novel genes that upregulate the Proteus mirabilis flhDC master operon controlling flagellar biogenesis and swarming. Mol. Microbiol. 29:741-751. [DOI] [PubMed] [Google Scholar]
- 26.Eberl, L., G. Christiansen, S. Molin, and M. Givskov. 1996. Differentiation of Serratia liquefaciens into swarm cells is controlled by the expression of the flhD master oepron. J. Bacteriol. 178:554-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Francis, N., V. Irikura, S. Yamaguchi, D. DeRosier, and R. Macnab. 1992. Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc. Natl. Acad. Sci. USA 89:6304-6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Francis, N. R., G. E. Sosinky, D. Thomas, and D. J. DeRosier. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235:1261-1270. [DOI] [PubMed] [Google Scholar]
- 29.Fraser, G. M., L. Claret, R. Furness, S. Gupta, and C. Hughes. 2002. Swarming-coupled expression of the Proteus mirabilis hpmBA haemolysin operon. Microbiology 148:2191-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fraser, G. M., and C. Hughes. 1999. Swarming motility. Curr. Opin. Microbiol. 2:630-635. [DOI] [PubMed] [Google Scholar]
- 31.Furness, R., G. Fraser, N. Hay, and C. Hughes. 1997. Negative feedback from a Proteus class II flagellum export defect to the flhDC master operon controlling cell division and flagellum assembly. J. Bacteriol. 179:5585-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Givaudan, A., and A. Lanois. 2000. flhDC, the flagellar master operon of Xenorhabdus nematophilus: requirement for motility, lipolysis, extracellular hemolysis, and full virulence in insects. J. Bacteriol. 182:107-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Givskov, M., L. Eberl, G. Christiansen, M. J. Benedik, and S. Molin. 1995. Induction of phospholipase and flagellar synthesis in Serratia liquefaciens is controlled by expression of the flagellar master operon flhD. Mol. Microbiol. 16:445-454. [DOI] [PubMed] [Google Scholar]
- 34.Gryllos, I., J. G. Shaw, R. Gavin, S. Merino, and J. M. Tomas. 2001. Role of flm locus in mesophilic Aeromonas species adherence. Infect. Immun. 69:65-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gygi, D., M. Bailey, C. Allison, and C. Hughes. 1995. Requirement for FlhA in flagella assembly and swarm-cell differentiation by Proteus mirabilis. Mol. Microbiol. 15:761-769. [DOI] [PubMed] [Google Scholar]
- 36.Gygi, D., G. Fraser, A. Dufour, and C. Hughes. 1997. A motile but non-swarming mutant of Proteus mirabilis lacks FlgN, a facilitator of flagella filament assembly. Mol. Microbiol. 25:597-604. [DOI] [PubMed] [Google Scholar]
- 37.Harshey, R. 1994. Bees aren't the only ones: swarming in gram-negative bacteria. Mol. Microbiol. 13:389-394. [DOI] [PubMed] [Google Scholar]
- 38.Harshey, R., and T. Matsuyama. 1994. Dimorphic transition in E. coli and S. typhimurium: surface-induced differentiation into hyperflagellate swarmer cells. Proc. Natl. Acad. Sci. USA 91:8631-8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jenal, U., J. M. White, and L. Shapiro. 1994. Caulobacter flagellar function, but not assembly, requires FliL, a non-polarly localized membrane protein present in all cell types. J. Mol. Biol. 243:227-244. [DOI] [PubMed] [Google Scholar]
- 40.Kearns, D. B., and R. Losick. 2003. Swarming motility in undomesticated Bacillus subtilis. Mol. Microbiol. 49:581-590. [DOI] [PubMed] [Google Scholar]
- 41.Khan, I. H., T. S. Reese, and S. Khan. 1992. The cytoplasmic component of the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 89:5956-5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim, D., B. Boylan, N. George, and S. Forst. 2003. Inactivation of ompR promotes precocious swarming and flhDC expression in Xenorhabdus nematophila. J. Bacteriol. 185:5290-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 44.Liu, X., and P. Matsumura. 1994. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J. Bacteriol. 176:7345-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCarter, L., M. Hilmen, and M. Silverman. 1988. Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell 54:345-351. [DOI] [PubMed] [Google Scholar]
- 46.McCarter, L., and M. Silverman. 1990. Surface-induced swarmer cell differentiation of Vibrio parahaemolyticus. Mol. Microbiol. 4:1057-1062. [DOI] [PubMed] [Google Scholar]
- 47.Minamino, T., and R. M. Macnab. 1999. Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 181:1388-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mobley, H., and R. Belas. 1995. Swarming and pathogenicity of Proteus mirabilis in the urinary tract. Trends Microbiol. 3:280-284. [DOI] [PubMed] [Google Scholar]
- 49.Mobley, H., R. Belas, V. Lockatell, G. Chippendale, A. Trifillis, D. Johnson, and J. Warren. 1996. Construction of a flagellum-negative mutant of Proteus mirabilis: effect on internalization into human renal epithelial cells and virulence in a mouse model of ascending urinary tract infection. Infect. Immun. 64:5332-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mobley, H. L., G. R. Chippendale, K. G. Swihart, and R. A. Welch. 1991. Cytotoxicity of the HpmA hemolysin and urease of Proteus mirabilis and Proteus vulgaris against cultured human renal proximal tubular epithelial cells. Infect. Immun. 59:2036-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muller, D., D. Lievremont, D. D. Simeonova, J.-C. Hubert, and M.-C. Lett. 2003. Arsenite oxidase aox genes from a metal-resistant β-proteobacterium. J. Bacteriol. 185:135-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pruss, B. M., J. W. Campbell, T. K. Van Dyk, C. Zhu, Y. Kogan, and P. Matsumura. 2003. FlhD/FlhC is a regulator of anaerobic respiration and the Enter-Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J. Bacteriol. 185:534-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pruss, B. M., X. Liu, W. Hendrickson, and P. Matsumura. 2001. FlhD/FlhC-regulated promoters analyzed by gene array and lacZ gene fusions. FEMS Microbiol. Lett. 197:91-97. [DOI] [PubMed] [Google Scholar]
- 54.Pruss, B. M., D. Markovic, and P. Matsumura. 1997. The Escherichia coli flagellar transcriptional activator flhD regulates cell division through induction of the acid response gene cadA. J. Bacteriol. 179:3818-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pruss, B. M., and P. Matsumura. 1997. Cell cycle regulation of flagellar genes. J. Bacteriol. 179:5602-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pruss, B. M., and P. Matsumura. 1996. A regulator of the flagellar regulon of Escherichia coli, flhD, also affects cell division. J. Bacteriol. 178:668-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raha, M., H. Sockett, and R. Macnab. 1994. Characterization of the fliL gene in the flagellar regulon of Escherichia coli and Salmonella typhimurium. J. Bacteriol. 176:2308-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rashid, M. H., and A. Kornberg. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rashid, M. H., N. N. Rao, and A. Kornberg. 2000. Inorganic polyphosphate is required for motility of bacterial pathogens. J. Bacteriol. 182:225-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rauprich, O., M. Matsushita, C. J. Weijer, F. Siegert, S. E. Esipov, and J. A. Shapiro. 1996. Periodic phenomena in Proteus mirabilis swarm colony development. J. Bacteriol. 178:6525-6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 62.Schmidtchen, A., I. M. Frick, E. Andersson, H. Tapper, and L. Bjorck. 2002. Proteinases of common pathogenic bacteria degrade and inactivate the antibacterial peptide LL-37. Mol. Microbiol. 46:157-168. [DOI] [PubMed] [Google Scholar]
- 63.Schoenhals, G., and R. Macnab. 1999. FliL is a membrane-associated component of the flagellar basal body of Salmonella. Microbiology 145:1769-1775. [DOI] [PubMed] [Google Scholar]
- 64.Silverman, M., and M. Simon. 1974. Flagellar rotation and the mechanism of bacterial motility. Nature 249:73-74. [DOI] [PubMed] [Google Scholar]
- 65.Simon, R., U. Priefer, and A. Puhler. 1982. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 66.Sockett, H., S. Yamaguchi, M. Kihara, V. M. Irikura, and R. M. Macnab. 1992. Molecular analysis of the flagellar switch protein FliM of Salmonella typhimurium. J. Bacteriol. 174:793-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stafford, G. P., T. Ogi, and C. Hughes. 2005. Binding and transcriptional activation of non-flagellar genes by the Escherichia coli flagellar master regulator FlhD2C2. Microbiology 151:1779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sturgill, G., and P. N. Rather. 2004. Evidence that putrescine acts as an extracellular signal required for swarming in Proteus mirabilis. Mol. Microbiol. 51:437-446. [DOI] [PubMed] [Google Scholar]
- 69.Toker, A. S., M. Kihara, and R. M. Macnab. 1996. Deletion analysis of the FliM flagellar switch protein of Salmonella typhimurium. J. Bacteriol. 178:7069-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ueno, T., K. Oosawa, and S. Aizawa. 1992. M ring, S ring and proximal rod of the flagellar basal body of Salmonella typhimurium are composed of subunits of a single protein, FliF. J. Mol. Biol. 227:672-677. [DOI] [PubMed] [Google Scholar]
- 71.Ueno, T., K. Oosawa, and S.-I. Aizawa. 1994. Domain structures of the MS ring component protein (FliF) of the flagellar basal body of i. J. Mol. Biol. 236:546-555. [DOI] [PubMed] [Google Scholar]
- 72.Uphoff, T. S., and R. A. Welch. 1990. Nucleotide sequencing of the Proteus mirabilis calcium-independent hemolysin genes (hpmA and hpmB) reveals sequence similarity with the Serratia marcescens hemolysin genes (shlA and shlB). J. Bacteriol. 172:1206-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Walker, K. E., S. Moghaddame-Jafari, C. V. Lockatell, D. Johnson, and R. Belas. 1999. ZapA, the IgA-degrading metalloprotease of Proteus mirabilis, is a virulence factor expressed specifically in swarmer cells. Mol. Microbiol. 32:825-836. [DOI] [PubMed] [Google Scholar]
- 74.Wang, Q., J. G. Frye, M. McClelland, and R. M. Harshey. 2004. Gene expression patterns during swarming in Salmonella typhimurium: genes specific to surface growth and putative new motility and pathogenicity genes. Mol. Microbiol. 52:169-187. [DOI] [PubMed] [Google Scholar]
- 75.Wang, Q., A. Suzuki, S. Mariconda, S. Porwollik, and R. M. Harshey. 2005. Sensing wetness: a new role for the bacterial flagellum. EMBO J. 24:2034-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Williams, F. D., and R. H. Schwarzhoff. 1978. Nature of the swarming phenomenon in Proteus. Annu. Rev. Microbiol. 32:101-122. [DOI] [PubMed] [Google Scholar]
- 77.Winson, M. K., S. Swift, L. Fish, J. P. Throup, F. Jorgensen, S. R. Chhabra, B. W. Bycroft, P. Williams, and G. S. A. B. Stewart. 1998. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 163:185-192. [DOI] [PubMed] [Google Scholar]
- 78.Winson, M. K., S. Swift, P. J. Hill, C. M. Sims, G. Griesmayr, B. W. Bycroft, P. Williams, and G. S. Stewart. 1998. Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini-Tn5 constructs. FEMS Microbiol. Lett. 163:193-202. [DOI] [PubMed] [Google Scholar]
- 79.Yamaguchi, S., S. Aizawa, M. Kihara, M. Isomura, C. J. Jones, and R. M. Macnab. 1986. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J. Bacteriol. 168:1172-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Young, G. M., M. J. Smith, S. A. Minnich, and V. L. Miller. 1999. The Yersinia enterocolitica motility master regulatory operon, flhDC, is required for flagellin production, swimming motility, and swarming motility. J. Bacteriol. 181:2823-2833.10217774 [Google Scholar]
- 81.Yu, J., and L. Shapiro. 1992. Early Caulobacter crescentus genes fliL and fliM are required for flagellar gene expression and normal cell division. J. Bacteriol. 174:3327-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]