Abstract
GGDEF domain-containing proteins have been implicated in bacterial signal transduction and synthesis of the second messenger molecule cyclic-di-GMP. A number of GGDEF proteins are involved in controlling the formation of extracellular matrices. AdrA (Salmonella enterica serovar Typhimurium) and HmsT (Yersinia pestis) contain GGDEF domains and are required for extracellular cellulose production and biofilm formation, respectively. Here we show that hmsT is able to restore cellulose synthesis to a Salmonella serovar Typhimurium adrA mutant and that adrA can replace hmsT in Y. pestis Hms-dependent biofilm formation. Like Y. pestis HmsT overproducers, Y. pestis cells carrying adrA under the control of an arabinose-inducible promoter produced substantial biofilms in the presence of arabinose. Finally, we demonstrate that HmsT is involved in the synthesis of cyclic di-GMP.
One of the most intriguing findings of large-scale sequencing of bacterial genomes was the discovery of highly abundant novel protein domain superfamilies. Two of the most prominent ones are the GGDEF and EAL domain families (12), which had 1,601 and 1,016 members, respectively, in the Pfam protein family database in July 2005. Proteins containing those domains are involved in motility and exopolysaccharide-biofilm production in a variety of bacteria (8, 22). Some GGDEF-domain proteins control biofilm formation and/or cell aggregation by enhancing the levels of the novel second messenger cyclic di(3′→5′)-guanylic acid (c-di-GMP) (38, 42).
The rdar (red, dry, and rough) morphotype, a multicellular behavior of Salmonella enterica serovar Typhimurium, produces the extracellular matrix (ECM) components cellulose and curli fimbriae (35, 45), both of which mediate different forms of biofilm formation and cell aggregation. When only cellulose is produced, the cells express a pdar (pink, dry, and rough) morphotype on Congo red (CR) agar plates and bind the dye Calcofluor, which leads to a fluorescent phenotype when stimulated with 366-nm light. Cellulose production requires the constitutively expressed bcsABZC operon, which contains the structural genes required for cellulose biosynthesis (45), among them bcsA, encoding the catalytic subunit of the cellulose synthase. Cellulose production is activated by the GGDEF-domain protein AdrA via production of the allosteric activator c-di-GMP (36, 38). Once AdrA is expressed no upstream components of the regulatory network of the rdar morphotype are required for cellulose production (45). Thus, although cellulose production is observed only at 28°C in wild-type Salmonella serovar Typhimurium ATCC14028 due to the temperature-regulated expression of the upstream regulator CsgD, temperature-independent expression of CsgD or expression of AdrA from a plasmid leads to temperature-independent cellulose production (35, 38).
The hmsHFRS operon, encoded within the 102-kb pgm locus, and hmsT are required for the Hms+ phenotype of Yersinia pestis-defined as the formation of greenish-brown or red “pigmented” colonies on hemin and CR plates, respectively. Cells grown at 35°C or higher do not exhibit this binding phenomenon and form white colonies on CR plates, unless they contain increased copy numbers of hmsHFRS and/or hmsT (20, 23, 24, 26, 29, 30, 32, 33, 40). We and others have recently shown that the Hms system is required for the temperature-dependent formation of a biofilm (6, 21, 24) that is probably responsible for the spread of plague from fleas to mammals. Earlier studies showed that Hms+ but not Hms− strains of Y. pestis were able to block the flea proventriculus, the valve which separates the esophagus from the midgut of the flea. Efficient transmission of plague from fleas to mammals is dependent upon blockage of the flea by biofilm formation (17, 21, 25, 34).
Temperature-dependent expression of the Hms phenotype is probably controlled by the degradation of select Hms proteins. The levels of HmsH, HmsR, and HmsT are significantly lower at 37°C than at 26°C (31). HmsH is an outer membrane protein of unknown function. HmsR possesses a glycosyltransferase domain and is likely an important component of the biofilm synthase. HmsT, like AdrA, belongs to the family of GGDEF proteins, and cells which express high levels of HmsT produce extensive biofilms (3, 12, 23, 24, 26).
Formation of the Y. pestis biofilm appears to be controlled by the actions of two gene products, HmsT and HmsP. HmsP was identified in a screening for mutants able to form red colonies on CR agar at 37°C. The EAL domain of HmsP has phosphodiesterase activity and is probably involved in the degradation of c-di-GMP. Y. pestis strains with mutations in hmsP produce robust biofilms. Thus, synthesis of the biofilm in Y. pestis may be controlled by the levels of c-di-GMP (5, 24, 31).
Here we show that the GGDEF domain-containing proteins of Salmonella serovar Typhimurium (AdrA) and Y. pestis (HmsT) are somewhat interchangeable. Thus, hmsT is able to restore cellulose synthesis to a Salmonella serovar Typhimurium adrA mutant. Similarly, adrA replaces the function of hmsT in Y. pestis Hms-dependent biofilm formation. Like Y. pestis HmsT overproducers, Y. pestis cells carrying adrA under the control of an arabinose-inducible promoter produced substantial biofilms in the presence of arabinose. Finally, we demonstrate that Y. pestis cells expressing hmsT from multicopy plasmids increased the levels of cellular c-di-GMP compared to cells of an hmsT mutant strain.
MATERIALS AND METHODS
Bacterial strains and cultivation.
All bacterial strains used in this study are listed in Table 1. Y. pestis cells were streaked onto CR plates from buffered glycerol stocks stored at −80°C and incubated at 26 to 30°C for 48 h. Where appropriate, ampicillin (Ap; 100 μg/ml), tetracycline (Tc; 6.25 μg/ml), chloramphenicol (Cm; 20 to 30 μg/ml), or 0.2% (wt/vol) arabinose was added to cultures. Individual colonies were inoculated onto Tryptose blood agar base (TBA; Difco Laboratories) slants and incubated at 26 to 30°C for 24 to 48 h. Cells were washed off the slants with heart infusion broth (Difco Laboratories) or a defined medium, PMH2 (14), and grown overnight with aeration in heart infusion broth or PMH2 at 26 or 37°C. Cell growth was monitored on a Spectronic Genesys5 spectrophotometer at 620 nm. CR plates (40) were also used for initial characterization of the Hms phenotype of recombinant Y. pestis strains. CR binding has been correlated with exopolysaccharide production and with biofilm formation in Y. pestis and other organisms (16, 24, 39, 44). Salmonella serovar Typhimurium was grown on plates of LB medium agar without salt as described previously (35), whereby CR or Calcofluor was added when appropriate.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| Salmonella serovar Typhimurium | ||
| MAE52 | PagfD101; rdar28/37 | 36 |
| MAE 103 | ΔcsgBA102 adrA101::MudJ; white | 36 |
| MAE 190 | ΔcsgBA102 bcsA101::MudJ; white | 45 |
| Y. pestisa | ||
| KIM6+ | Pgm+ Hms+ Pla+ | 11 |
| KIM6-2051+ | Kmr Hms− (hmsT2051::mini-kan) Pla+ | 23 |
| Plasmids | ||
| pAHMS14 | 6.9 kb, Tcr, hmsT+; 3.3-kb EcoRI-PstI fragment from pAHMS8 ligated into pBR322 | 23 |
| pAHMS16 | 4.4 kb, Apr, hmsT+; 1.9-kb kpnI-FspI fragment from pAHMS14 ligated into pUC18 | 23 |
| pAHMS16.1 | 5.6 kb, Tcr, hmsT+; 1.9-kb KpnI-FspI fragment from pAHMS14 ligated into pBR322 | 23 |
| pAHMS16.3 | 10.1 kb, Apr, hmsT+; EcoRI linkers added to 1.9-kb KpnI-FspI fragment from pAHMS16 and ligated into pLC8.2 | 23 |
| pBAD30 | 4.9 kb, Apr, araC+, araBAD promoter upstream of multiple cloning site | 15 |
| pBADHmsT6xH | 6.1 kb, Apr, arabinose-inducible hmsT with C-terminal His tag; 1.2-kb SmaI-EcoRI PCR product from pAHMS16 ligated into pBAD30 | This study |
| pGFPmut3.1 | 3.3 kb, Apr; GFP expression vector | Clontech |
| pGFPmut3.1cat | 3.5 kb, Cmr; modified GFP expression vector | This study |
| pLC8.2 | 8.2 kb, Apr; single-copy-number cloning vector derived from pLC682 | 23 |
| pWJB30 | 6.0 kb, Apr, arabinose-inducible adrA with C-terminal His tag | 38 |
All Y. pestis strains are avirulent due to lack of the low-calcium response plasmid pCD1. Strains with a plus sign possess an intact 102-kb pgm locus containing the genes for the hemin storage (hms) and yersiniabactin (Ybt) iron transport systems: All other Y. pestis strains have either a pgm deletion or a mutation within the pgm locus. Abbreviations: Apr, Cmr, and Tcr, resistance to ampicillin, chloramphenicol, and tetracycline, respectively.
Plasmids, protein, and recombinant DNA techniques.
All the plasmids used in this study are listed in Table 1. Plasmids were purified from overnight cultures by alkaline lysis (4) or with a Qiaprep spin Miniprep kit (QIAGEN) and further purified when necessary by polyethylene glycol precipitation (18). Y. pestis and Salmonella serovar Typhimurium cells were transformed by electroporation as previously described (10, 35). To construct an arabinose-inducible HmsT with a C-terminal His tag, the hmsT sequence was amplified from pAHMS16 by use of Platinum Pfx DNA polymerase (Invitrogen) and primers HmsT-His1 (5′-CGGAATTCTAAGGAGGTTTCTAATGCAGAGTAAATTGAATATG-3′) and HmsT-His2 (5′-TCCCCCGGGTCAATGATGATGATGATGA TGATGATGAGGGGAAGACTGTACATTTG-3′). After digestion with SmaI and EcoRI, the PCR products were cloned into the corresponding sites of pBAD30 and transformed into KIM6-2051+ (hmsT2051::mini-kan). Plasmids were isolated from colonies that were red on CR plates containing 0.2% arabinose. For one plasmid, designated pBADHmsT6xH, sequencing by Elim Biopharmaceuticals confirmed that the insert contained no PCR errors.
Western blot analysis of HmsT-6xH and AdrA-6xH protein levels after expression in Y. pestis KIM6-2051+ by use of antibodies against the His tag (BD Biosciences) was performed as previously described (24).
Crystal violet staining assays.
Cells attached to glass test tubes were detected with crystal violet staining essentially as described by O'Toole et al. (27). Briefly, Y. pestis cells grown overnight on TBA slants were used to inoculate PMH2, containing either 0.2% (wt/vol) glucose or arabinose, to an optical density at 620 nm of 0.1 and grown in test tubes for 16 to 18 h in a shaking water bath at 26°C or 37°C. The cultures were incubated with 0.01% crystal violet for 15 to 20 min. The liquid culture was discarded, and test tubes containing stained, attached cells were washed three times with water. The retained crystal violet was solubilized with a mixture of 80% ethanol and 20% acetone. The amount of dye bound, representing the mass of attached bacterial cells, was monitored by measuring the absorbance at 570 nm on a Spectronic Genesys5 spectrophotometer. S. enterica serovar Typhimurium biofilms were similarly processed except that 100% dimethyl sulfoxide was used to dissolve the crystal violet. All results shown are the averages of two or more independent experiments.
Confocal laser scanning microscopy.
Y. pestis strains containing either pGFPmut3.1 or pGFPmut3.1cat were grown overnight on TBA slants and used to inoculate PMH2, containing either 0.2% glucose or 0.2% arabinose, to an optical density at 620 nm of 0.1. Five milliliters of each culture was placed into a 50-ml conical tube containing a glass coverslip and incubated overnight at 26°C or 37°C in a shaking water bath. The growth rates and final yields of Hms+ and Hms− cells are equivalent in PMH2. The coverslips from three independent experiments were rinsed well with distilled water and examined with a Leica TCS laser scanning confocal microscope system. The samples were viewed with a 63X1.2 HCX PL APO objective on a Leica DM RXE microscope equipped with an argon laser emitting at 488 nm.
Detection of c-di-GMP.
c-di-GMP was detected as described by Simm et al. (38) using modified running conditions for high-performance liquid chromatography (HPLC) as stated below. Briefly, 300 mg of cells were harvested from two independently grown cultures. For Salmonella serovar Typhimurium, cells were harvested from high-salt agar plates after growth for 20 h at 28°C whereas Y. pestis cells were from shaken, liquid cultures grown overnight at 26°C. Nucleotides were extracted by heat as described previously (2, 38). Afterwards, the extract was lyophilized, resuspended in 500 μl of water, and filtered (0.2 μm pore size). Extracts equivalent to 10 mg (wet weight) of cells were subjected to HPLC separation on a 250- by 4.6-mm reverse-phase column (Hypersil ODS 5u; Hypersil-Keystone) at room temperature and detected at 260 and 280 nm on an Äktabasic apparatus (Amersham-Pharmacia). Runs were carried out in 0.10 M triethyl ammonium acetate buffer (pH 6.0) at 1 ml/min, using a multi-step gradient of acetonitrile. Relevant fractions of 1 ml were collected, lyophilized, and resuspended in 10 μl of water.
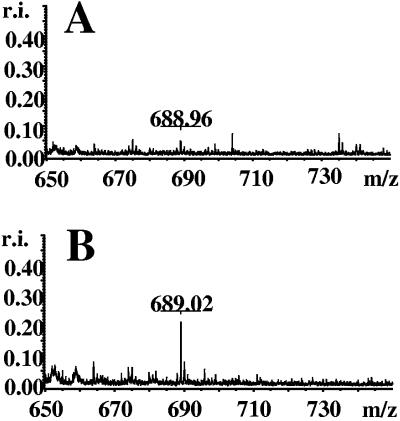
Relevant fractions collected by HPLC (0.5-μl aliquots, or appropriate dilutions thereof) were applied to an anchor chip mixed plate by the fast evaporation method (matrix, α-cyano 4-hydroxycinnamic acid) and allowed to dry. Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis was performed on a Bruker Reflex II (Bruker-Franzen-Analytik) mass spectrometer using the negative-ion mode.
RESULTS
Complementation of Y. pestis hmsT2051::mini-kan.
The plasmid pBAD30 (vector) or pWJB30 (adrA+) was electroporated into Y. pestis KIM6-2051+, which has a mini-kan insert that disrupts hmsT (23). Recombinant and control strains were streaked onto CR plates in the presence or absence of arabinose and incubated at 26°C. KIM6-2051(pWJB30)+ but not KIM6-2051(pBAD30)+ formed red colonies on CR plates containing arabinose at 26°C. Both strains formed white colonies at 26°C on CR agar that lacked arabinose. This result indicates that Salmonella serovar Typhimurium AdrA can substitute for HmsT in Y. pestis Hms-dependent biofilm formation.
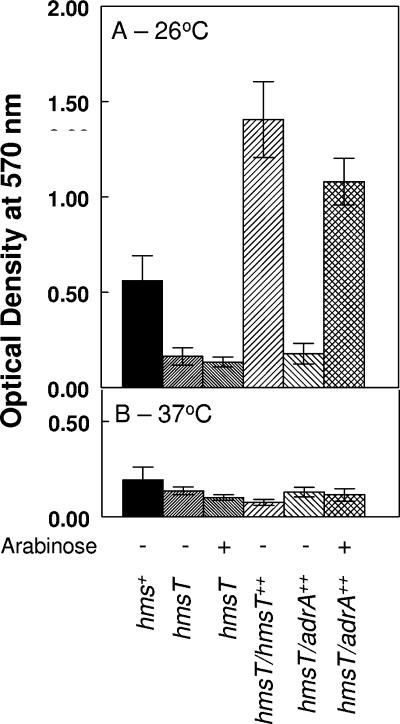
As a quantitative assay for biofilm formation, a crystal violet assay was performed on cells grown overnight at 26 and 37°C in PMH2 with glucose or arabinose (Fig. 1). This assay has been used to assess the bacterial mass attached to a surface as a measure of biofilm formation in a variety of bacterial species (27). At 26°C, very few if any cells of the mutant lacking hmsT (KIM6-2051+) bound to the glass test tube compared to its hms+ parent, KIM6+. As previously described, KIM6-2051(pAHMS14)+, which overexpresses HmsT, showed enhanced attachment compared to KIM6+ (Fig. 1) (24). When grown at 26°C with arabinose but not with glucose, Y. pestis strains expressing adrA under the control of an arabinose-inducible promoter attached to the glass surface almost as well as the HmsT-overexpressing strain. Growth in the presence of arabinose had no effect on biofilm production by KIM6-2051+. At 37°C, there were no significant differences in the low levels of crystal violet staining among the Y. pestis derivatives (Fig. 1). These results provide further evidence that AdrA can substitute for HmsT in Y. pestis biofilm formation.
FIG. 1.
Crystal violet staining of Y. pestis cells attached to borosilicate glass test tubes after growth at 26°C (A) or 37°C (B) in media containing either glucose or arabinose. After staining with crystal violet was performed, the dye was solubilized with ethanol-acetone and optical densities were measured at 570 nm. KIM6+, hms+; KIM6-2051(pBAD30)+, hmsT; KIM6-2051(pAHMS14)+, hmsT/hmsT++; KIM6-2051(pWJB30)+, hmsT/adrA++. Values are averages from two or more independent experiments, with error bars indicating standard deviations.
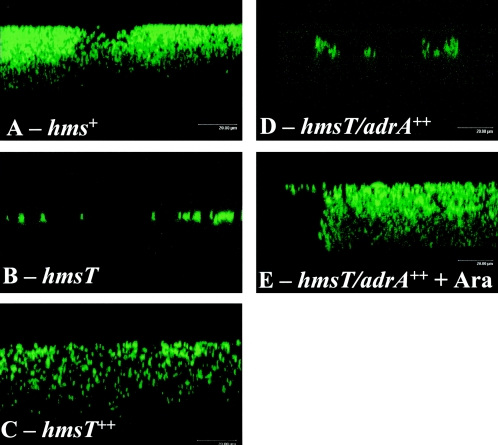
Cells in Hms-dependent biofilms were visualized using confocal laser scanning microscopy. Cells expressing green fluorescent protein were incubated overnight at 26°C in a shaking water bath in medium containing a glass coverslip. The structures which remained attached to the coverslip after washing are shown in Fig. 2. As previously observed (24), KIM6+ cells formed a structure that resembles a biofilm (Fig. 2A) while only a few single cells of KIM6-2051+ (hmsT2051::mini-kan) remained bound to the coverslip (Fig. 2B). In contrast, the KIM6-2051+ strain complemented with hmsT or adrA produced a biofilm (Fig. 2C and 2E). As expected, arabinose was required for biofilm formation in strains carrying adrA under the control of the araBAD promoter (Fig. 2D and 2E). While this method visualizes bacterial cells and does not accurately depict the extent of the extracellular matrix surrounding them, it is clear that Salmonella serovar Typhimurium AdrA can substitute for Y. pestis HmsT in controlling biofilm formation in Y. pestis.
FIG. 2.
Confocal laser scanning microscopy images of green fluorescent protein-expressing Y. pestis KIM6+ (hms+), KIM6-2051+ (hmsT), KIM6-2051(pAHMS14)+ (hmsT++), and KIM6-2051(pWJB30)+ (hmsT/adrA++) cells attached to a glass coverslip after overnight growth at 26°C in the absence (A to D) or presence (E) of arabinose. Representative areas from one of three independent replicate studies are shown in the xzy plane.
Complementation of a Salmonella serovar Typhimurium adrA mutant with HmsT.
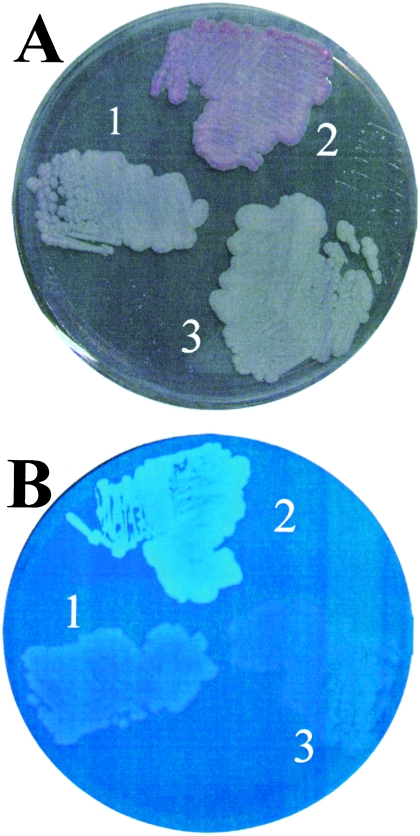
Since Salmonella serovar Typhimurium AdrA can substitute for HmsT in Y. pestis for biofilm formation, the complementation of an adrA mutant of Salmonella serovar Typhimurium by HmsT was assessed. The hmsT gene, under the control of its own promoter on low-, medium-, or high-copy-number plasmids pAHMS16.3, pAHMS16.1, and pAHMS16, was electroporated into Salmonella serovar Typhimurium MAE103. In all instances, cellulose production was stimulated at 28°C, as observed by the appearance of pink, dry, and rough colonies (pdar) on CR agar plates and fluorescent colonies on Calcofluor plates (Fig. 3) and the intensity of the phenotype increased with the copy number of the plasmid (data not shown). In the case of the medium- and high-copy-number plasmids pAHMS16.1 and pAHMS16, stimulation of cellulose production was also observed at 37°C. The change in colony morphology was due to cellulose biosynthesis, since Salmonella serovar Typhimurium MAE190, in which bcsA encoding the catalytic subunit of the cellulose synthase was mutated, showed neither the pdar morphotype nor fluorescence upon expression of HmsT (Fig. 3).
FIG. 3.
Cellulose expression by Salmonella serovar Typhimurium MAE103 (ΔadrA csgBA) containing HmsT. Cellulose expression was observed on CR plates (A) and Calcofluor plates (B). 1, Salmonella serovar Typhimurium MAE103 (pLC8.2; HmsT−); 2, Salmonella serovar Typhimurium MAE103 (pAHMS16.3; HmsT+); 3, Salmonella serovar Typhimurium MAE190 (pAHMS16.3; HmsT+).
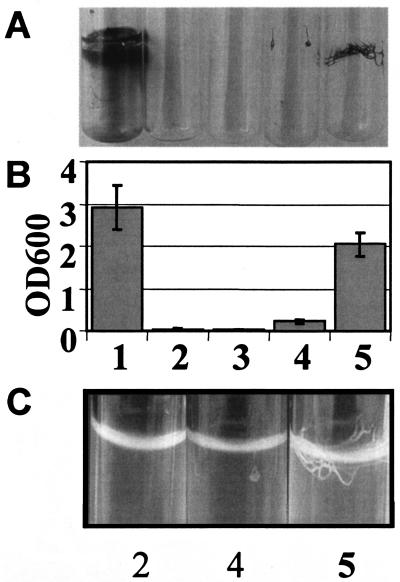
Stimulation of cellulose production by HmsT was observed not only in LB without salt medium but also under high-salt conditions as well as on M9 minimal medium agar plates. Cellulose production was more pronounced on high-salt LB and M9 minimal medium than on LB medium without salt, and the HmsT-mediated phenotype was not temperature regulated when expressed from medium- to high-copy-number plasmids (data not shown). When biofilm production and cell aggregation of Salmonella serovar Typhimurium mediated by HmsT were assessed in liquid culture, a temperature-independent phenotype was observed (Fig. 4 and data not shown). While the results of crystal violet staining (Fig. 4A and 4B) confirm that HmsT can complement the Salmonella serovar Typhimurium adrA mutant, the extent of biofilm formation is not entirely reflected by the spatial distribution of cells. Fig. 4C shows that expression of hmsT from a higher-copy-number plasmid causes the production of thick cell bundles originating from cells attached to the wall of the well at the air-liquid interface. Consequently, there is a high biomass spatially concentrated, visually giving the impression of a minor biofilm (Fig. 4A). However, quantification of the crystal violet staining revealed a biofilm mass equal to that of the positive control (Fig. 4B).
FIG. 4.
Biofilm phenotype of a Salmonella serovar Typhimurium ΔadrA mutant expressing HmsT. The pattern (A), quantification (B), and structure (C) of adherence of Salmonella serovar Typhimurium MAE103 (adrA csgBA) to glass are shown. Growth was in M9 minimal medium at 37°C with shaking for 24 h. Note the thick fibrous biofilm produced by Salmonella serovar Typhimurium MAE103 complemented with HmsT on a multiple-copy-number plasmid (panel C, lane 5), which leads to enhanced crystal violet binding comparable to that of the positive control (panel B, lane 1). Lanes: 1, Salmonella serovar Typhimurium MAE52 (pBAD30), positive control; 2, Salmonella serovar Typhimurium MAE103 (pLC8.2); 3. Salmonella serovar Typhimurium MAE190 (pAHMS16.3; HmsT+); 4, Salmonella serovar Typhimurium MAE103 (pAHMS16.3; HmsT+); 5, Salmonella serovar Typhimurium MAE103 (pAHMS16.1; HmsT+). Values are averages from two independent experiments, with error bars indicating standard deviations.
HmsT enhances c-di-GMP production in vivo.
Recently, GGDEF-domain proteins, including AdrA, were shown to mediate diguanylate cyclase activity in vivo or in vitro (28, 37, 38, 41, 42). Thus we investigated the ability of HmsT to stimulate c-di-GMP production in vivo. Salmonella serovar Typhimurium cells were grown on high-salt LB medium at 28°C, conditions under which cellulose production was most pronounced. However, even with improved running conditions for HPLC, significantly enhanced c-di-GMP levels could only be observed by MALDI-TOF analysis and only when HmsT was expressed from a high-copy-number plasmid (pAHMS16) but not from the low-copy-number plasmid (pAHMS16.3) (Fig. 5 and data not shown). Even with the high-copy-number plasmid, the c-di-GMP concentration was much lower than that achieved by AdrA overexpression (38). However, HPLC analysis allowed detection and quantitation of the low levels of c-di-GMP produced by HmsT in Y. pestis that are higher than levels detected in the uncomplemented Y. pestis hmsT mutant (Fig. 6 and data not shown). Despite this low level of c-di-GMP attributable to HmsT, HmsT replaces AdrA in a Salmonella serovar Typhimurium adrA mutant when expressed from low-, medium-, and high-copy-number plasmids.
FIG. 5.
Detection of c-di-GMP. MALDI-TOF analysis of relevant HPLC fractions run in the negative-ion detection mode. c-di-GMP is detected at a mass-to-charge ratio (m/z) of 689 [M-H]. Fractions were derived from Salmonella serovar Typhimurium MAE103 (pLC8.2) (control) (A) and Salmonella serovar Typhimurium MAE103 (pAHMS16, HmsT+) (B). r.i., relative intensity.
FIG. 6.
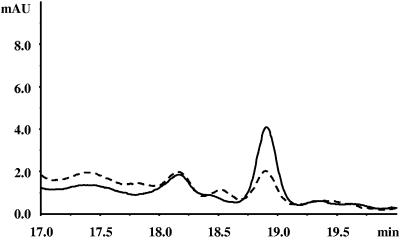
Detection of c-di-GMP in nucleotide extracts from Y. pestis by HPLC analysis. An overlay of the A254 curves from reverse-phase HPLC analysis of extracts from Y. pestis KIM6-2051(pBR322)+ and Y. pestis KIM6-2051(pAHMS16.1)+ grown in PMH2 medium at 26°C overnight in a shaken culture is shown. The peak at 18.9 min represents c-di-GMP, as determined from its correspondence to the retention time of synthetic c-di-GMP and by MALDI-TOF analysis (data not shown). This representative run used 10 times more sample than usual for the determination of cyclic di-GMP. Vector control, dashed line; HmsT+, solid line.
Since expression levels of HmsT (native Y. pestis promoter) and AdrA (arabinose-inducible promoter) could help account for the large observed differences in cellular c-di-GMP levels, we constructed pBADHmsT6xH. This expression vector has the same promoter and ribosome binding site as that of the AdrA expression vector, pWJB30. Both proteins have C-terminal His tags, which were used to detect protein levels with an anti-His-tag antiserum. Despite the similarities in these expression vectors, Western blot analysis showed much lower levels of HmsT compared to AdrA after expression in Y. pestis. There are no obvious reasons why expression levels of these proteins or their anti-His-tag antibody affinities should differ. One possibility is that HmsT is inherently more susceptible to proteolytic degradation even when expressed in Y. pestis.
Cellular levels of c-di-GMP were also much lower in Y. pestis KIM6-2051+ cells complemented with pBADHmsT6xH compared to those with pWJB30 (Table 2). However, the low levels of c-di-GMP in KIM6-2051(pBADHmsT6xH)+ were significantly higher than those in the uncomplemented strain (Table 2). Thus, in vivo c-di-GMP synthesis increases with HmsT expression.
TABLE 2.
c-di-GMP concentration for Y. pestis cells containing plasmids encoding the GGDEF domain containing proteins HmsT and AdrA
| Strain | [c-di-GMP] (pmol/mg of cells) |
|---|---|
| Y. pestis KIM6-2051(pBAD30)+ (HmsT−) | 0.031 ± 0.005 |
| Y. pestis KIM6-2051(pBADHmsT6xH)+ (HmsT+) | 0.082 ± 0.022 |
| Y. pestis KIM6-2051(pWJB30)+ (AdrA+) | 35.0 ± 17 |
DISCUSSION
In this study we have demonstrated that the GGDEF-domain proteins HmsT of Y. pestis and AdrA of Salmonella serovar Typhimurium serve similar functions and can restore expression of the heterologous extracellular matrixes—cellulose production in Salmonella serovar Typhimurium and biofilm formation in Y. pestis. Y. pestis does not encode enzymes essential for cellulose synthesis; instead, the Hms proteins show similarities (83 to 34%) to the PGA proteins of E. coli which synthesize a poly-β-1,6-N-acetyl-d-glucosamine (9, 23, 43). Recently, an enzyme which specifically degrades this polysaccharide linkage was shown to prevent Y. pestis biofilm formation but not to disperse a preformed biofilm (19). Overexpression of AdrA restored biofilm formation in a Y. pestis hmsT mutant as assessed by CR binding, a crystal violet attachment assay, and confocal laser scanning microscopy. Conversely, HmsT expressed from its native promoter complemented a Salmonella serovar Typhimurium adrA mutant, restoring CR and Calcofluor binding as well as attachment as measured by the crystal violet assay. Here the level of complementation was dependent upon the copy number of the recombinant plasmid. Complementation of biofilm formation in a Y. pestis hmsT mutant was temperature dependent, occurring only at 26°C. This corresponds to the normal temperature dependency of Y. pestis biofilm formation, which naturally occurs at 26 to 34°C but not at 37°C. It is unlikely that reduced levels of HmsH and HmsR at 37°C (24 31) account for the inability of strains expressing AdrA to form biofilms, since overexpression of HmsT confers a CR-binding phenotype to Y. pestis cells grown at 37°C. Perhaps there is increased turnover of AdrA or its product, c-di-GMP, in Y. pestis at 37°C. In Salmonella serovar Typhimurium, HmsT complemented an adrA mutation in a temperature-dependent manner but only when expressed from a low-copy-number plasmid. When expressed from higher-copy-number plasmids, HmsT restored cellulose biosynthesis at both 26°C and 37°C.
We have demonstrated the ability of HmsT to function in Salmonella serovar Typhimurium and AdrA to function in Y. pestis. The ability of GGDEF-domain-containing proteins to function in heterologous systems has been demonstrated in other bacteria. For example, CelR2 (Rhizobium leguminosarum), DGC1 (Gluconacetobacter xylinus), and “YhcK” (Escherichia coli; likely YcdT of E. coli MG1655) complemented cellulose synthesis defects in R. leguminosarum and enhanced cellulose production in Agrobacterium tumefaciens (8). Similarly, the GGDEF domain of Pseudomonas fluorescens WspR functionally replaces the GGDEF domain of Caulobacter crescentus PleD (1) and expression of Salmonella serovar Typhimurium AdrA or a constitutively active P. fluorescens WspR restores biofilm formation in Pseudomonas aeruginosa (7, 38). Indeed, individual overexpression of four other GGDEF-domain proteins of Salmonella serovar Typhimurium was recently shown to complement a Salmonella serovar Typhimurium adrA mutant in cellulose biosynthesis; one of these, STM4551 (GcpB), appears to be an orthologue of HmsT (13). Y. pestis KIM strains do not possess an apparent orthologue of AdrA (9, 23).
Proteins containing GGDEF domains are now commonly assumed to synthesize c-di-GMP from two molecules of GTP. This has been demonstrated in vitro or in vivo for an increasing number of proteins, including DGC1-3 of G. xylinus, PleD of C. crescentus, and AdrA of Salmonella serovar Typhimurium, and recently for proteins randomly chosen from six species from various positions in the phylogenetic tree (28, 37, 38, 41, 42). Here we have presented in vivo evidence that expression of HmsT increases cellular levels of c-di-GMP, suggesting it has diguanylate cyclase activity. This supports previous findings that individual substitutions in the GGEE residues of HmsT cause a loss of biofilm formation (24). Thus, a functional HmsT protein likely needs diguanylate cyclase activity to stimulate biofilm synthesis.
The observations that proteins containing functional GGDEF domains likely synthesize c-di-GMP and that many bacterial genomes possess multiple open reading frames (ORFs) with putative GGDEF domains raises several interesting questions. One of these concerns the apparent specificity of some GGDEF-domain proteins. While the Salmonella serovar Typhimurium genome has 12 ORFs with GGDEF domains, none of these chromosomal genes compensate for mutations in adrA (13, 36, 38). Similarly, none of the other five ORFs with GGDEF domains in the Y. pestis KIM10+ genome can substitute for hmsT in biofilm formation (9, 23), although a basal c-di-GMP level is present which is only approximately two- to threefold enhanced when HmsT is overexpressed (Table 2). Given the large difference in cellular levels of c-di-GMP associated with AdrA versus HmsT, it is clear the c-di-GMP levels are not the only controlling factor in plague biofilm synthesis. One possibility is that protein-protein interactions between HmsT and its target enzyme (perhaps HmsR) compensate for the observed lower c-di-GMP levels. Additional experimentation will be required to determine whether protein-protein interactions are occurring and to demonstrate the in vitro diguanylate cyclase activities of HmsT and AdrA.
Acknowledgments
R.S. and U.R. thank Mats Andersson for generous access to the HPLC apparatus. J.D.F. and R.D.P. thank Alex Bobrov and Olga Kirillina for preparation of Y. pestis cell extracts. Studies by J.D.F. and R.D.P. are supported by Public Health Service grant AI25098 from the U.S. National Institutes of Health. The work of U.R. was supported by an “Elitforskartjänst” from the Karolinska Institutet.
REFERENCES
- 1.Aldridge, P., R. Paul, P. Goymer, P. Rainey, and U. Jenal. 2003. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol. Microbiol. 47:1695-1708. [DOI] [PubMed] [Google Scholar]
- 2.Amikam, D., O. Steinberger, T. Shkolnik, and Z. Ben-Ishai. 1995. The novel cyclic dinucleotide 3′-5′ cyclic diguanylic acid binds to p21ras and enhances DNA synthesis but not cell replication in the Molt 4 cell line. Biochem. J. 311:921-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausmees, N., R. Mayer, H. Weinhouse, G. Volman, D. Amikam, M. Benziman, and M. Lindberg. 2001. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol. Lett. 204:163-167. [DOI] [PubMed] [Google Scholar]
- 4.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bobrov, A. G., O. Kirillina, and R. D. Perry. 2005. The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol. Lett. 247:123-130. [DOI] [PubMed] [Google Scholar]
- 6.Darby, C., J. W. Hsu, N. Ghori, and S. Falkow. 2002. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417:243-244. [DOI] [PubMed] [Google Scholar]
- 7.D'Argenio, D. A., M. W. Calfee, P. B. Rainey, and E. C. Pesci. 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol. 184:6481-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Argenio, D. A., and S. I. Miller. 2004. Cyclic di-GMP as a bacterial second messenger. Microbiology 150:2497-2502. [DOI] [PubMed] [Google Scholar]
- 9.Deng, W., V. Burland, G. Plunkett III, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome Sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fetherston, J. D., J. W. Lillard, Jr., and R. D. Perry. 1995. Analysis of the pesticin receptor from Yersinia pestis: role in iron-deficient growth and possible regulation by its siderophore. J. Bacteriol. 177:1824-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fetherston, J. D., P. Schuetze, and R. D. Perry. 1992. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol. Microbiol. 6:2693-2704. [DOI] [PubMed] [Google Scholar]
- 12.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 13.García, B., C. Latasa, C. Solano, F. Garcia-del Portillo, C. Gamazo, and I. Lasa. 2004. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol. Microbiol. 54:264-277. [DOI] [PubMed] [Google Scholar]
- 14.Gong, S., S. W. Bearden, V. A. Geoffroy, J. D. Fetherston, and R. D. Perry. 2001. Characterization of the Yersinia pestis Yfu ABC iron transport system. Infect. Immun. 67:2829-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartzell, P. L., J. Millstein, and C. LaPaglia. 1999. Biofilm formation in hyperthermophilic Archaea. Methods Enzymol. 310:335-349. [DOI] [PubMed] [Google Scholar]
- 17.Hinnebusch, B. J., R. D. Perry, and T. G. Schwan. 1996. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273:367-370. [DOI] [PubMed] [Google Scholar]
- 18.Humphreys, G. O., G. A. Willshaw, and E. S. Anderson. 1975. A simple method for the preparation of large quantities of pure plasmid DNA. Biochim. Biophys. Acta 383:457-463. [DOI] [PubMed] [Google Scholar]
- 19.Itoh, Y., X. Wang, B. J. Hinnebusch, J. F. Preston III, and T. Romeo. 2005. Depolymerization of β-1,6-N-acetyl-d-glucosamine disrupts the integrity of diverse bacterial biofilms. J. Bacteriol. 187:382-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson, S., and T. W. Burrows. 1956. The pigmentation of Pasteurella pestis on a defined medium containing haemin. Br. J. Exp. Pathol. 37:570-576. [PMC free article] [PubMed] [Google Scholar]
- 21.Jarrett, C. O., E. Deak, K. E. Isherwood, P. C. Oyston, E. R. Fischer, A. R. Whitney, S. D. Kobayashi, F. R. DeLeo, and B. J. Hinnebusch. 2004. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J. Infect. Dis. 190:783-792. [DOI] [PubMed] [Google Scholar]
- 22.Jenal, U. 2004. Cyclic di-guanosine-monophosphate comes of age: a novel secondary messenger involved in modulating cell surface structures in bacteria? Curr. Opin. Microbiol. 7:185-191. [DOI] [PubMed] [Google Scholar]
- 23.Jones, H. A., J. W. Lillard, Jr., and R. D. Perry. 1999. HmsT, a protein essential for expression of the haemin storage (Hms+) phenotype of Yersinia pestis. Microbiology 145:2117-2128. [DOI] [PubMed] [Google Scholar]
- 24.Kirillina, O., J. D. Fetherston, A. G. Bobrov, J. Abney, and R. D. Perry. 2004. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol. Microbiol. 54:75-88. [DOI] [PubMed] [Google Scholar]
- 25.Kutyrev, V. V., A. A. Filippov, O. S. Oparina, and O. A. Protsenko. 1992. Analysis of Yersinia pestis chromosomal determinants Pgm+ and Psts associated with virulence. Microb. Pathog. 12:177-186. [DOI] [PubMed] [Google Scholar]
- 26.Lillard, J. W., Jr., J. D. Fetherston, L. Pedersen, M. L. Pendrak, and R. D. Perry. 1997. Sequence and genetic analysis of the hemin storage (hms) system of Yersinia pestis. Gene 193:13-21. [DOI] [PubMed] [Google Scholar]
- 27.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 28.Paul, R., S. Weiser, N. C. Amiot, C. Chan, T. Schirmer, B. Giese, and U. Jenal. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pendrak, M. L., and R. D. Perry. 1991. Characterization of a hemin-storage locus of Yersinia pestis. Biol. Metals 4:41-47. [DOI] [PubMed] [Google Scholar]
- 30.Pendrak, M. L., and R. D. Perry. 1993. Proteins essential for expression of the Hms+ phenotype of Yersinia pestis. Mol. Microbiol. 8:857-864. [DOI] [PubMed] [Google Scholar]
- 31.Perry, R. D., A. G. Bobrov, O. Kirillina, H. A. Jones, L. L. Pedersen, J. Abney, and J. D. Fetherston. 2004. Temperature regulation of the hemin storage (Hms+) phenotype of Yersinia pestis is posttranscriptional. J. Bacteriol. 186:1638-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perry, R. D., T. S. Lucier, D. J. Sikkema, and R. R. Brubaker. 1993. Storage reservoirs of hemin and inorganic iron in Yersinia pestis. Infect. Immun. 61:32-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perry, R. D., M. L. Pendrak, and P. Schuetze. 1990. Identification and cloning of a hemin storage locus involved in the pigmentation phenotype of Yersinia pestis. J. Bacteriol. 172:5929-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pollitzer, R. 1954. Plague. W.H.O. Monogr. Ser. 22:1-698. [Google Scholar]
- 35.Römling, U., Z. Bian, M. Hammar, W. D. Sierralta, and S. Normark. 1998. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180:722-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Römling, U., M. Rohde, A. Olsén, S. Normark, and J. Reinköster. 2000. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 36:10-23. [DOI] [PubMed] [Google Scholar]
- 37.Ryjenkov, D. A., M. Tarutina, O. V. Moskvin, and M. Gomelsky. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187:1792-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simm, R., M. Morr, A. Kader, M. Nimtz, and U. Römling. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123-1134. [DOI] [PubMed] [Google Scholar]
- 39.Spiers, A. J., S. G. Kahn, J. Bohannon, M. Travisano, and P. B. Rainey. 2002. Adaptive divergence in experimental populations of Pseudomonas fluorescens. I. Genetic and phenotypic bases of wrinkly spreader fitness. Genetics 161:33-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Surgalla, M. J., and E. D. Beesley. 1969. Congo red-agar plating medium for detecting pigmentation in Pasteurella pestis. Appl. Microbiol. 18:834-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tal, R., H. C. Wong, R. Calhoon, D. Gelfand, A. L. Fear, G. Volman, R. Mayer, P. Ross, D. Amikam, H. Weinhouse, A. Cohen, S. Sapir, P. Ohana, and M. Benziman. 1998. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol. 180:4416-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tischler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, X., J. F. Preston III, and T. Romeo. 2004. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186:2724-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wood, P. J. 1980. Specificity in the interaction of direct dyes with polysaccharides. Carbohydr. Res. 85:271-287. [Google Scholar]
- 45.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Römling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452-1463. [DOI] [PubMed] [Google Scholar]