Abstract
Bacillus anthracis, a gram-positive, spore-forming bacterium, is the etiological agent of anthrax. It belongs to the Bacillus cereus group, which also contains Bacillus cereus and Bacillus thuringiensis. Most B. anthracis strains are sensitive to phage γ, but most B. cereus and B. thuringiensis strains are resistant to the lytic action of phage γ. Here, we report the identification of a protein involved in the bacterial receptor for the γ phage, which we term GamR (Gamma phage receptor). It is an LPXTG protein (BA3367, BAS3121) and is anchored by the sortase A. A B. anthracis sortase A mutant is not as sensitive as the parental strain nor as the sortase B and sortase C mutants, whereas the GamR mutant is resistant to the lytic action of the phage. Electron microscopy reveals the binding of the phage to the surface of the parental strain and its absence from the GamR mutant. Spontaneous B. anthracis mutants resistant to the phage harbor mutations in the gene encoding the GamR protein. A B. cereus strain that is sensitive to the phage possesses a protein similar (89% identity) to GamR. B. thuringiensis 97-27, a strain which, by sequence analysis, is predicted to harbor a GamR-like protein, is resistant to the phage but nevertheless displays phage binding.
Bacillus anthracis, the etiological agent of anthrax, belongs to the Bacillus cereus group which also comprises Bacillus cereus and Bacillus thuringiensis. B. cereus, a ubiquitous soil bacterium, is generally considered an opportunistic pathogen, and some strains cause serious infections. B. thuringiensis is an entomopathogen. On a genetic basis, these three bacteria are specialized variants of a single species (12). One of the criteria used, before the advent of molecular biology, to differentiate B. anthracis from the other members of the group was sensitivity to the phage γ. Indeed, almost all B. anthracis isolates are γ phage sensitive, whereas most B. cereus and B. thuringiensis strains are γ phage resistant. The γ phage was first reported in 1955 as a variant of phage W isolated in 1951 (6, 22). The γ phage has an icosahedral head and long, sheathless, noncontractile tail (30, 43).
The use of the γ phage for B. anthracis identification led to the search for its receptor. There has been less work on bacterial phage receptors in gram-positive than in gram-negative bacteria. In Bacillus subtilis, teichoic acid has been described as the receptor for bacteriophage SP50, but other cell wall components are also involved in phage-bacterium interaction (2). In Lactobacillus lactis, saccharides are involved in initial phage binding. In some cases, this is followed by protein-dependent secondary binding (9, 42). Contradictory conclusions have been proposed in different papers concerning the γ phage receptor. Lantos and Ivanovics reported that γ phage binding was lost after a treatment with 5% trichloracetic at 90°C and then trypsin, suggesting that the receptor might be a protein (15). Watanabe and Shiomi suggested that one or a combination of the following substances, meso-Dap, d-glucosamine, d-galactosamine, d-mannosamine, constituents of the peptidoglycan or the associated polysaccharide (23), and l-lysine, constitute the phage receptor site (44). They excluded the protein hypothesis due to maintenance of phage binding after solvent treatment. However, the treatments they used would only affect membrane proteins, not peptidoglycan-anchored proteins, which were not known at that time.
Published data are therefore consistent with the γ phage receptor being a peptidoglycan-anchored protein. A cell-wall-protein-anchoring mechanism has been described in B. anthracis, involving a polysaccharide, composed of galactose, N-acetylglucosamine, and N-acetylmannosamine (23). This polysaccharide is covalently anchored to the peptidoglycan stricto sensu. For proteins to bind to it, the polysaccharide has to be pyruvylated, and mutants unable to pyruvylate the polysaccharide, and thus to anchor the corresponding proteins, have been isolated. These mutations map to an operon termed csa (23). The proteins that bind to this polysaccharide do so by a domain termed SLH, for S-layer homology. This domain was initially described in S-layer proteins as a triple repetition in their N termini of a 50-amino-acid motif (17). It has since been found in many gram-positive cell-wall-anchored proteins. Another anchoring mechanism for gram-positive bacterial surface proteins involves sortases. Sortase catalyzes covalent anchoring of LPXTG-harboring proteins to the peptide moiety of peptidoglycan (19, 33). The sequence of the B. anthracis genome suggests that it has three sortases and 14 LPXTG proteins (27, 29). The existence of more than one sortase has been suggested in other bacteria, and experimentally demonstrated in some (3-5, 21, 26, 27, 38). It has been proposed that the sortases be divided into four classes, A to D, on basis of sequence (8). Recent data for various gram-positive organisms suggest that there is a major sortase, SrtA, that generally anchors many LPXTG proteins, and that the other sortases have a more restricted subset of substrates, seemingly linked to a particular function and to specific LPXTG motif sequence. For example, sortase B probably only anchors iron acquisition proteins (5, 21). Other non-A non-B sortases seem dedicated to pilus assembly, or morphological differentiation (3, 8, 21, 38). The genes encoding these sortases and their substrates are often clustered, and are sometimes in the same operon. The LPXTG proteins have an LPXTG motif followed by a hydrophobic tail in their C-terminal region. While the protein translocation is arrested in the membrane due to the hydrophobic region, the sortase covalently links the threonine in the LPXTG motif to an amino-group of a peptidoglycan precursor amino-acid in a two-step transpeptidation reaction. The resulting molecule is covalently incorporated into the peptidoglycan (19, 37). In a sortase A mutant, the LPXTG proteins that depend on it for anchoring are found in the supernatant and associated with the membrane (14)
Here, we tried to identify the receptor of the phage γ. The sensitivity of a sortase A mutant was lower than that of the parental strain. An LPXTG-harboring protein was identified as being required for phage sensitivity and its involvement in phage binding was confirmed by electron microscopy.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Escherichia coli TG1 (31) was used as a host for the cloning experiments. E. coli HB101 harboring pRK24 was used for mating experiments. The B. anthracis strains and plasmids used or constructed in this work are listed Table 1. E. coli cells were cultured in Luria (L) broth or on L agar plates. B. anthracis, B. thuringiensis and B. cereus strains were grown in brain-heart infusion (BHI; Difco Laboratories) or on BHI agar plates. To induce pag expression, bacteria were grown on CAP plates in a 5% CO2 atmosphere (35). Antibiotics were used as previously described (36).
TABLE 1.
B. anthracis strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Genotype or description | Source or reference |
|---|---|---|
| Bacillus anthracis | ||
| 7702 | Sterne strain; pXO1+ | Laboratory stock |
| 7SBON30 | 7702 ΔsrtA (pSON30) | This work |
| 7SBON40 | 7SBON30 srtA complemented (pSRTA40) | This work |
| 7SDG30 | 7702 ΔgamR (pGAR30) | This work |
| 7SDG30(pGAR40) | Transconjugant 7SDG30 strain harboring pGAR40; (ΔgamR + gamR) Kanr | This work |
| 7SBTR30 | 7702 ΔsrtB (pSTR30) | This work |
| 7SBTO30 | 7702 ΔsrtC (pSTO30) | This work |
| 7SBONTO | 7702 ΔsrtA ΔsrtC (pSON30, pSTO30E) | This work |
| 7SBTRTO | 7702 ΔsrtB ΔsrtC (pSTR30, pSTO30E) | This work |
| SM96 | 9131 (plasmidless derivative of 7702) ΔcsaA ΔcsaB | 23 |
| B. cereus WW3 | B. cereus γ phage-sensitive strain | 30 |
| B. thuringiensis 97-27 | B. thuringiensis strain harboring a gamR homologue | Laboratory stock |
| Plasmid | ||
| pUC19 | Cloning vector | 45 |
| pGEM-T-easy | Cloning vector | Qiagen |
| pAT113 | Conjugative suicide plasmid used for gene inactivation in B. anthracis Kanr, Ermr | 40 |
| pAT187 | Conjugative replicating plasmid used for complementation in B. anthracis Kanr | 39 |
| pAT28 | Conjugative replicating plasmid used for complementation in B. anthracis | 41 |
| pACP1 | PUC harboring pag gene | 7 |
| pACP50 | pAT28 harboring the pag gene with NdeI overlapping ATG | Laboratory stock |
| pBAK | Conjugative suicide plasmid used for gene inactivation in B. anthracis Kanr | 24 |
| pSpecH+1 | pUC19 carrying a Spc cassette ending with an rbs and an ATG | 23 |
| pSpecH+2 | pUC19 carrying a Spc cassette ending with an rbs and an ATG | 23 |
| pUC1318Spc | PUC1318 carrying a Spc cassette | 25 |
| pUC1318Erm | PUC1318 carrying an Erm cassette | 24 |
| pPPA10 | Integrative vector harboring the pag promoter region | This work |
| pSON10 | pUC19 carrying the srtA gene | This work |
| pSON20 | pUC19 carrying the inactivated srtA gene Spcr | This work |
| pSON30 | pAT113 carrying the inactivated srtA gene Spcr | This work |
| pSTR10 | pUC19 carrying the srtB gene | This work |
| PSTR20 | pUC19 carrying the inactivated srtB gene Spcr | This work |
| PSTR30 | pAT113 carrying the inactivated srtB gene Spcr | This work |
| pSTO10 | pUC19 carrying the srtC gene | This work |
| PSTO20 | pUC19 carrying the inactivated srtC gene Spcr | This work |
| PSTO30 | pAT113 carrying the inactivated srtC gene Spcr | This work |
| PSTO20E | pUC19 carrying the inactivated srtC gene Ermr | This work |
| PSTO30E | pBAK carrying the inactivated srtC gene Ermr | This work |
| pSRTA10 | pGEM carrying the sortA gene with an NdeI site | This work |
| pSRTA40 | pPPA10 carrying the srtA gene under the control of the pag promoter | This work |
| pGAR10 | pGEM-T-easy carrying the gamR gene | This work |
| pGAR20 | pGEM-T-easy carrying the inactivated gamR gene Spcr | This work |
| pGAR30 | pAT113 carrying the inactivated gamR gene Spcr | This work |
| pGAR40 | pAT187 carrying the gamR gene | This work |
| Oligonucleotide | ||
| pU | CGCCAGGGTTTTCCCAGTCACGAC | |
| pR | AGCGGATAACAATTTCACACAGGA | |
| SortA-5′ | CATATGAATAAGCAAAGAATTTATAGTATA | |
| srt1-5′ | GGTATTGTATCGACTGTTCTTTATAAAGTT | |
| srt1-3′ | GGAATGATTATCGGACAAATTTTATTTACA | |
| Sort2-5′ | CCGATCTGTGCAATAGCACTTTTAACAGTA | |
| Sort2-3′ | CGCTAATATTTCAAATTCTGTATGTGTTAA | |
| Sort3-5′ | CCTCGCAAACGGATGAAGATAGAGAAGCGA | |
| Sort3-3′ | GGAAACAATGCCAGGCTTTTTAGGATTAGA | |
| Prebas5′ | GTCGCCAACCTTTTCGGTCTTTTGGAAAGA | |
| Bas5′ | GGATCCGTCATGGGGACAAGTAGAAGCTGAAAC | |
| Bas3′ | AAGCTTAGCTGCTTTACGTCTCCATAATACATATGC | |
| Postbas3′ | AAGCTTTACAGCTGCCTCTTCACATCCGGC |
DNA manipulation.
Plasmid extraction, endonuclease digestion, ligation and agarose gel electrophoresis were carried out as described by Maniatis et al. (18). PCR amplifications were carried out with long-range, high-fidelity Taq, according to the manufacturer's instructions (Roche), or with Vent polymerase. Sequences of the plasmids were determined either from PCR products after an amplification step or directly from the plasmid DNA. All sequencing reactions were carried out by Genome Express Sequencing. The B. anthracis mutant gamR genes and the B. cereus gamR homologue gene were sequenced after PCR amplification from the chromosomes using the Bas5′ and Bas3′ oligonucleotides (Table 1).
Construction of recombinant strains.
Recombinant plasmids were transferred from E. coli to B. anthracis, B. thuringiensis, or B. cereus by heterogramic conjugation (39). Allelic exchange was carried out as described previously (28).
Genetic constructions.
The 1.85-, 1.54-, and 1.68-kb DNA fragments carrying the srtA (BA0688, BAS0654), the srtB (BA4783, BAS4708), and the srtC (BA5069, BAS4438) genes and that of 2.53 kb carrying the gamR (BA3367, BAS3121) gene were obtained by PCR from B. anthracis 7702 chromosomal DNA, using the primers (Table 1) srt1-5′ and sort1-3′ for the srtA gene, Sort3-5′ and Sort3-3′ for the srtB gene, Sort2-5′ and Sort2-3′ for the srtC gene, and prebas5′ and bas3′ for the gamR gene.
The srtA fragment was inserted into pUC19 giving pSON10. pSON20 was obtained by replacing the 800-bp BclI-SnaB1 fragment, from nucleotides (nt) 795 (+1 corresponding to the A of the translational initiation codon) to the end of the srtA fragment, with the spectinomycin resistance cassette from pUC1318Spc. The ΔsrtA construction was excised from pSON20, by digesting with EcoRI-HindIII, and inserted into the conjugative suicide vector pAT113 giving pSON30.
To complement the ΔsrtA mutant, the srtA gene was placed under pag promoter control as follows. First, an integrative plasmid harboring a NdeI cloning site 3′ to the pag promoter was constructed. An NdeI site was introduced overlapping the pag ATG codon by directed mutagenesis of the 6-kb BamHI fragment in pACP1 (7). The 3.4-kb Xho1-BamHI fragment was ligated into the replicative vector pAT28, giving rise to pACP50. The 2-kb SphI fragment was then inserted into the SphI site of pAT113 giving pPPA10. Then, the srtA gene was amplified by PCR using the primers sortA-5′ and srt1-3′. The primer sortA-5′ contains a NdeI site overlapping the ATG initiation codon of the srtA gene. The DNA fragment was inserted into pGemT-easy giving pSRTA10. The srtA gene was excised from pSRTA10, digested with NdeI and HindIII and inserted into pPPA10, giving pSRTA40.
The srtB fragment was inserted into pUC19 giving pSTR10. pSTR20 was obtained by replacing the 479 bp Acc651-Acc1 fragment, from nt-45 (+1 corresponding to the A of the translational initiation codon) to 220 nt 5′ from the end of the srtB fragment, with the spectinomycin-resistance cassette from pUC1318Spc. The ΔsrtB construction was excised from pSTR20, by digesting with EcoRI-HindIII, and inserted into the conjugative suicide vector pAT113 giving pSTR30.
The srtC fragment was inserted into pUC19 giving pSTO10. pSTO20 was obtained by replacing the 542-bp BamHI-HpaI fragment, from nt 33 (+1 corresponding to the A of the translational initiation codon) to 55 nt 5′ from the end of srtC gene, with the non-polar spectinomycin-resistance cassette from pSpecH + 2 (23). The ΔsrtC construction was excised from pSTO20, by digesting with HindIII-EcoRI and inserted into the conjugative suicide vector pAT113 giving pSTO30. pSTO20E was obtained by replacing the same BamHI-HpaI fragment with the Erm cassette from pUC1318Erm. The pSTO20E ΔsrtC construction was amplified using the pU pR oligonucleotides and inserted into the conjugative suicide vector pBAK digested by SmaI, giving pSTO30E.
The DNA fragment carrying the gamR sequence was inserted into pGemT-easy giving pGAR10. pGAR20 was obtained by replacing the 1.1-kb Bcl1-Cla1 fragment in pGAR10, from nt 317 (+1 corresponding to the A of the translational initiation codon) to 302 nt 5′ from the end of the gamR gene with the non polar spectinomycin-resistance cassette from pSpecH + 1. The ΔgamR construction was excised from pGAR20, by digesting with SphI-EcoRI, and inserted into pAT113, yielding pGAR30.
To complement the ΔgamR mutant strain, a 2.5-kb DNA fragment harboring the gamR gene with its own promoter was amplified by PCR with Vent polymerase from B. anthracis strain 7702 chromosomal DNA, using primers prebas5′ and postbas3′. This fragment was inserted into the SmaI site of the self-replicating shuttle vector, pAT187, giving pGAR40.
Bacteriophages.
A γ phage stock active against B. anthracis was obtained from PCB Turnbull (NMRC, U.S. Navy). Phage was amplified by spreading 100 μl of a log-phase culture of B. anthracis 7702 and 100 μl of the phage preparation onto BHI Petri dishes and incubating for 16 h at 37°C. The lawns were then taken up in one ml of BHI and centrifuged at 4°C for 20 min at 6,000 rpm. The supernatants were filtered through a 0.45-μm-pore-size membrane. These filtrates usually had titers of approximately 5 × 109 PFU per ml, as assayed on B. anthracis strain Sterne (7702).
Sensitivity test.
Exponential cultures of B. anthracis cells were spread on BHI petri dishes and 20 μl of eight consecutive 10-fold dilutions (from non-diluted to 10−7) were spotted onto the surface. The plaques were observed after 16 h of incubation at 37°C. The sensitivity of B. anthracis wild-type or mutant strains was defined by determining the number of plaques at the dilution not giving rise to confluent lysis on that strain. For some strains, sensitivity was also determined during growth in liquid medium. BHI medium was inoculated with the bacteria, and when the optical density at 600 nm (OD600) reached 0.5, the culture was divided into two halves. To one-half, 2 × 107 PFUs were added per ml and incubation resumed. The OD600 was followed for another 7 h.
Resistant mutant selection.
To obtain spontaneous γ phage-resistant mutants, undiluted phage preparations were spotted on a lawn of B. anthracis 7702. Colonies appearing in the lysis zone were isolated.
Electron microscopy.
Bacteria were grown to an OD600 of 0.2. Approximately 106 phage particles were then added to 100 μl of culture and left to adsorb at 37°C for 30 min. The samples were then maintained at 0°C until further treatment. Ten-μl drops of these preparations were applied to grids, and the grids rinsed twice with 150 μl drops of 10 mM Tris-HCl, pH 7.5, to eliminate remaining salts. Grid processing and negative staining were as previously described (25).
RESULTS
A cell wall-anchored protein is required for sensitivity to γ phage.
The γ phage receptor is probably a cell wall-associated protein. Indeed, some of the substances described as determinants of the phage receptor site are constituents of the peptidoglycan or of the associated polysaccharide (23, 44). To identify the γ phage receptor, we assayed the resistance of B. anthracis mutants deficient in cell wall protein anchoring. The first mutant tested was strain SM96, a csa mutant which cannot pyruvylate the cell-wall-associated polysaccharide of B. anthracis and is thus deficient in SLH-harboring-protein anchoring (23). This mutant had the same sensitivity to the lytic action of the γ phage as the parental strain (data not shown). The γ phage bacterial receptor is therefore not an SLH-harboring protein.
We then assayed the resistance of B. anthracis mutants deficient in LPXTG cell wall protein anchoring. A mutant was constructed in the sortase gene, srtA, encoding the sortase with the greatest similarity to that involved in the anchoring of most LPXTG-proteins in other bacteria (27). This allowed testing the involvement of as many LPXTG-proteins as possible with a single mutant. The srtA mutant was less sensitive than the parental strain to the lytic action of phage γ on solid medium (Table 2), but was not resistant. The srtA mutant was complemented by an integrative plasmid harboring the srtA gene under the control of the pag promoter. The strain was grown under conditions inducing pag expression, and sensitivity to phage γ lysis was restored. Indeed, with the same phage stock, the sensitivity score for the complemented strain was 7.5 × 109 PFU/ml and that for the parental strain 7702 was 5 × 109 PFU/ml. This implicates an LPXTG protein in phage γ lysis. LPXTG proteins are surface proteins, and therefore an LPXTG protein could be, or be part of, the phage γ binding site.
TABLE 2.
γ phage lytic activity on various Bacillus strains
| Plasmid | Strain
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| B. aa parental strain | B. a ΔsrtA mutant | B. a ΔgamR mutant | B. a ΔsrtB mutant | B. a ΔsrtC mutant | B. a ΔsrtA ΔsrtC mutant | B. a ΔsrtB ΔsrtC mutant | B. ca WW3 | B. ta 97-27 | |
| / | 5 × 109b | 5 × 107 | NL | 5 × 109 | 3.5 × 109 | 2 × 107 | 2 × 109 | 1.5 × 107 | NL |
| pGAR40a | ND | 1010 | 8 × 109 | ND | ND | 5 × 109 | 1010 | 2 × 107 | ND |
B. a, B. c and B. t stand for B. anthracis, B. cereus and B. thuringiensis respectively. pGAR40, gamR-harboring plasmid.
The numbers correspond to the phage plating efficiency on the given strain. All plaques were of the same size NL: not lysed ND: not done.
BA3367 (BAS3121) is required for sensitivity to γ phage.
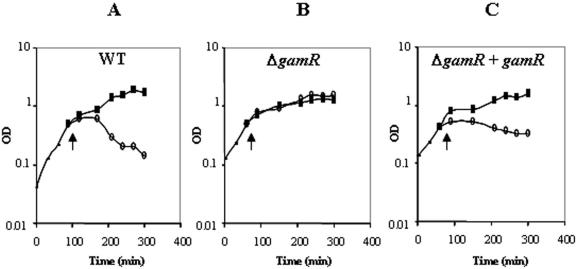
Proteins of a given sortase repertoire contain a defined LPXTG motif sequence. We analyzed the B. anthracis genome in silico and found six to eight putative LPXTG proteins possibly anchored by sortase A. During revision, another in silico analysis reported that B. anthracis sortase A could anchor 7 LPXTG proteins (10). γ phage lysis is nearly B. anthracis specific, so we searched for putatively sortase A-dependent LPXTG proteins only found in B. anthracis. When this work was being carried out, B. cereus ATCC 14579 was the closest relative of B. anthracis for which the entire sequence was available (13). The BA3367 (BAS3121) open reading frame encodes an LPXTG protein that is absent from the ATCC 14579 genome. Other B. cereus strains have been sequenced since, and the BA3367 open reading frame is, in fact, not unique to B. anthracis. Neverthless, for the reasons explained above, we deleted this gene and tested the mutant, 7SDG30, for phage γ resistance (Fig. 1; Table 2). 7SDG30 was resistant to phage γ lytic action in tests on solid medium (Table 2). It was also tested for phage γ lytic action during growth in liquid medium (Fig. 1A and B). The OD600 of cultures of the parental strain declined in the presence of the phage, but the mutant strain grew equally well in its presence and absence (Fig. 1A and B). Thus, the mutant is totally resistant to the lytic action of phage γ. The mutant was complemented using a replicative plasmid carrying BA3367 (BAS3121) with its own promoter. The complemented strain was assayed by both methods for its sensitivity to γ phage (Fig. 1C; Table 2): it was as sensitive as the parental strain to the lytic action of phage γ. The resistance of the BA3367 (BAS3121) mutant is therefore a consequence of the deletion. Presumably, BA3367 (BAS3121) encodes a protein that is, or part of, the receptor for the γ phage. The protein was named GamR (Gamma phage receptor).
FIG. 1.
The γ phage requires the presence of BA3367 (BAS3121) to exert its lytic action. γ Phage particles were added (arrows, open circles) or not added (closed squares) during exponential growth to the control 7702 strain (A), the BA3367 (BAS3121) (gamR)-deleted mutant (7DG30) (B), and the BA3367 (BAS3121)-deleted mutant complemented with the gamR gene [7DG30 (pGAR40)] (C), and the OD600 was monitored for another 7 h.
A plasmid harboring gamR was introduced into the srtA mutant, and the strain tested for γ phage sensitivity (Table 2). The complemented strain was more sensitive than the srtA mutant, and was as sensitive as the parental and ΔgamR complemented strains. Presumably, GamR was not anchored in the srtA mutant strain but nevertheless overproduction of nonanchored GamR protein conferred sensitivity.
GamR is anchored by the sortase A.
The GamR mutant is totally resistant to the lytic action of phage γ, whereas the sortase A mutant is only partially resistant. Either GamR serves as a receptor despite not being anchored, or it could be anchored by another sortase. To test this possibility, single mutants were constructed in each of the other two sortases, namely sortase B and sortase C. Both sortase B and sortase C single mutants displayed the same sensitivity as the wild-type strain (Table 2). However, efficient sortase A anchoring of GamR could mask the absence of a weak anchoring by another sortase. We therefore tested double mutants. The sensitivity of the double sortase A sortase C mutant was the same as that of the sortase A mutant (Table 2). Furthermore, the overproduction of GamR in that strain and in the Δ srtA strain gave similar sensitivity scores. Thus, sortase C is not responsible for residual phage sensitivity of the sortase A mutant strain. The sortase B sortase C double mutant was as sensitive as the wild-type strain and the overproduction of GamR did not modify the sensitivity. This strongly suggested that sortase B is not involved in GamR anchoring either (Table 2). This suggested that GamR is exclusively sortaseA dependent.
GamR is required for γ phage binding.
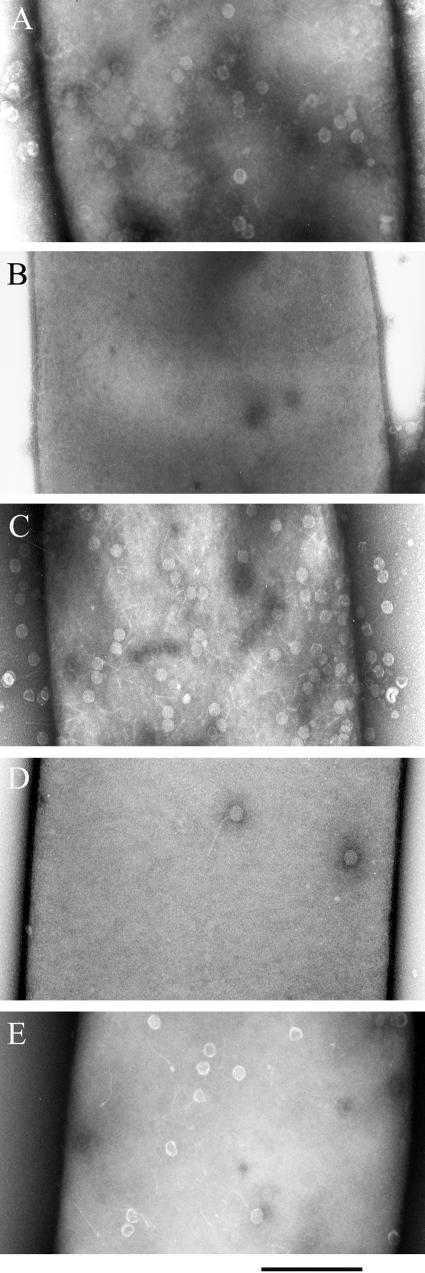
We studied γ phage binding to the bacteria by electron microscopy (Fig. 2). The γ phage bound uniformly over the surface of the parental strain (Fig. 2A). There was no detectable phage binding to the GamR mutant strain (Fig. 2B), but phage binding was restored in the complemented strain (Fig. 2C). This confirms that GamR is required for γ phage binding. In the complemented strain, the gamR gene was under its own promoter on a multi-copy plasmid, and therefore there was probably more GamR protein than in the parental strain. Consistent with this, more phage particles were bound to the surface of the complemented strain than to that of the parental strain (Fig. 2A versus 2C). This indicates a direct link between the number of GamR protein molecules anchored at the surface of the bacterium and the number of bound phage particles. The gamR gene was similarly introduced into the srtA mutant strain (Fig. 2D and 2E). The ΔsrtA mutant bound phage particles but bound fewer than the parental strain, consistent with the sensitivities (Fig. 2D). The ΔsrtA mutant overproducing GamR bound more phage particles than the control ΔsrtA mutant (Fig. 2D versus 2E), but fewer than the GamR mutant strain complemented with GamR (Fig. 2C versus 2E).
FIG. 2.
Binding of γ phage particles to the surface of B. anthracis. γ phage was added to exponentially growing cultures of strains 7702 (A), 7DG30 (gamR-deleted mutant) (B), 7DG30 (pGAR40) (gamR-deleted mutant complemented with the gamR gene) (C), 7SBON30 (srtA-deleted mutant) (D), and 7SBON30 (pGAR40) (srtA-deleted mutant complemented with the gamR gene) (E). The bound particles were visualized by electron microscopy. Bar, 500 nm.
GamR integrity, γ phage binding, and bacterial sensitivity.
The susceptibility of a bacterium to phage infection is primarily dependent on bacteriophage attachment to receptors (16). Spontaneous resistant mutants of the B. anthracis parental strain were therefore sought and indeed, colonies grew in γ phage lysis areas. Four mutants, from two independent lysis zones, were studied (see Materials and Methods). The gamR gene (595 codons in the parental strain) in each was amplified and sequenced. In all the four mutants, gamR had a non-sense mutation (Fig. 3). Two mutations (clones 1 and 3) were 1-bp deletions near the start of the sequence. These mutations give rise to stop codons at the 94th and 104th codons (Fig. 3, clone 3, clone 1). The other two mutations were near the 3′ end of the gene (Fig. 3, clone 2, clone 4). Both modify the LPKTG motif, yielding proteins that, if stable, cannot be anchored to the peptidoglycan by sortase A. Furthermore, the mutations generating either subsitutions followed by a stop codon in the membranous domain (clone 2) or a stop codon (clone 4) before that domain, these two mutant proteins are expected to be devoid of the membranous domain. In clones 1, 2, and 3, the mutations were single bp deletions. However, clone 4 is more complex with four consecutive bp substituted, followed by a 2-bp insertion. These mutants were tested for complementation with the wild-type gamR gene, and γ phage sensitivity was restored in all cases (data not shown).
FIG. 3.
Mutations in the gamR gene and deduced GamR proteins harbored by the spontaneous B. anthracis γ phage-resistant clones. The gamR wild-type sequence (wt) and the mutant sequences are given from nt 244 to 325 and 1651 to 1732 to show mutations in clones 1 and 3, and 2 and 4, respectively. The deduced translated sequences are indicated below the nucleotide sequences. The mutations are highlighted in bold underlined characters in the wild-type sequences. Where a bp is deleted, the surrounding nucleotides are underlined in the mutant sequence; the substitution/insertion is also underlined.
Phage γ is nearly specific to B. anthracis strains. However, some B. cereus strains are sensitive to its lytic action. In our hands, B. cereus WW3 was lysed by the phage (Table 2), but was not as sensitive as B. anthracis: with the same phage stock the sensitivity score was 1.5 × 107 PFU/ml for B. cereus WW3 and 5 × 109 PFU/ml for B. anthracis (Table 2). We tested WW3 for a GamR protein by amplifying a DNA fragment and sequencing (accession number bankit705279 DQ057580). A fragment of 1.6 kb, which is the expected size, was obtained. The deduced amino acid sequence shows 89% identity with GamR; it includes a 21-amino-acid insertion at the 210th codon but is otherwise 93% identical to GamR. We studied γ phage particle binding to WW3 (Fig. 4A). Note that B. cereus and B. thuringiensis, unlike B. anthracis, harbor appendages (Fig. 4B and D). Phage particles bound to the surface (Fig. 4A). The B. anthracis gamR gene was introduced into WW3 (Table 2), but this did not enhance the sensitivity of the strain. Other strains from the B. cereus group, with sequences similar to GamR have been described since this work was started. For example, B. thuringiensis 97-27 possesses an LPXTG protein sharing 96.8% identity with GamR (BT9727_3108). However, γ phage was unable to lyse B. thuringiensis strain 97-27 (Table 2). Interestingly, electron microscopy analysis revealed that phage bound to the surface of strain 97-27 (Fig. 4C, D) although fewer particles bound to strain 97-27 than to the sensitive bacteria.
FIG. 4.
Binding of γ phage particles to the surface of a sensitive B. cereus WW3 strain (A) and a resistant B. thuringiensis 97-27 strain (C). The same strains are shown without addition of phage particles (B and D, respectively). The experiment was carried out as in Fig. 2. Bar, 500 nm.
DISCUSSION
We deleted a single gene from B. anthracis and this rendered it resistant to γ phage lytic action. The deleted gene encodes GamR, a surface-anchored protein. Moreover, the phage did not adsorb onto the surface of this mutant strain. Four spontaneous B. anthracis γ-phage-resistant mutants were analyzed and all harbor mutations in the gamR gene. GamR overproduction in the ΔgamR mutant strain led to more phage particles binding to the bacterial surface. This indicates that GamR is directly involved in the γ phage receptor. Interestingly, sequence analysis suggests that BA3367 (BAS3121), encoding GamR, is the first gene of an operon putatively involved in cobalt transport. Indeed, BA3366 to BA3363 encode proteins with similarities to lipoproteins, cobalt transport protein (ABC type), ABC ATPase, and a membrane protein with six transmembrane-domains, respectively. That the phage receptor is part of a transporter is not unusual.
The difference in sensitivity between the ΔsrtA and the ΔgamR mutants could be the consequence of the protein being present in the ΔsrtA mutants, whereas it is totally absent from the ΔgamR mutants. The B. anthracis sortase repertoires have not yet been defined, but available knowledge about sortases suggests that a given LPXTG-harboring protein is anchored by a given sortase (8) (and references herein). The involvement of sortase B in GamR anchoring is improbable: the plating efficiency of the phage was unaffected in ΔsrtB and ΔsrtB ΔsrtC mutants; sortase B in other species is extremely specific for the NPQTN motif and GamR has a classical LPKTG motif (20); and sortase B, is specialized in iron uptake in all bacteria in which it has been studied (5). The sequences of the locus to which gamR belongs do not suggest iron acquisition proteins. We constructed ΔsrtC mutants to test the involvement of sortase C. The ΔsrtC, and ΔsrtA ΔsrtC mutants were as sensitive to the γ phage as the parental and the ΔsrtA strains, respectively, indicating that sortase C is not involved in GamR anchoring. Thus, GamR is anchored by sortase A.
In the srtA mutant, GamR like other LPXTG proteins in sortase mutants, is presumably not covalently linked to the peptidoglycan but only transiently associated with the membrane (14). A direct consequence is that at any given time, fewer GamR molecules are exposed at the bacterial surface. The phage may still bind, but the instability of the non-anchored proteins at the surface might reduce the efficiency of infection, lowering the plating efficiency. It is probable that the overproduction of GamR in the srtA mutant partially compensates for this effect. Indeed, the plating efficiency is restored in the GamR-overproducing strain. However, it bound fewer phage particles than did the ΔgamR strain complemented with GamR as assessed by electron microscopy. This reinforces the view that fewer GamR molecules are exposed at the surface of a ΔsrtA mutant strain than at that of a sortase A wild-type strain. Plating efficiency may be proportional to the binding efficiency only up to a certain threshold, where lysis becomes independent of the binding of additional particles. Note that two of the gamR mutants we obtained (clones 2 and 4) carried substitutions in the LPKTG and substitutions and/or a stop codon in the following hydrophobic sequences. These mutants are unsurprisingly resistant because, not only are the putative proteins not properly anchored (lacking the LPKTG motif), but also they are not even expected to linger in the membrane.
The adsorption of phages onto the bacterial surface has been suggested to proceed in two steps (1). The first is reversible binding to a recognition molecule, followed by an irreversible step, probably the DNA ejection step or an irreversible adsorption. This second step can be phage dependent as is the case for φ29 (11). It can also depend on molecular aggregates, or on a bacterial protein, as described for B. subtilis YueB which is required for irreversible binding of phage SPP1 (16, 32). In our model, GamR is the recognition protein involved in the first step. The four spontaneous mutants analyzed were all GamR mutants. This suggests that either there is no other host protein involved in γ phage infection of B. anthracis, or that mutants in the bacterial component(s) other than in GamR are more difficult to obtain, as would be the case for essential or metabolically important proteins.
A B. cereus strain and a B. thuringiensis strain were tested for their sensitivity to the lytic action of phage γ. B. cereus WW3 was confirmed to be sensitive and B. thuringiensis 97-27 was shown to be resistant. Interestingly, both possess a GamR-like protein very similar but not identical to GamR (89 and 96% identity, respectively). Electron microscopy showed that γ phage binds to the surface of both strains. Thus, in both cases the protein is present at the surface. However, fewer phages were adsorbed by these strains than by the B. anthracis strain. This could be due to less GamR-like protein being present at the surface of the bacteria. Alternatively, the sequence differences could affect the recognition site and modify the affinity. Furthermore a second component, necessary for efficient bacterium-γ phage interaction, such as B. subtilis YueB (32), may be divergent in B. cereus. The complete insensitivity of the B. thuringiensis 97-27 strain suggests that this putative second component is completely absent. Alternatively, a protein of phage origin could be necessary for phage γ to exert its lytic activity. An obvious candidate is PlyG (34), a γ-phage-encoded lysin that mediates lawn clearance when added to B. anthracis but not B. cereus or B. thuringiensis cultures; its hydrolytic activity specificity reflects that of the γ phage.
Our data also indicate that at least some LPXTG proteins do not have to be properly anchored to display some activity. It may therefore be possible to delete genes encoding sortases that catalyze the anchoring of essential proteins without abolishing viability. Similarly, virulence could be more affected by LPXTG protein deletions than by that of the sortases.
Acknowledgments
P. C. B. Turnbull (NMRC, US Navy) is thanked for the gift of the B. cereus strain WW3 and γ phage stock. We are grateful to M. Moya for the in silico analysis and to J.-L. Ranck for help with the figures.
This work was supported in part by AIP “Microbiologie” INRA-IP 2003/P00244. T.C. was funded by MNRT.
REFERENCES
- 1.Adams, M. H. 1959. Bacteriophages. Wiley Interscience, New York, N.Y.
- 2.Archibald, A. R., K. Glassey, R. S. Green, and W. K. Lang. 1989. Cell wall composition and surface properties in Bacillus subtilis: anomalous effect of incubation temperature on the phage-binding properties of bacteria containing varied amounts of teichoic acid. J. Gen. Microbiol. 135:667-673. [DOI] [PubMed] [Google Scholar]
- 3.Barnett, T. C., A. R. Patel, and J. R. Scott. 2004. A novel sortase, SrtC2, from Streptococcus pyogenes anchors a surface protein containing a QVPTGV motif to the cell wall. J. Bacteriol. 186:5865-5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnett, T. C., and J. R. Scott. 2002. Differential recognition of surface proteins in Streptococcus pyogenes by two sortase gene homologs. J. Bacteriol. 184:2181-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bierne, H., C. Garandeau, M. G. Pucciarelli, C. Sabet, S. Newton, F. Garcia-del Portillo, P. Cossart, and A. Charbit. 2004. Sortase B, a new class of sortase in Listeria monocytogenes. J. Bacteriol. 186:1972-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, E. R., W. Cherry. 1955. Specific identification of Bacillus anthracis by means of a variant bacteriophage. J. Infect. Dis. 96:34-39. [DOI] [PubMed] [Google Scholar]
- 7.Cataldi, A., E. Labruyere, and M. Mock. 1990. Construction and characterization of a protective antigen-deficient Bacillus anthracis strain. Mol. Microbiol. 4:1111-1117. [DOI] [PubMed] [Google Scholar]
- 8.Dramsi, S., P. Trieu-Cuot, and H. Bierne. 2005. Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res. Microbiol. 156:289-297. [DOI] [PubMed] [Google Scholar]
- 9.Dupont, K., F. K. Vogensen, H. Neve, J. Bresciani, and J. Josephsen. 2004. Identification of the receptor-binding protein in 936-species lactococcal bacteriophages. Appl. Environ Microbiol. 70:5818-5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaspar, A. H., L. A. Marraffini, E. M. Glass, K. L. Debord, H. Ton-That, and O. Schneewind. 2005. Bacillus anthracis sortase A (SrtA) anchors LPXTG motif-containing surface proteins to the cell wall envelope. J. Bacteriol. 187:4646-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez-Huici, V., M. Salas, and J. M. Hermoso. 2004. The push-pull mechanism of bacteriophage O29 DNA injection. Mol. Microbiol. 52:529-540. [DOI] [PubMed] [Google Scholar]
- 12.Helgason, E., O. A. Okstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and Kolsto. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis-one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. D. Ehrlich, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87-91. [DOI] [PubMed] [Google Scholar]
- 14.Lalioui, L., E. Pellegrini, S. Dramsi, M. Baptista, N. Bourgeois, F. Doucet-Populaire, C. Rusniok, M. Zouine, P. Glaser, F. Kunst, C. Poyart, and P. Trieu-Cuot. 2005. The SrtA Sortase of Streptococcus agalactiae is required for cell wall anchoring of proteins containing the LPXTG motif, for adhesion to epithelial cells, and for colonization of the mouse intestine. Infect. Immun. 73:3342-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lantos, J., and G. Ivanovics. 1961. The phage receptors of Bacillus anthracis. Acta. Microbiol. Acad. Sci. Hung. 8:379-388. [PubMed] [Google Scholar]
- 16.Lindberg, A. A. 1973. Bacteriophage receptors. Annu. Rev. Microbiol. 27:205-241. [DOI] [PubMed] [Google Scholar]
- 17.Lupas, A., H. Engelhardt, J. Peters, U. Santarius, S. Volker, and W. Baumeister. 1994. Domain structure of the Acetogenium kivui surface layer revealed by electron crystallography and sequence analysis. J. Bacteriol. 176:1224-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Mazmanian, S. K., G. Liu, H. Ton-That, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760-763. [DOI] [PubMed] [Google Scholar]
- 20.Mazmanian, S. K., H. Ton-That, and O. Schneewind. 2001. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 40:1049-1057. [DOI] [PubMed] [Google Scholar]
- 21.Mazmanian, S. K., H. Ton-That, K. Su, and O. Schneewind. 2002. An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. USA 99:2293-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCloy, E. W. 1951. Studies on a lysogenic Bacillus strain. I. A bacteriophage specific for Bacillus anthracis. J. Hyg. 49:114-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mesnage, S., T. Fontaine, T. Mignot, M. Delepierre, M. Mock, and A. Fouet. 2000. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 19:4473-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mesnage, S., E. Tosi-Couture, and A. Fouet. 1999. Production and cell surface anchoring of functional fusions between the SLH motifs of the Bacillus anthracis S-layer proteins and the Bacillus subtilis levansucrase. Mol. Microbiol. 31:927-936. [DOI] [PubMed] [Google Scholar]
- 25.Mesnage, S., E. Tosi-Couture, M. Mock, P. Gounon, and A. Fouet. 1997. Molecular characterization of the Bacillus anthracis main S-layer component: evidence that it is the major cell-associated antigen. Mol. Microbiol. 23:1147-1155. [DOI] [PubMed] [Google Scholar]
- 26.Osaki, M., D. Takamatsu, Y. Shimoji, and T. Sekizaki. 2002. Characterization of Streptococcus suis genes encoding proteins homologous to sortase of gram-positive bacteria. J. Bacteriol. 184:971-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pallen, M. J., A. C. Lam, M. Antonio, and K. Dunbar. 2001. An embarrassment of sortases - a richness of substrates? Trends Microbiol. 9:97-102. [DOI] [PubMed] [Google Scholar]
- 28.Pezard, C., P. Berche, and M. Mock. 1991. Contribution of individual toxin components to virulence of Bacillus anthracis. Infect. Immun. 59:3472-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 30.Redmond, C., I. Henderson, P. C. B Turnbull, and J. Bowen. 1996. Phage from different strains of Bacillus anthracis. Salisbury Med. Bull. 87:60-63. [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Sao-Jose, C., C. Baptista, and M. A. Santos. 2004. Bacillus subtilis operon encoding a membrane receptor for bacteriophage SPP1. J. Bacteriol. 186:8337-8346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneewind, O., P. Model, and V. A. Fischetti. 1992. Sorting of protein A to the staphylococcal cell wall. Cell 70:267-281. [DOI] [PubMed] [Google Scholar]
- 34.Schuch, R., D. Nelson, and V. A. Fischetti. 2002. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 418:884-889. [DOI] [PubMed] [Google Scholar]
- 35.Sirard, J. C., C. Guidi-Ronlani, A. Fouet, and M. Mock. 2000. Characterization of a plasmid region involved in Bacillus anthracis toxin production and pathogenesis. Int. J. Med. Microbiol. 290:313-316. [DOI] [PubMed] [Google Scholar]
- 36.Sylvestre, P., E. Couture-Tosi, and M. Mock. 2002. A collagen-like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol. Microbiol. 45:169-178. [DOI] [PubMed] [Google Scholar]
- 37.Ton-That, H., S. K. Mazmanian, L. Alksne, and O. Schneewind. 2002. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Cysteine 184 and histidine 120 of sortase form a thiolate-imidazolium ion pair for catalysis. J. Biol. Chem. 277:7447-7452. [DOI] [PubMed] [Google Scholar]
- 38.Ton-That, H., and O. Schneewind. 2003. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 50:1429-1438. [DOI] [PubMed] [Google Scholar]
- 39.Trieu-Cuot, P., C. Carlier, P. Martin, and P. Courvalin. 1987. Plasmid transfer by conjugation from Escherichia coli to Gram-positive bacteria. FEMS Microbiol. Lett. 48:289-294. [Google Scholar]
- 40.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1991. An integrative vector exploiting the transposition properties of Tn1545 for insertional mutagenesis and cloning of genes from gram-positive bacteria. Gene 106:21-27. [DOI] [PubMed] [Google Scholar]
- 41.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1990. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in gram-positive bacteria. Nucleic Acids Res. 18:4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valyasevi, R., W. E. Sandine, and B. L. Geller. 1991. A membrane protein is required for bacteriophage c2 infection of Lactococcus lactis subsp. lactis C2. J. Bacteriol. 173:6095-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe, T., A. Morimoto, and T. Shiomi. 1975. The fine structure and the protein composition of gamma phage of Bacillus anthracis. Can. J. Microbiol. 21:1889-1892. [DOI] [PubMed] [Google Scholar]
- 44.Watanabe, T., and T. Shiomi. 1975. Inhibiting materials for gamma phage adsorption to the cell wall of Bacillus anthracis, strain Pasteur No. 2-H. Jpn. J. Microbiol. 19:115-121. [DOI] [PubMed] [Google Scholar]
- 45.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]