Abstract
Certain type II restriction modification gene systems can kill host cells when these gene systems are eliminated from the host cells. Such ability to cause postsegregational killing of host cells is the feature of bacterial addiction modules, each of which consists of toxin and antitoxin genes. With these addiction modules, the differential stability of toxin and antitoxin molecules in cells plays an essential role in the execution of postsegregational killing. We here examined in vivo stability of the EcoRI restriction enzyme (toxin) and modification enzyme (antitoxin), the gene system of which has previously been shown to cause postsegregational host killing in Escherichia coli. Using two different methods, namely, quantitative Western blot analysis and pulse-chase immunoprecipitation analysis, we demonstrated that both the EcoRI restriction enzyme and modification enzyme are as stable as bulk cellular proteins and that there is no marked difference in their stability. The numbers of EcoRI restriction and modification enzyme molecules present in a host cell during the steady-state growth were estimated. We monitored changes in cellular levels of the EcoRI restriction and modification enzymes during the postsegregational killing. Results from these analyses together suggest that the EcoRI gene system does not rely on differential stability between the toxin and the antitoxin molecules for execution of postsegregational cell killing. Our results provide insights into the mechanism of postsegregational killing by restriction-modification systems, which seems to be distinct from mechanisms of postsegregational killing by other bacterial addiction modules.
A type II restriction-modification (RM) gene system is typically composed of a gene encoding a DNA endonuclease (restriction enzyme [R]), which cleaves double-stranded DNA at a specific recognition sequence, and a gene encoding a DNA methyltransferase (modification enzyme [M]), which specifically transfers a methyl group to a base within the recognition sequence, thereby preventing the cognate restriction enzyme from cleaving DNA there. It has generally been believed that RM gene systems are functional in cellular defenses against invading foreign DNA and that they have been selected and maintained during evolution for this benefit they provide to the host bacteria. However, a hypothesis was proposed that certain type II RM gene systems may represent selfish genetic elements in the sense that they can maintain and increase their copy number even when they do not confer any advantageous phenotype on their host cells (21, 23, 32). This hypothesis was based on the observation that certain type II RM gene systems on a plasmid can increase stability of the plasmid by selectively killing cells that failed to retain the plasmid, thereby causing an increase in their relative frequency in the viable bacterial population. Similar resistance to loss from a host cell was observed with a chromosomally located RM gene system that was threatened by an allelic DNA (15, 42).
Analyses of various bacterial genomes provided evidence that RM gene systems can move between bacterial genomes, which seems to be consistent with their behavior as selfish DNA elements (21, 23, 31, 35, 36). Their association with genome rearrangements was inferred from comparisons of closely related bacterial genomes (2, 36) and was demonstrated experimentally (15). The selfish-gene hypothesis was given strong support when an RM gene system on a chromosome was found to multiply in tandem in a manner dependent on a functional restriction gene (42). This was reminiscent of the induction of the replication of prophage genomes.
Indeed, type II RM gene systems show some similarity to viruses in their regulation of gene expression (23). When they enter a new host, they have to establish themselves in the host without excessive killing of the host cells, just as temperate bacteriophages establish themselves in host cells as prophages. It was postulated that RM gene systems express the modification activity before restriction activity to protect the host chromosome by methylation. The methyltransferases of certain type II RM systems, including the SsoII and EcoRII RM systems, were shown to function as transcription regulators that are required for the coordinated expression of the restriction and modification enzymes (20, 45). In some type II RM systems, a third regulatory protein, called C protein, plays the role of delaying expression of their restriction enzymes (33, 47). After establishment, type II RM systems are expected to tightly regulate their gene expression to maintain constant cellular levels of restriction enzyme and modification enzyme to prevent attack on the host, just as prophages do until critical events such as gene loss happens to trigger the attack.
In the case of the EcoRI RM system, which had previously been shown to cause postsegregational host killing (13, 25, 32) and is the subject of the present study, little is known about regulation of gene expression. The EcoRI system is composed of the ecoRIR (R) and ecoRIM (M) genes, which encode EcoRI R and M proteins, respectively (11, 34). The R gene is located upstream of the M gene. It has been proposed that the R and M genes constitute an operon, in which expression of the two genes is coordinately controlled by a promoter located immediately upstream of the R gene (40). In addition, a specific promoter for the M gene has been proposed to be present within the R gene (11, 37). This postulated M gene-specific promoter, which should allow expression of the M gene in the absence of the R gene, might play a role for sequential expression of modification activity and restriction activity when the EcoRI gene system enters a new host cell (11). However, regulation of the two promoter activities for EcoRI RM gene expression remains unexplained.
The mechanism for plasmid stabilization by RM gene systems appears very similar to the mechanism for plasmid stabilization by several gene complexes known as addiction modules found in many naturally occurring plasmids (10, 22). As with the above type II RM gene systems, these addiction modules on a plasmid enhance the stability of maintenance of the plasmid by killing plasmid-free segregants, a process called postsegregational killing. An addiction module of the type called a classical proteic killer system or toxin-antitoxin system is generally composed of two adjacent genes, one for toxin protein and the other for antitoxin protein. The toxin inhibits cell growth or, in some cases, kills cells by inhibiting important cellular processes, such as replication and translation. The cognate antitoxin counteracts the toxin action through direct interaction. The toxin is stable, whereas the cognate antitoxin is metabolically unstable because of degradation by a specific protease (1, 4, 27, 28, 48). Such differential stability was shown to be critically important for postsegregational host cell killing. When cells fail to retain the addiction module, degradation of the unstable antitoxin leads to imbalance between the concentrations of the toxin and antitoxin in cells because the antitoxin can no longer be replenished. This imbalance would result in release of the toxin to attack its cellular target. Differential stability of the toxin and the antitoxin is also the strategy employed by the third type of postsegregational killing system programmed by the antisense RNA-regulated addiction modules (9). Expression of a toxin from its mRNA is prevented by its antisense RNA, an antitoxin in a sense, which is metabolically unstable. Loss of the gene system results in preferential decay of the antisense RNA, which leads to translation of the toxin mRNA and, eventually, to cell death.
On the other hand, a plausible sequence of molecular events underlying RM gene system-mediated postsegregational killing has been proposed on the basis of experimental results (13, 14, 32). The modification enzyme protects the host chromosome from lethal cleavage by the restriction enzyme in cells carrying the RM gene system (on the chromosome or on a resident plasmid). Once the RM gene system is eliminated from a cell, the cellular level of modification enzyme molecules is gradually decreased through dilution by cell growth to a certain threshold point, beyond which its concentration is not sufficient for complete protection of the newly replicated host chromosome from lethal cleavage by the remaining molecules of the restriction enzyme. It should be noted here that, while the restriction enzyme has to cleave the chromosome at only a few recognition sites to kill the cell (unless the breakage is repaired [13, 14]), the modification enzyme likely has to modify almost all of the recognition sites on the host chromosome to protect the cell from lethal cleavage by the restriction enzyme.
Although this simple scheme, or dilution model, seems plausible, could it sufficiently explain how the EcoRI gene system undergoes the transition from the autoregulation mode to the host-killing mode after the gene loss? How can the RM systems trigger cell death only at appropriate times, namely, only when they are eliminated from the host cells, while they suppress cell killing in cells that retain the gene systems? To address this problem, the classical proteic addiction modules and the antisense RNA-regulated addiction modules employ a common strategy of differential stability between the toxin (or its mRNA) and the antitoxin, in addition to mechanisms regulating gene expression (7, 10, 22). The RM systems might also depend on the same strategy to solve the problem.
The recent findings that toxin proteins of several toxin-antitoxin modules (that is, RelE, MazF, and PemK) are endoribonucleases with specific recognition sequences (38, 51, 52) also make it extremely interesting to see whether RM gene systems and other toxin-antitoxin modules have experienced similar constraints in function and evolution. Since regulatory mechanisms of postsegregational killing by RM gene systems likely reflect such functional and evolutionary constraints, their elucidation would provide insights into this issue.
Thus, we initiated an analysis of the regulatory mechanism for EcoRI RM gene system-mediated postsegregational cell killing with a focus on the stability of restriction and modification enzymes. We found that both the EcoRI restriction enzyme and the modification enzyme are stable in vivo. These findings strongly suggest that, unlike the other two classes of addiction modules, the EcoRI RM system does not rely on differential stability between toxin (R) and antitoxin (M) for regulation of postsegregational host killing.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, DNA manipulations, and nomenclature for restriction modification enzymes.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were grown in LB medium (29) or M9 minimal medium (29). When required, antibiotics were added at the following concentrations: ampicillin, 50 μg/ml; tetracycline, 15 μg/ml; kanamycin, 30 μg/ml; chloramphenicol, 20 μg/ml (10 μg/ml for minimal medium); oxacillin, 100 μg/ml. Recombinant DNA manipulations were performed according to standard procedures (43). Generalized transduction with phage P1vir was conducted as described previously (29). The new nomenclature for restriction modification enzymes was followed (39).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Source | Reference |
|---|---|---|---|
| E. coli | |||
| MC1061 | araD139 Δ(ara-leu)7696 galE15 galK16 Δ(lac)X74 rpsL hsdR2 mcrA mcrB1 | Laboratory collection | 5 |
| JH139 | dinD1::MudI1734(Kmrlac)a | J. Heitman | 18 |
| BIK5607b | MC1061 dinD1::MudI1734(Kmrlac) | This study | |
| DH5 | endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 Δ(lacZYA-argF)U169 deoR | N. Yamaguchi | 12 |
| DH5α | endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 Δ(lacZYA-argF)U169 deoR (φ80dlacΔ(lacZ)M15) | D. Hanahan | 49 |
| BL21(DE3) | ompT lon hsdSBgal dcm λDE3 lysogen | Novagen | |
| Plasmids | |||
| pMB4 | EcoRI r+ m+ plasmid; ColE1 origin; Apr | F. Stahl | 3 |
| pIK163 | pBR322 carrying EcoRI r+ m+ genes; Apr | K. Kusano | 25 |
| pIK164 | pBR322 carrying EcoRI r− m+ genes; Apr | K. Kusano | 25 |
| pIK166 | pBR322 carrying EcoRI r+ m+ genes; Apr | K. Kusano | 25 |
| pIK167 | pBR322 carrying EcoRI r− m+ genes; Apr | K. Kusano | 25 |
| pHSG415 | Vector derived from pSC101; temperature-sensitive for replication; Apr, Kmr, Cmr | J. Kato | 17 |
| pIK172 | pHSG415 carrying EcoRI r+ m+ genes; Apr, Cmr | K. Kusano | 32 |
| pIK173 | pHSG415 carrying EcoRI r− m+ genes; Apr, Cmr | K. Kusano | 32 |
| pIK179 | Aps derivative of pIK173; Cmr | K. Kusano | 25 |
| pET-15b | Expression vector; ColE1 origin; Apr | Novagen | |
| pET-EcoRIR | pET-15b carrying EcoRI r+ gene; Apr | This study | |
| pET-EcoRIM | pET-15b carrying EcoRI m+ gene; Apr | This study | |
| pMAN885EH | Vector containing the arabinose PBAD promoter and its regulator; araC; p15A origin; Cmr | H. Tokuda | 50 |
| pMAN-RM | pMAN885EH carrying EcoRI r+ m+ genes under the control of the PBAD promoter; Cmr | This study |
MudI1734(Kmr lac) is a derivative of Mud(Apr lac), but devoid of Mu A and Mu B genes, which are required for transposition and replication. Thus, unlike E. coli strains carrying the original Mud(Apr lac), E. coli strains carrying MudI1734(Kmr lac) are stable at 42°C.
BIK5607 was constructed by transducing the dinD1::MudI1734(Kmr lac) allele from JH139 into MC1061.
Assay for the EcoRI restriction and modification activity in vivo.
EcoRI restriction activity in vivo was measured by determining the efficiency of plating (efficiency of plaque formation) of unmodified λvir phage on the Escherichia coli strains to be tested. EcoRI modification activity in vivo was assessed by growing λvir phage on the E. coli strains to be tested and then determining the efficiency of plaque formation of the phage on an EcoRI r+ m+ strain [DH5(pIK166)] relative to that on an EcoRI r− m+ strain [DH5(pIK167)].
Construction of plasmids.
Plasmids pET-EcoRIR and pET-EcoRIM were constructed as follows. The coding sequence of the EcoRI R gene was amplified by PCR from pIK166 using the following primer pair: R-N-Nde (5′-CCGGGATTTCCATATGTCTAATAAAAAACAGTCAAATA-3′) and R-C-Bam (5′-CCGCGGGATCCTCACTTAGATGTAAGCTGTTCA-3′). The italic letters indicate the start and termination codons, respectively, of the EcoRI R gene. These primers contain an NdeI or BamHI site (underlined). The PCR-amplified fragment was digested with NdeI and BamHI and ligated with the corresponding sites of pET-15b (Novagen). The resulting plasmid pool was transformed into E. coli DH5α carrying pIK179, which encodes the EcoRI modification enzyme and is compatible with pET-15b. Ampicillin-chloramphenicol-resistant transformants were selected at 30°C and then screened for restriction activity in vivo using a phage spot assay. A plasmid recovered from one of the transformants with EcoRI restriction activity was designated pET-EcoRIR and used in this study. The coding sequence of the EcoRI modification enzyme was similarly amplified from pIK166 using the following primer pair: M-N-Nde (5′-CCGGGATTTCCATATGGCTAGAAATGCAACAAACA-3′) and M-C-Bam (5′-CCGCGGGATCCTTACTTTTGTAATCGTTTGTTTTTT-3′). The italic letters indicate the ATG start and termination codons, respectively, of the EcoRI M gene. These primers contain an NdeI or BamHI site (underlined). The amplified fragment was digested with NdeI and BanHI, cloned into the corresponding sites of pET-15b, and transformed into E. coli strain DH5α. Ampicillin-resistant transformants were selected and then screened for EcoRI modification activity in vivo using a phage spot assay. A plasmid recovered from one of the transformants that exhibited EcoRI modification activity was designated pET-EcoRIM and used in this study. Plasmids pET-EcoRIR and pET-EcoRIM encode His-tagged EcoRI R and M proteins, respectively, with an N terminus extensions of 20 amino acid residues including a stretch of six consecutive histidine residues.
Plasmid pMAN-RM was constructed as follows. Plasmid pIK164 was digested with PstI and HindIII. The resulting 1.27-kb PstI-HindIII fragment, which contained the entire coding sequence for the EcoRI M protein as well as a part of coding sequence for the EcoRI R protein, was subcloned into the corresponding sites of pMAM885EH to construct pMAN-M. Plasmid pIK163 was digested with NdeI, blunt ended with T4 DNA polymerase, and further digested with PstI to generate a 0.65-kb PstI-NdeI (blunt ended) fragment. This fragment, which contained a part of the coding sequence for the EcoRI R protein, was ligated with the large PstI-SmaI fragment of pMAN-M to construct pMAN-RM, in which the complete coding sequences for EcoRI RM proteins were placed under the control of the arabinose-inducible PBAD promoter.
Analytical methods for proteins.
The method of Laemmli (26) was used for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Unless otherwise specified, the gels contained 15% acrylamide and 0.5% bisacrylamide. Protein concentrations were determined by the bicinchoninic acid (BCA) method (44) with a Micro-BCA protein assay kit (Pierce).
Expression and purification of EcoRI RM proteins.
E. coli strain BL21(DE3)(pIK179) carrying pET-EcoRIR or BL21(DE3) carrying pET-EcoRIM was inoculated into LB broth containing ampicillin, oxacillin, and chloramphenicol or into LB broth containing ampicillin and oxacillin, respectively, and grown at 30°C with aeration to exponential phase (optical density at 600 nm [OD600] of 0.4). Expression of His-tagged recombinant proteins was induced by adding isopropyl-β-d-thiogalactopyranoside (final concentration, 1 mM) to the culture. The culture was incubated with aeration at 30°C for 4 h for BL21(DE3)(pIK179)(pET-EcoRIR) or at 30°C for 6 h for BL21(DE3)(pET-EcoRIM). Purification of the His-tagged proteins was performed by nickel affinity chromatography under denaturing conditions according to the pET system manual (Novagen). Briefly, cell extracts were prepared by thawing frozen cell pellets and sonicating them in TSPB buffer (20 mM Tris-HCl [pH 7.9]-0.5 M NaCl-1 mM phenylmethylsulfonyl fluoride-1 mM benzamidine) containing 5 mM imidazole and 6 M urea. The resulting cell extract was applied to a precharged nickel affinity column (His-Bind Ni resin column; Novagen). The column was washed with TSPB buffer containing 15 mM imidazole and 6 M urea, and the purified His-tagged proteins were eluted with TSPB buffer containing 300 mM imidazole and 6 M urea.
Antibody preparation and Western blot analysis.
The purified preparations of His-tagged EcoRI R and M proteins were separately resolved on SDS-PAGE gels. The gels were lightly stained with 0.05% Coomassie brilliant blue R-250 for 10 min as described previously (16). The stained protein band was cut out. The frozen slices of the gel containing protein were sent to Panapharm Laboratories Co., Ltd. (Kumamoto, Japan), and were used to raise polyclonal antibodies against EcoRI R or M protein in rabbits. For Western blot analysis, cellular proteins were separated by SDS-PAGE (15% polyacrylamide) and then blotted to a polyvinylidene fluoride membrane (Immobilon-P; Millipore Corp.) using a semidry blotting apparatus. Detection of the EcoRI RM proteins with specific antibodies was carried out by the enhanced chemiluminescent method with an ECL Plus Western blot detection reagent system (Amersham Biosciences) according to the manufacturer's instructions. The polyclonal antibody against EcoRI R or M protein was used at a 1:10,000 dilution. Anti-rabbit polyclonal antibody from donkey linked with horseradish peroxidase (Amersham Biosciences) was used as a secondary antibody at a 1:300,000 dilution. Chemiluminescent signals on the blots were visualized on Hyperfilm ECL (Amersham Biosciences). The quantitation of bands was performed by analyzing blots with an FLA-5100 image analyzer (Fuji film) and Image Gauge software (Fuji film) or by analyzing scanned film images using ImageJ software (W.S. Rasband, National Institutes of Health, Bethesda, Md.; http://rsb.info.nih.gov/ij/).
Analysis of stability of EcoRI R and M proteins in vivo.
Two cultures of MC1061 (pMB4) were grown with aeration at 30°C in LB broth containing ampicillin to early exponential phase (OD600 of 0.2). One of the two cultures was shifted to 42°C, while the other culture was kept at 30°C. After incubation for 30 min, spectinomycin was added to both cultures at a concentration of 200 μg/ml to block protein synthesis, and the cultures were further incubated. A portion of the cultures was taken at intervals, and the cellular proteins were precipitated with 10% trichloroacetic acid (TCA) in the cold. Pellets were washed with cold 80% acetone solution and resuspended in 10 mM sodium phosphate buffer (pH 7.2) containing 1% SDS. The resulting suspensions were boiled for 5 min, and protein concentrations were determined using a Micro-BCA protein assay kit (Pierce) with bovine serum albumin as the standard. Samples containing equal amounts (50 μg) of cellular proteins were analyzed by Western blot analysis. Chemiluminescent signals were visualized on films. Quantitation of protein bands was performed by the use of an FLA-5100 image analyzer equipped with Image Gauge software.
Estimation of number of molecules per cell for EcoRI restriction enzyme and modification enzyme.
E. coli strains BIK5607(pIK172) and BIK5607(pMB4) were grown with aeration at 30°C in LB broth containing ampicillin and oxacillin to exponential phase (OD600 of 0.3). Samples were taken and added to cold TCA solution (final concentration, 10% [wt/vol]) to precipitate proteins. The precipitated proteins were washed with cold 80% acetone solution, resuspended in 10 mM sodium phosphate buffer (pH 7.2) containing 1% SDS, and then boiled for 5 min. After protein concentrations were measured, a portion of each sample containing equal amounts (28 μg) of cellular proteins was loaded per lane, resolved by SDS-PAGE with 15% polyacrylamide gel, and subjected to Western blot analysis. Different amounts of denatured purified His-tagged EcoRI RM proteins mixed with 28 μg of denatured total cellular proteins prepared from the EcoRI r− m− strain MC1061 were used as the quantitation standards on the same gel. Chemiluminescent signals on the blots were visualized on films. Quantitation of bands was performed using ImageJ software. The numbers of cells in the cultures at the sampling time point were determined by direct cell counting under a microscope. Numbers of EcoRI R and M protein molecules per cell were calculated from the amounts of EcoRI RM proteins determined by Western blot analysis, using the molecular masses of the two proteins, Avogadro's number (6.02 ×1023 molecules per mol), and numbers of cells equivalent to the amounts of protein analyzed by Western blot analysis as described previously (8).
Measurement of cellular EcoRI RM protein levels upon induction of postsegregational cell killing.
Strains BIK5607(pIK172) and BIK5607(pIK173) were grown with aeration in LB broth containing ampicillin and oxacillin to exponential phase (OD600 of 0.3). Postsegregational killing was induced by recovering cells by low-speed centrifugation and resuspending them in LB broth prewarmed at 42°C, a nonpermissive temperature for replication of these plasmids. The cultures were then incubated at 42°C and diluted whenever the OD600 of the cultures reached about 0.3. Samples were taken at intervals for measurement of the cellular levels of the EcoRI RM proteins, bacterial mass (OD600), and viable cell count. Total cellular proteins of BIK5607(pIK172) at each sampling point were prepared by adding cold TCA solution to each sample (final concentration, 10% [wt/vol]), washing the resulting pellets with cold 80% acetone solution, and resuspending the proteins in 10 mM sodium phosphate buffer (pH 7.2) containing 1% SDS. The samples were boiled for 5 min, and protein concentrations were determined. A portion of the samples containing equal amounts of cellular proteins was analyzed by Western blot analysis. Different dilutions of the denatured cellular proteins prepared from BIK5607(pIK172) at time zero were mixed with denatured cellular proteins from the EcoRI r− m− strain MC1061 so that the total protein concentration was kept the same between the mixtures. These mixtures were used as the quantitation standards in Western blot analysis. Chemiluminescent signals on the blots were visualized on films. Quantitation of bands was performed using ImageJ software. Viable cells were counted by spreading dilutions of the cultures on LB plates and counting colonies after overnight incubation at 30°C.
Pulse-labeling and immunoprecipitation.
MC1061(pMAN-RM) and MC1061(pMAN885EH) were grown aerobically at 37°C in M9 medium supplemented with glycerol (0.2%), MgSO4 (1 mM), thiamine (10 μg/ml), all of the 20 amino acids (20 μg/ml each) except methionine and cysteine, and chloramphenicol (10 μg/ml) to log-phase (OD600 of 0.2), and then arabinose was added to the cultures (final concentration, 0.002%) to induce the expression of the EcoRI RM genes. After 35 min of incubation at 37°C, the cultures were pulse labeled for 5 min by the addition of a mixture of [35S]methionine and [35S]cysteine (Redivue PRO-MIX Cell Labeling Mix; a mixture of 70% [35S]methionine and 30% [35S]cysteine [1,000 Ci/mmol]) (Amersham Biosciences) to a final concentration of 100 μCi/ml and then chased with an excess of unlabeled methionine and cysteine (final concentration, 1 mg/ml [each]). After 3 min of incubation at 37°C, 250 μl of the culture was taken (time zero), and additional 250-μl samples were taken after 1 and 3 h of incubation with aeration at 37°C. The EcoRI RM proteins in each sample were immunoprecipitated with specific antibodies according to the method of Ito et al. (19), with a slight modification as follows. Cellular proteins were precipitated with 5% TCA in the cold. The pellets were washed with cold 80% acetone solution, resuspended in 50 μl of 50 mM Tris-HCl (pH 8.0)-1% SDS-1 mM EDTA, and then boiled for 3 min. A total of 30 μl of the solution was mixed with 1 ml of Triton buffer (50 mM Tris-HCl [pH 8.0]-150 mM NaCl-2% Triton X-100-1 mM EDTA), and the precipitates were removed by centrifugation at 13,000 × g for 5 min. The antibody (2 μl) against EcoRI R protein or M protein was added to the supernatant, and the mixture was incubated overnight at 4°C. To this mixture, 50 μl of 10% (wt/vol) suspension of IgGsorb (The Enzyme Center, Inc.) in the Triton buffer was added, and the mixture was kept at 4°C for 1 h. After centrifugation at 10,000 × g for 5 min, the pellet was washed with 1 ml of the Triton buffer and then with 1 ml of 10 mM Tris-HCl (pH 8.0). The washed pellet was resuspended in 30 μl of a sample buffer for SDS-PAGE and then boiled for 3 min. The supernatant (10 μl) was subjected to SDS-PAGE (12.5% acrylamide and 0.42% bisacrylamide). The gels were dried and exposed to X-ray films (Kodak X-Omat) for autoradiography.
RESULTS
In vivo stability of the EcoRI restriction modification enzyme proteins.
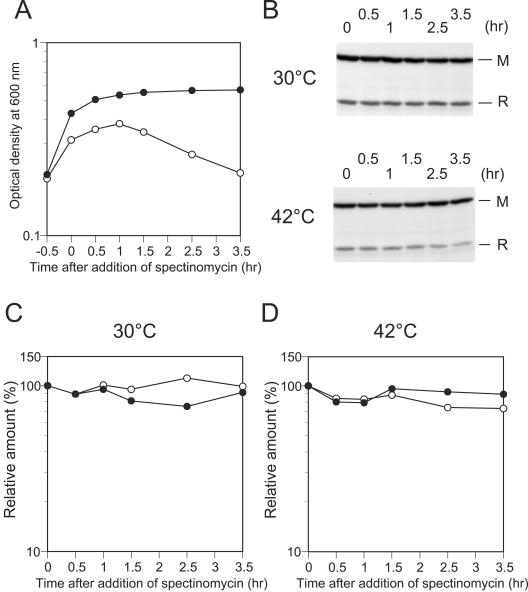
To gain insight into the regulation of postsegregational host cell killing by the EcoRI RM gene system, we first examined the stability of the EcoRI restriction and modification enzymes in vivo. We examined their stability both at 30°C and 42°C for comparison with plasmid loss experiments with thermosensitive plasmids (see below). By Western blot analysis, we monitored cellular levels of the two proteins after blockage of protein synthesis. Two cultures of MC1061 carrying pMB4 (Table 1, EcoRI r+ m+) were grown at 30°C to early log phase, and then the two cultures were incubated at 30°C and 42°C, respectively. After 30 min of incubation (time zero), spectinomycin was added to the cultures to block protein synthesis, after which a portion of each culture was taken at intervals (Fig. 1A). The same amount of cellular proteins was applied to each lane. Amounts of the two proteins were not decreased significantly during 3 h of incubation at 30°C or at 42°C after the inhibition of protein synthesis (Fig. 1B, C, and D). This indicates that the two proteins are as stable as bulk cellular proteins and that there is no striking difference in stability between the two proteins under the conditions used.
FIG. 1.
Change in cellular levels of EcoRI R and M proteins after inhibition of protein synthesis. Two cultures of MC1061(pMB4) were grown at 30°C to early log phase (OD600 of 0.2). One culture was shifted to 42°C while the other culture was kept at 30°C. After a 30-min incubation at the respective temperature, spectinomycin was added to the two cultures to inhibit protein synthesis. Samples were taken just before the addition of spectinomycin (time zero) and 0.5, 1, 1.5, 2.5, and 3.5 h later. The samples were processed and examined for amounts of the EcoRI R and M proteins by Western blot analysis as described in Materials and Methods. (A). Change in optical density at 600 nm of the two cultures incubated at 30°C (open circle) or 42°C (filled circle) after blockage of protein synthesis. (B) Levels of EcoRI RM proteins in cells at each sampling point as determined by Western blot analysis. Samples containing the same amounts (50 μg) of cellular proteins were analyzed in each lane. Chemiluminescent signals visualized on film are shown. (C and D) Changes in levels of the EcoRI R protein (open circle) and M protein (filled circle) in cells incubated at 30°C or 42°C after blockage of protein synthesis. The amount of the EcoRI R or M proteins at each sampling point was normalized to the amount at time zero.
These findings suggested the possibility that the mechanism of postsegregational host cell killing by the EcoRI RM gene system is different from the mechanisms of postsegregational host cell killing by other toxin-antitoxin addiction modules, which rely on differential stability between toxin and antitoxin molecules for execution of postsegregational host cell killing.
Estimation of cellular amounts of the EcoRI restriction modification enzymes.
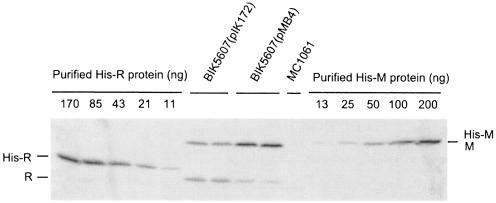
To further understand how the EcoRI RM gene system regulates postsegregational host cell killing, we attempted to learn how many molecules of the EcoRI restriction enzyme and modification enzyme are present in cells before and after the induction of postsegregational cell killing. To determine the levels of the two proteins within cells growing under the steady-state condition, we estimated the numbers of molecules per cell for the EcoRI RM proteins in two E. coli strains, BIK5607(pMB4) and BIK5607(pIK172), which carry the EcoRI genes on different plasmids (Table 1). Plasmid pMB4 is very closely related to pMB1, a naturally occurring EcoRI r+ m+ plasmid obtained from the original clinical isolate of E. coli with the EcoRI host specificity (3). Thus, the cellular levels of EcoRI RM proteins in BIK5607(pMB4) were expected to reflect the regulation of EcoRI RM gene expression from the naturally occurring EcoRI r+ m+ plasmid. Another plasmid, pIK172, constructed by subcloning the EcoRI genes from pMB4 into pHSG415 (vector with a temperature-sensitive replication machinery) (32), was used in the previous experiments in our laboratory (13, 14, 25, 32) and in the experiments described in the next section, in which plasmid loss was artificially induced by a temperature shift.
Strains BIK5607(pMB4) and BIK5607(pIK172) were grown at 30°C to exponential phase in LB broth containing selective antibiotics. Their total cellular proteins as well as different amounts of purified His-tagged EcoRI RM proteins were analyzed by quantitative Western blot analysis using specific antibodies against EcoRI R or M proteins (Fig. 2). On the assumption that antibodies react similarly with the His-tagged recombinant proteins and with the native proteins, we estimated amounts of the EcoRI RM proteins and calculated their numbers per cell. In BIK5607(pIK172), the number of molecules per cell was estimated to be 4,000 and 8,000 for EcoRI R protein and M protein, respectively. In BIK5607(pMB4), estimated numbers of molecules per cell were 1,000 and 16,000 for EcoRI R protein and M protein, respectively. It should be noted that the EcoRI restriction enzyme is functional as a dimer, whereas a monomer is the active form for the EcoRI modification enzyme.
FIG. 2.
Estimation of numbers of EcoRI R and M protein molecules per cell. BIK5607(pIK172) and BIK5607(pMB4) were grown at 30°C to log phase. Equal amounts (28 μg) of cellular proteins prepared from independent cultures of each strain were analyzed by Western blotting. Different amounts of purified His-tagged EcoRI R and M proteins mixed with cellular proteins prepared from MC1061 were used as the standards. A sample containing 28 μg of cellular proteins from MC1061 was also included in the Western blot analysis as a negative control. Chemiluminescent signals were visualized on film.
Change in cellular levels of the EcoRI RM proteins during postsegregational host cell killing.
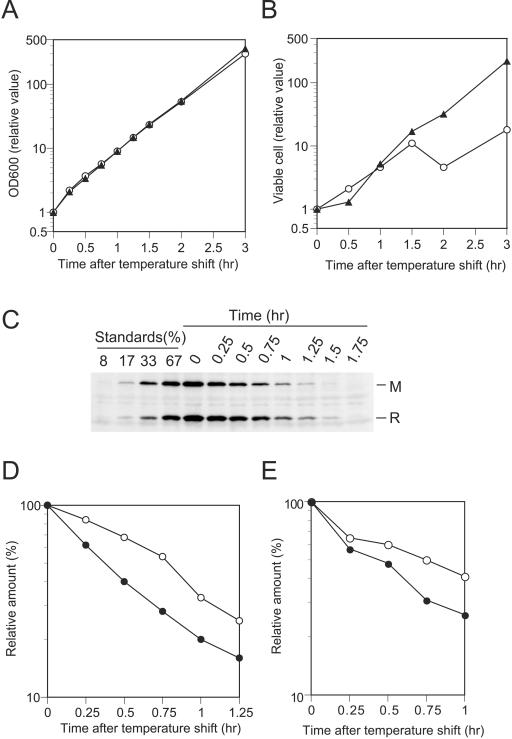
Our laboratory has previously established an experimental system in which postsegregational host cell killing by the EcoRI RM gene system can be artificially induced by shifting the growth temperature of bacterial cultures (25, 32). In this experimental system, a temperature shift from 30°C to 42°C causes replication arrest of pIK172, a plasmid that is temperature sensitive for replication and carries the EcoRI RM genes, resulting in loss of the plasmid from the cells. We used this experimental system to investigate how cellular levels of the EcoRI RM proteins change after loss of their genes.
As shown in Fig. 3B, the temperature shift blocked an increase in viable cell counts of the cells carrying the EcoRI restriction gene but not cells carrying a restriction-negative control plasmid. The change in optical density of the two cultures did not differ within the limits of detection (Fig. 3A). Western blot analysis showed that cellular levels of the two proteins gradually decreased after the temperature shift (Fig. 3C, D, and E). We followed the change in cellular levels of the two proteins up to 1.25 h (experiment 1) (Fig. 3C and D) or 1 h (experiment 2) (Fig. 3E) after the temperature shift. A portion of cells in the culture of BIK5607(pIK172) began to die about 1 to 1.5 h after the temperature shift, as revealed by viable cell counts (Fig. 3B). At 1 h after the temperature shift, the cellular amount of R protein decreased to 33% of that at the time of the temperature shift, whereas the cellular amount of M protein decreased to 20% in experiment 1 (Fig. 3D). In experiment 2, cellular amounts of the R and M proteins at 1 h after temperature shift were 44% and 26% of their amounts at the time of the temperature shift, respectively (Fig. 3E).
FIG. 3.
Change in cellular levels of EcoRI R and M proteins after induction of plasmid loss by temperature shift. BIK5607(pIK172) and BIK5607(pIK173) were grown at 30°C to log phase. Plasmid loss was induced by temperature shift to 42°C. The cultures were then incubated at 42°C and diluted whenever their optical density at 600 nm reached about 0.3. Samples were taken just before the temperature shift (time zero) and 0.25, 0.5, 0.75, 1, 1.25, 1.5, and 1.75 h later. The samples were examined for amounts of the EcoRI R and M proteins by Western blot analysis as described in Materials and Methods. (A) Growth curves of BIK5607(pIK172) (restriction positive) (open circle) and BIK5607(pIK173) (restriction negative) (filled triangle). Optical densities at 600 nm at indicated time points were normalized to the value at time zero using dilution factors. (B) Change in viable cell counts. Viable cells were counted for BIK5607(pIK172) (open circle) and BIK5607(pIK173) (filled triangle) on LB agar. Viable cell numbers at indicated time points were normalized to the values at time zero using dilution factors. (C) Levels of EcoRI R and M proteins in BIK5607(pIK172) at each sampling point as determined by Western blot analysis. The sample taken just before the temperature shift (time zero) was diluted with a sample containing cellular proteins from the EcoRI r− m− strain MC1061 so that the total amount of protein in each dilution was kept the same. (D and E) Change in cellular levels of EcoRI R (open circle) and M (filled circle) proteins after temperatureshift. Cellular amounts of EcoRI R and M proteins were estimated from the standard curve generated with the standards. Results from quantitative analysis of the signals shown in panel C (experiment 1) are presented in panel D. Results from an independent experiment (experiment 2) are shown in panel E. Samples containing 51 μg of cellular proteins were analyzed in experiment 1, while samples containing a smaller amount (24 μg) were analyzed in experiment 2.
If we assume that neither degradation nor synthesis of the EcoRI R M proteins took place after the temperature shift so that their cellular levels were determined solely by dilution through cell growth, cellular levels of R and M proteins at 1 h after the temperature shift can be calculated to be 17% of their levels at time zero, using the doubling time of bacterial cultures after temperature shift in experiment 1 [23 min for BIK5607(pIK172)] (Fig. 3A). The cellular levels observed in our experiments were higher than these predicted values for both EcoRI R and M proteins. This suggests that both proteins are synthesized after the temperature shift and also provides further support for the notion that EcoRI RM proteins are not degraded markedly during postsegregational cell killing. However, it should be noted that the cellular level of R protein decreased more slowly than that of M protein after the induction of plasmid loss by temperature shift in our two independent experiments (Fig. 3D and E).
Pulse-chase analysis of in vivo stability of EcoRI RM proteins.
The observations that the cellular amount of EcoRI M protein decreased faster than that of R protein after induction of plasmid loss (Fig. 3D and E) raised the possibility that M protein was selectively degraded in the cells. Although we could not detect significant degradation of M protein at 30°C or 42°C in our stability assay after blockage of protein synthesis with spectinomycin (Fig. 1B, C, and D), it may still be possible that degradation of M protein (and/or R protein) takes place when cells are allowed to grow in the absence of spectinomycin.
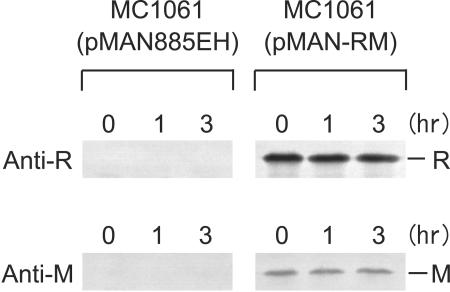
To address this question, we examined the stability of the EcoRI RM proteins in cells growing in the absence of spectinomycin by pulse-chase analysis. In order to obtain sufficient signals, we placed EcoRI r+ m+ genes under the control of PBAD promoter (Table 1, pMAN-RM). After induction with arabinose, cultures of MC1061(pMAN-RM) and MC1061(pMAN885EH) (vector, as control) were pulse-labeled with [35S]-labeled methionine and cysteine and then chased with an excess of unlabeled methionine and cysteine. The labeled EcoRI R and M proteins in the cells were separately immunoprecipitated with antibody against EcoRI R or M proteins and, after SDS-PAGE, visualized by autoradiography.
EcoRI R and M proteins were specifically detected with each antibody with MC1061(pMAN-RM) but not with MC1061(pMAN885EH) (Fig. 4), confirming the specificity of these antibodies under the conditions used. EcoRI R and M proteins were not degraded markedly during 3 h of incubation at 37°C in these growing cells (Fig. 4). Consistent with the results after blockage of protein synthesis (Fig. 1B, C, and D), these observations provide additional support for the notion that EcoRI RM proteins are relatively stable in vivo and that there is no marked difference in their stability.
FIG. 4.
Pulse-chase analysis of stability of EcoRI RM proteins. MC1061(pMAN-RM) and MC1061(pMAN885EH) (vector only, as control) were grown at 37°C in the supplemented M9 minimal medium containing glycerol, chloramphenicol, and all of the 20 amino acids except methionine and cysteine. Expression of the EcoRI RM genes was induced by adding arabinose to their log-phase cultures. After 35 min of incubation at 37°C, the cultures were labeled with a mixture of [35S]methionine and [35S]cysteine for 5 min and then chased with an excess of unlabeled methionine and cysteine. Samples were taken after 3 min of incubation (time zero), and additional samples were taken after 1 and 3 h of incubation at 37°C. The labeled EcoRI R and M proteins in the samples were separately immunoprecipitated with the anti-EcoRI R or anti-EcoRI M antibody and, after SDS-PAGE, visualized by autoradiography (exposure time, 24 h for R and 64 h for M) as described in Materials and Methods.
DISCUSSION
In the present study, we examined the in vivo stability of EcoRI RM proteins by two independent methods, namely, Western blot analysis after blockage of protein synthesis and pulse-chase labeling followed by immunoprecipitation analysis. The results demonstrated that both of the proteins are as stable as bulk intracellular proteins and that there is no striking difference in stability between them, at least under the conditions used. In the classical proteic toxin-antitoxin modules examined so far, that is, PemKI of R100 plasmid (48), CcdAB of F plasmid (28), MazEF of E. coli (1), Phd/Doc of P1 phage (27), and epsilon/zeta of Streptococcus pyrogenes plasmid pSM19035 (4), the toxin is stable, whereas the antitoxin is quite unstable. Their differential stability was shown to be important in the execution of postsegregational host cell killing; upon elimination of a toxin-antitoxin module from cells, degradation of the unstable antitoxin molecules results in release of lethal activity of the stable toxin. Similar instability of the antisense RNA of the toxin gene (an antitoxin) is implicated in the action of antisense RNA-mediated postsegregational killing systems (9). The observed in vivo stability of the EcoRI RM proteins strongly suggests that the regulation of postsegregational host cell killing by the EcoRI RM gene system is distinct from regulation of cell killing by the two other types of postsegregational systems.
It has been argued that dilution of cellular RM proteins by cell growth should be able to induce postsegregational killing even in the absence of degradation of the M protein (antitoxin) (21, 32). If this is the case, the balance between cellular levels of the R and M proteins should be precisely regulated so that the RM gene system can suppress uncontrolled cell death under steady-state growth conditions, while it can efficiently trigger cell killing once its genes are eliminated from the cells. Our analysis showed that there was a larger number of M protein molecules than R protein molecules in cells before induction of plasmid loss (Fig. 2 and 3C) and that cellular levels of the two proteins decreased gradually after the induction of plasmid loss by temperature shift (Fig. 3C, D, and E). About 1 to 1.5 h after the induction of plasmid loss, a portion of cells carrying an EcoRI r+ m+ plasmid in the culture began to lose viability (Fig. 3B). This suggests that around this time point, the cellular level of M protein becomes insufficient to protect newly replicated chromosomes from lethal attack by remaining R molecules. These findings are consistent with the notion that dilution of RM proteins by cell growth contributes to induction of EcoRI RM gene system-mediated postsegregational cell killing.
In addition to the absolute and relative cellular levels of EcoRI R and M proteins, their catalytic efficiency should represent a critically important factor that determines whether chromosomal DNA is cleaved by the restriction enzyme or whether such cleavage is blocked by the modification enzyme. Reported turnover numbers for the EcoRI restriction enzyme (four double-strand scissions/min/dimer at 37°C in vitro) (30) and modification enzyme (three methyl transfers/min/monomer at 37°C in vitro) (41) are similar. In addition, direct competition experiments using a 775-bp fragment as a substrate showed that the catalytic efficiency of the EcoRI modification enzyme was slightly better than that of the EcoRI restriction enzyme in site location and catalysis (46). Other potentially important factors in the choice between death and survival include the replication rate of the target restriction sites in growing cells and the capacity of the host cells to repair the restriction breakage (13, 14, 25). It is difficult to extrapolate from these observations to an ideal ratio for these proteins to achieve protection and/or restriction of the chromosome in vivo.
Despite some experiment-to-experiment variation, we consistently observed that the cellular level of the R protein decreased more slowly than that of the M protein after the induction of plasmid loss (Fig. 3D and E). Since cell growth should dilute cellular pools of the two proteins similarly, this difference should be explained by factor(s) other than dilution of the proteins through cell growth. It seems unlikely that selective degradation of the M protein by cellular protease(s) is responsible for the difference, because we could not detect significant degradation of the M protein at 30°C or 42°C in our protein stability assay after blockage of protein synthesis with spectinomycin (Fig. 1B, C, and D) as well as in our pulse-chase labeling experiments in the absence of such blockage (Fig. 4). However, we cannot completely exclude the possibility that degradation of the M protein takes place in BIK5607(pIK172) under the conditions used in experiments for Fig. 3, because in vivo stability of the EcoRI RM proteins could vary between strains carrying different plasmids or between strains with different physiological states.
A change in cellular levels of the two proteins could reflect not only their degradation and their dilution by cell growth but also their synthesis. It is reasonable to assume that transcription of the EcoRI gene on the remaining plasmid copies and translation of the RM proteins from the remaining transcripts continue in cells, at least transiently, even after the temperature shift to induce plasmid loss. Thus, an alternative explanation for the difference in rates of decrease of the RM proteins (Fig. 3D and E) is that a larger amount of the R protein is synthesized than the M protein after the induction of plasmid loss. Such differential synthesis of toxin and antitoxin after induction of gene loss might be important for regulation of postsegregational cell killing by the EcoRI RM gene system.
Our analysis showed that cellular levels of the EcoRI RM proteins and the ratio between cellular levels of the two proteins are different between the two strains examined, BIK5607(pIK172) and BIK5607(pMB4) (Fig. 2). The basis for these differences is unclear, although one interesting possibility is that genetic alternations introduced in the upstream and downstream regions of the EcoRI RM genes during construction of pIK172 from pMB4 (14, 32) might affect expression of the two genes. Because pMB4 is very closely related to pMB1, a naturally occurring EcoRI r+ m+ plasmid (3), the cellular amounts of the RM proteins estimated for BIK5607(pMB4) are expected to reflect the regulation of EcoRI RM gene expression in the naturally occurring plasmid. The cellular amount of the EcoRI R protein has previously been estimated to be 2,000 monomer equivalents per cell in an E. coli strain RY13, which carries pMB3, a plasmid closely related to pMB4 (30). This value is comparable to our estimate in BIK5607(pMB4). Given that the balance between the cellular levels of the two proteins should be important for regulation of postsegregational cell killing, strains with different cellular levels of the RM proteins might behave differently after plasmid loss. Indeed, the SOS induction level was found to be markedly higher in BIK5607(pIK172) than in BIK5607(pMB4) when the strains were grown at 30°C, a permissive temperature for replication of pIK172 (data not shown). Our previous analysis has revealed that double-stranded breaks on the host chromosome generated by the EcoRI gene system during postsegregational killing induce an SOS response in cells and that homologous recombination is involved in repair of those breaks (13). Thus, the elevated level of SOS induction observed in BIK5607(pIK172) suggests that chromosome cleavage by the EcoRI R protein occurs in plasmid-containing cells when cellular levels of the two proteins are somehow distorted. In this sense we should be cautious when we try to extrapolate experimental results obtained with particular plasmid constructions to behavior of RM gene systems under natural conditions.
Although our present results strongly suggest that the EcoRI gene system is distinct from the other types of bacterial addiction modules in the regulatory mechanism for postsegregational cell killing, more work is needed to clarify the mechanism. In particular, it is currently not clear whether factor(s) other than dilution of the RM proteins by cell growth contribute to enabling the lethal activity of the R protein. In addition to the EcoRI RM gene system, several other type II RM gene systems have been demonstrated to cause postsegregational host killing (6, 24, 32, 33). It will also be interesting to determine how these RM gene systems control the timing and extent of postsegregational cell killing. Such investigation should give us insight into functional and evolutionary similarities and differences between RM gene systems and other types of bacterial addiction systems (22).
Acknowledgments
We thank Joseph Heitman for providing strain JH139, Hajime Tokuda for pMAN885EH, Hai Ping Cheng for advice on antigen preparation, and Noriko Sato for advice on Western blotting. We are grateful to Shin-ichi Matsuyama and Yoshitaka Tago for many helpful discussions.
This work was supported by grants from MEXT of Japan (Kiban-Evolution, Kiban-Genome, Genome Homeostasis, Protein 3000, and 21COE: Genome language). A.I. was supported by grants from MEXT of Japan to I.K.
REFERENCES
- 1.Aizenman, E., H. Engelberg-Kulka, and G. Glaser. 1996. An Escherichia coli chromosomal “addiction module” regulated by 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 93:6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 3.Betlach, M., V. Hershfield, L. Chow, W. Brown, H. Goodman, and H. W. Boyer. 1976. A restriction endonuclease analysis of the bacterial plasmid controlling the EcoRI restriction and modification of DNA. Fed. Proc. 35:2037-2043. [PubMed] [Google Scholar]
- 4.Camacho, A. G., R. Misselwitz, J. Behlke, S. Ayora, K. Welfle, A. Meinhart, B. Lara, W. Saenger, H. Welfle, and J. C. Alonso. 2002. In vitro and in vivo stability of the ɛ2ζ2 protein complex of the broad host-range Streptococcus pyogenes pSM19035 addiction system. Biol. Chem. 383:1701-1713. [DOI] [PubMed] [Google Scholar]
- 5.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 6.Chinen, A., Y. Naito, N. Handa, and I. Kobayashi. 2000. Evolution of sequence recognition by restriction-modification enzymes: selective pressure for specificity decrease. Mol. Biol. Evol. 17:1610-1619. [DOI] [PubMed] [Google Scholar]
- 7.Engelberg-Kulka, H., and G. Glaser. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53:43-70. [DOI] [PubMed] [Google Scholar]
- 8.Feng, G., H.-C. T. Tsui, and M. E. Winkler. 1996. Depletion of the cellular amounts of the MutS and MutH methyl-directed mismatch repair proteins in stationary-phase Escherichia coli K-12 cells. J. Bacteriol. 178:2388-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerdes, K., A. P. Gultyaev, T. Franch, K. Pedersen, and N. D. Mikkelsen. 1997. Antisense RNA-regulated programmed cell death. Annu. Rev. Genet. 31:1-31. [DOI] [PubMed] [Google Scholar]
- 10.Gerdes, K., J. S. Jacobsen, and T. Franch. 1997. Plasmid stabilization by post-segregational killing. Genet. Eng. 19:49-61. [DOI] [PubMed] [Google Scholar]
- 11.Greene, P. J., M. Gupta, H. W. Boyer, W. E. Brown, and J. M. Rosenberg. 1981. Sequence analysis of the DNA encoding the EcoRI endonuclease and methylase. J. Biol. Chem. 256:2143-2153. [PubMed] [Google Scholar]
- 12.Hanahan, D. 1985. Techniques for transformation in E. coli, p. 109-135. In D. M. Glover (ed.), DNA cloning, vol. 1. IRL Press, Oxford, United Kingdom. [Google Scholar]
- 13.Handa, N., A. Ichige, K. Kusano, and I. Kobayashi. 2000. Cellular responses to postsegregational killing by restriction-modification genes. J. Bacteriol. 182:2218-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Handa, N., and I. Kobayashi. 1999. Post-segregational killing by restriction modification gene complexes: observations of individual cell deaths. Biochimie 81:931-938. [DOI] [PubMed] [Google Scholar]
- 15.Handa, N., Y. Nakayama, M. Sadykov, and I. Kobayashi. 2001. Experimental genome evolution: large-scale genome rearrangements associated with resistance to replacement of a chromosomal restriction-modification gene complex. Mol. Microbiol. 40:932-940. [DOI] [PubMed] [Google Scholar]
- 16.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Hashimoto-Gotoh, T., F. C. Franklin, A. Nordheim, and K. N. Timmis. 1981. Specific-purpose plasmid cloning vectors. I. Low copy number, temperature-sensitive, mobilization-defective pSC101-derived containment vectors. Gene 16:227-235. [DOI] [PubMed] [Google Scholar]
- 18.Heitman, J., and P. Model. 1991. SOS induction as an in vivo assay of enzyme-DNA interactions. Gene 103:1-9. [DOI] [PubMed] [Google Scholar]
- 19.Ito, K., P. J. Bassford, Jr., and J. Beckwith. 1981. Protein localization in E. coli: is there a common step in the secretion of periplasmic and outer-membrane proteins? Cell 24:707-717. [DOI] [PubMed] [Google Scholar]
- 20.Karyagina, A., I. Shilov, V. Tashlitskii, M. Khodoun, S. Vasil'ev, P. C. K. Lau, and I. Nikolskaya. 1997. Specific binding of SsoII DNA methyltransferase to its promoter region provides the regulation of SsoII restriction-modification gene expression. Nucleic Acids Res. 25:2114-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi, I. 2001. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 29:3742-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi, I. 2004. Genetic addiction: a principle in symbiosis of genes in a genome, p. 105-144. In B. E. Funnel and G. J. Phillips (ed.), Plasmid biology. ASM Press, Washington, D.C.
- 23.Kobayashi, I. 2004. Restriction-modification systems as minimal forms of life, p. 19-62. In A. Pingoud (ed.), Restriction endonucleases. Springer-Verlag, Berlin, Germany.
- 24.Kulakauskas, S., A. Lubys, and S. D. Ehrlich. 1995. DNA restriction-modification systems mediate plasmid maintenance. J. Bacteriol. 177:3451-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kusano, K., T. Naito, N. Handa, and I. Kobayashi. 1995. Restriction-modification systems as genomic parasites in competition for specific sequences. Proc. Natl. Acad. Sci. USA 92:11095-11099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Lehnherr, H., and M. B. Yarmolinsky. 1995. Addiction protein Phd of plasmid prophage P1 is a substrate of the ClpXP serine protease of Escherichia coli. Proc. Natl. Acad. Sci. USA 92:3274-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melderen, L. V., P. Bernard, and M. Couturier. 1994. Lon-dependent proteolysis of CcdA is the key control for activation of CcdB in plasmid-free segregant bacteria. Mol. Microbiol. 11:1151-1157. [DOI] [PubMed] [Google Scholar]
- 29.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Modrich, P., and D. Zabel. 1976. EcoRI endonuclease. Physical and catalytic properties of the homogenous enzyme. J. Biol. Chem. 251:5866-5874. [PubMed] [Google Scholar]
- 31.Naderer, M., J. R. Brust, D. Knowle, and R. M. Blumenthal. 2002. Mobility of a restriction-modification system revealed by its genetic contexts in three hosts. J. Bacteriol. 184:2411-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naito, T., K. Kusano, and I. Kobayashi. 1995. Selfish behavior of restriction-modification systems. Science 267:897-899. [DOI] [PubMed] [Google Scholar]
- 33.Nakayama, Y., and I. Kobayashi. 1998. Restriction-modification gene complexes as selfish gene entities: roles of a regulatory system in their establishment, maintenance, and apoptotic mutual exclusion. Proc. Natl. Acad. Sci. USA 95:6442-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newman, A. K., R. A. Rubin, S. H. Kim, and P. Modrich. 1981. DNA sequences of structural genes for EcoRI DNA restriction and modification enzymes. J. Biol. Chem. 256:2131-2139. [PubMed] [Google Scholar]
- 35.Nobusato, A., I. Uchiyama, and I. Kobayashi. 2000. Diversity of restriction-modification gene homologues in Helicobacter pylori. Gene 259:89-98.11163966 [Google Scholar]
- 36.Nobusato, A., I. Uchiyama, S. Ohashi, and I. Kobayashi. 2000. Insertion with long target duplication: a mechanism for gene mobility suggested from comparison of two related bacterial genomes. Gene 259:99-108.11163967 [Google Scholar]
- 37.O'Connor, C. D., and G. O. Humphreys. 1982. Expression of the EcoRI restriction-modification system and the construction of positive-selection cloning vectors. Gene 20:219-229. [DOI] [PubMed] [Google Scholar]
- 38.Pedersen, K., A. V. Zavialov, M. Y. Pavlov, J. Elf, K. Gerdes, and M. Ehrenberg. 2003. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112:131-140. [DOI] [PubMed] [Google Scholar]
- 39.Roberts, R. J., M. Belfort, T. Bestor, A. S. Bhagwat, T. A. Bickle, J. Bitinaite, R. M. Blumenthal, S. Degtyarev, D. T. Dryden, K. Dybvig, K. Firman, E. S. Gromova, R. I. Gumport, S. E. Halford, S. Hattman, J. Heitman, D. P. Hornby, A. Janulaitis, A. Jeltsch, J. Josephsen, A. Kiss, T. R. Klaenhammer, I. Kobayashi, H. Kong, D. H. Krüger, S. Lacks, M. G. Marinus, M. Miyahara, R. D. Morgan, N. E. Murray, V. Nagaraja, A. Piekarowicz, A. Pingoud, E. Raleigh, D. N. Rao, N. Reich, V. E. Repin, E. U. Selker, P.-C. Shaw, D. C. Stein, B. L. Stoddard, W. Szybalski, T. A. Trautner, J. L. Van Etten, J. M. B. Vitor, G. G. Wilson, and S.-Y. Xu. 2003. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 31:1805-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenberg, J. M., H. W. Boyer, and P. Greene. 1981. The structure and function of the EcoRI restriction endonuclease. Gene Amplif. Anal. 1:131-164. [PubMed] [Google Scholar]
- 41.Rubin, R. A., and P. Modrich. 1977. EcoRI methylase. Physical and catalytic properties of the homogeneous enzyme. J. Biol. Chem. 252:7265-7272. [PubMed] [Google Scholar]
- 42.Sadykov, M., Y. Asami, H. Niki, N. Handa, M. Itaya, M. Tanokura, and I. Kobayashi. 2003. Multiplication of a restriction-modification gene complex. Mol. Microbiol. 48:417-427. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 45.Som, S., and S. Friedman. 1993. Autogenous regulation of the EcoRII methylase gene at the transcriptional level: effect of 5-azacytidine. EMBO J. 12:4297-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Surby, M. A., and N. O. Reich. 1996. Contribution of facilitated diffusion and processive catalysis to enzyme efficiency: implications for the EcoRI restriction-modification system. Biochemistry 35:2201-2208. [DOI] [PubMed] [Google Scholar]
- 47.Tao, T., J. C. Bourne, and R. M. Blumenthal. 1991. A family of regulatory genes associated with type II restriction-modification systems. J. Bacteriol. 173:1367-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsuchimoto, S., Y. Nishimura, and E. Ohtsubo. 1992. The stable maintenance system pem of plasmid R100: degradation of PemI protein may allow PemK protein to inhibit cell growth. J. Bacteriol. 174:4205-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yakushi, T., T. Tajima, S. Matsuyama, and H. Tokuda. 1997. Lethality of the covalent linkage between mislocalized major outer membrane lipoprotein and the peptidoglycan of Escherichia coli. J. Bacteriol. 179:2857-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, J., Y. Zhang, L. Zhu, M. Suzuki, and M. Inouye. 2004. Interference of mRNA function by sequence-specific endoribonuclease PemK. J. Biol. Chem. 279:20678-20684. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, Y., J. Zhang, K. P. Hoeflich, M. Ikura, G. Qing, and M. Inouye. 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12:913-923. [DOI] [PubMed] [Google Scholar]