Abstract
In Escherichia coli, envelope stress can be overcome by three different envelope stress responses: the σE stress response and the Bae and Cpx two-component systems. The Cpx envelope stress response is controlled by the sensor kinase CpxA, the response regulator CpxR, and the novel periplasmic protein CpxP. CpxP mediates feedback inhibition of the Cpx pathway through a hypothetical interaction with the sensing domain of CpxA. No informative homologues of CpxP are known, and thus it is unclear how CpxP exerts this inhibition. Here, we identified six cpxP loss-of-function mutations using a CpxP-β-lactamase (CpxP′-′Bla) translational fusion construct. These loss-of-function mutations identified a highly conserved, predicted α-helix in the N-terminal domain of CpxP that affects both the function and the stability of the protein. In the course of this study, we also found that CpxP′-′Bla stability is differentially controlled by the periplasmic protease DegP in response to inducing cues and that mutation of degP diminishes Cpx pathway activity. We propose that the N-terminal α-helix is an important functional domain for inhibition of the Cpx pathway and that CpxP is subject to DegP-dependent proteolysis.
Constantly changing environments are a critical situation to which all bacteria must adapt in order to survive. In gram-negative bacteria, the envelope, consisting of the outer membrane, periplasm, and inner membrane, is in constant contact with the environment. Escherichia coli has at least three regulatory pathways, the σE stress response and the CpxRA and the BaeSR two-component systems, which are activated by and mediate adaptation to different envelope stresses (1, 2, 30, 34).
The Cpx signal transduction pathway is a typical two-component system with a membrane-bound histidine kinase, CpxA, and a cytoplasmic response regulator, CpxR (15, 42). The activating cues of the Cpx pathway include alterations in extracellular pH (9, 27), accumulation of enterobacterial common antigen intermediate lipid II (7), overexpression of NlpE (38), overexpression of P pilus subunits in the absence of their periplasmic chaperone PapD (18), and overexpression of the enteropathogenic Escherichia coli type IV bundle-forming pilus subunit BfpA (28). Each of these activating cues is expected to lead to the accumulation of misfolded and/or mislocalized proteins associated with the envelope, which are likely a component of the activating signal for the Cpx pathway.
When activated, CpxA acts as a histidine autokinase (33). The phosphorylated CpxA then transfers the phosphate to a conserved aspartate on CpxR (33). Phosphorylated CpxR has enhanced ability to bind to consensus sequences and increase transcription of the Cpx regulon (8, 29, 33), which contains numerous envelope protein folding and degrading factors, and a variety of other genes whose roles in responding to envelope stress are not understood (13, 34). Among the envelope protein folding and degrading factors induced are the periplasmic endoprotease DegP (6, 25), two peptidyl-prolyl-isomerases, PpiA and PpiD (11, 26, 29), and DsbA, the major periplasmic disulfide oxidase (3, 8, 20, 29). Along with increased transcription of the protein folding and degrading factors, a small periplasmic inhibitor protein, CpxP, is also expressed at elevated levels, together with the cpxRA genes (9, 31, 32). Thus, a major role of the Cpx response appears to be maintaining envelope proteins under adverse conditions.
cpxP was first identified as a pH-regulated locus which encodes a periplasmic protein that helps overcome extracytoplasmic protein-mediated toxicity (9). Danese and Silhavy (9) identified cpxP as a lacZ operon fusion that was up-regulated by NlpE in a CpxA-dependent manner. Furthermore, CpxP is involved in signal transduction, since overexpression of CpxP causes a three- to fivefold reduction in Cpx-mediated gene expression via the periplasmic sensing domain of CpxA (31, 32). An inner membrane-tethered maltose-binding protein-CpxP fusion protein can maintain Cpx inhibition in the presence of spheroplasting, a strong Cpx-activating signal, while a maltose-binding protein-CpxP fusion localized to the periplasm does not, suggesting that the CpxA-CpxP interaction is direct (31). Currently it is thought that in the absence of envelope stress, CpxP interacts with the sensing domain of CpxA, maintaining the pathway in an off state. Upon activation, CpxP inhibition would be relieved, allowing induction of the response. However, CpxP is not required for signal transduction, since in either the absence of, or presence of overexpression of, CpxP, the Cpx pathway can still be induced further (14, 32). Thus, the hypothesized role of CpxP is in fine-tuning the response.
In this study we address the question of how CpxP-mediated inhibition might occur and be relieved. Since CpxP has no informative homologues, we set out to identify possible functional domains in CpxP that are important for signal transduction. Using a translational CpxP′-′Bla fusion construct, we identified a highly conserved, predicted α-helix in the N-terminal domain of CpxP that affects both the inhibitory function and stability of the protein. Diminished levels of some of the loss-of-function mutants relative to the wild-type CpxP′-′Bla protein suggested that proteolysis might affect CpxP-mediated inhibition. Indeed, we noted that the levels of the mutant CpxP′-′Bla proteins could be returned to, or elevated above, normal in the absence of DegP. DegP proteolysis is likely important for controlling CpxP levels in response to inducing cues since degP mutation simultaneously abrogates the disappearance of CpxP′-′Bla and diminishes pathway activation at elevated pH. We propose that the predicted N-terminal α-helix is important for the CpxA-dependent inhibition of the pathway and that CpxP levels are controlled by DegP-dependent proteolysis.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The stains and plasmids used in this study are listed in Table 1. All strains were constructed using standard genetic techniques (36). PCR primers are described in Table 2.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| Strains | ||
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 (Strr) relA1 flbB5301 decC1 ptsF25 rbsR | 5 |
| TR50 | MC4100 λRS88 (cpxP′-lacZ+) | 33 |
| TR757 | MC4100 λRS88 (cpxP′-lacZ+) degP::Tn10 | This study |
| TR930 | TR50(pBBR1MCS) | This study |
| TR932 | TR50(pPB) | This study |
| DB4 | TR757(pPB) | This study |
| DB8 | TR50(pCpxP) | This study |
| DB10 | TR50(pPB1) | This study |
| DB12 | TR50(ptrc99A) | This study |
| DB30 | TR50(pPB3) | This study |
| DB31 | TR50(pPB2) | This study |
| DB32 | TR50(pPB4) | This study |
| DB33 | TR50(pPB5) | This study |
| DB40 | TR50(pPB6) | This study |
| DB58 | TR50(pDegP) | This study |
| DB59 | TR50(pDsbA) | This study |
| DB60 | TR50(pPpiA) | This study |
| DB61 | TR50(ptrc99:Spy) | This study |
| DB70 | TR757(pPB1) | This study |
| DB71 | TR757(pPB2) | This study |
| DB72 | TR757(pPB3) | This study |
| DB73 | TR757(pPB4) | This study |
| DB74 | TR757(pPB5) | This study |
| DB75 | TR757(pPB6) | This study |
| DB115 | TR50(pCpxPU) | This study |
| DB116 | TR50(pCpxPD61EU) | This study |
| DB117 | TR50(pCpxPQ55PU) | This study |
| DB127 | TR50(pCpxPD61E) | This study |
| DB128 | TR50(pCpxPQ55P) | This study |
| Plasmids | ||
| pBBR1MCS | Expression vector with a multiple cloning site following a lac promoter (Camr) | 22 |
| pCxpP | cpxP overexpression vector (Ampr) | 32 |
| pCpxPD61E | cpxPD61E gene amplified from pCpxPD61EU using the CpxPKpn and CpxP5′Eco primers and cloned into the Kpn and EcoRI sites of ptrc99A (Ampr) | This study |
| pCpxPD61EU | Site-directed mutagenesis (SDM) was carried out on pCpxPU using the D-61-E1 and D-61-E2 primers; refer to Materials and Methods for complete protocol (Ampr) | This study |
| pCpxPQ55P | cpxPQ55P gene was amplified from pCpxPQ55PU using CpxPKpn and CpxP5′Eco and cloned into ptrc99A using the Kpn and EcoRI sites (Ampr) | This study |
| pCpxPQ55PU | SDM was carried out on the pCpxPU using the Q-55-P1 and Q-55-P2 primers; refer to Materials and Methods for complete protocol (Ampr) | This study |
| pCpxPU | cpxP gene and flanking upstream region was amplified using CpxPBam and CpxPKpn and cloned into pUC18 BamHI and Kpn sites (Ampr) | This study |
| pDegP | degP open reading frame was PCR amplified using the primers DegPKpn and DegPBam and cloned into the KpnI and BamHI sites of ptrc99A (Ampr) | This study |
| pDsbA | dsbA gene was amplified using the DsbAKpn and DsbABam primers and cloned into the KpnI and BamHI sites of ptrc99A (Ampr) | This study |
| pPB | CpxP′-′Bla translational fusion overexpression vector; the cpxP gene was amplified from MC4100 using CpxP5Kpn and CpxPBla primers, and cloned into pBBR1MCS using KpnI and BamHI; β-lactamase gene from pUC19 was amplified using BlaCpxP and Bla3Bam primers, digested with BamHI and cloned in-frame downstream of cpxP (Ampr Camr) | This study |
| pPB1 | pPB encoding a CpxPD61E′-′Bla mutation (Ampr Camr) | This study |
| pPB2 | pPB encoding a CpxPQ55P′-′Bla mutation (Ampr Camr) | This study |
| pPB3 | pPB encoding a CpxPR60Q′-′Bla mutation (Ampr Camr) | This study |
| pPB4 | pPB encoding a CpxPD61V′-′Bla mutation (Ampr Camr) | This study |
| pPB5 | pPB encoding a CpxPM59T′-′Bla mutation (Ampr Camr) | This study |
| pPB6 | pPB encoding a CpxPQ128H′-′Bla mutation (Ampr Camr) | This study |
| pPpiA | ppiA gene was amplified using PpiAKpn and PpiABam primers and cloned into the KpnI and BamHI sites of ptrc99A (Ampr) | This study |
| ptrc99:Spy | spy open reading frame was amplified using the spy5′Eco and spy3′Bam primers and cloned into ptrc99A using EcoRI and BamHI (Ampr) | This study |
| ptrc99A | High-copy-number expression vector with a multiple cloning site following an IPTG-inducible trc promoter (Ampr) | Pharmacia |
| pUC18 | Cloning vector (Ampr) | Invitrogen |
TABLE 2.
Primers used for PCR
| Primer | Sequence |
|---|---|
| Bla3Bam | 5′-CGG GAT CCT CAC GTT AAG GGA TTT TGG TC-3′ |
| BlaCpxP | 5′-CGG GAT CCA CGC TGG TGA AAG TAA AAG A-3′ |
| CpxPBam | 5′-CGG GAT CCT GTG CCA GCA AAT AGA GCA G-3′ |
| CpxPBla | 5′-CGG GAT CCC TGG GAA CGT GAG TTG CTA C-3′ |
| CpxP5Kpn | 5′-GGG GTA CCT CGC GAC AGA AAG ATT TTG G-3′ |
| CpxPKpn | 5′-GGG GTA CCG GCA AGG AAA ACA GGG TTT A-3′ |
| CpxP5′Eco | 5′-GGA ATT CCC TCT CTA TCG TTG AAT CGC G-3′ |
| DegPBam | 5′-CGG GAT CCG CAC GGC TTA GCA TAA GGA A-3′ |
| DegPKpn | 5′-GGG GTA CCC GAA TCT GAA GAA CAC AGC AA-3′ |
| DsbABam | 5′-CGG GAT CCT GAA TAC TCA CGG GCT TTA TG-3′ |
| DsbAKpn | 5′-GGG GTA CCG TGG TTA ACC GGG GAA GAT T-3′ |
| D-61-E1 | 5′-TCA GCA GAT GCG AGA ACT TAT GCA ACA GGC CC-3′ |
| D-61-E2 | 5′-GGG CCT GTT GCA TAA GTT CTC GCA TCT GCT GA-3′ |
| PpiABam | 5′-CGG GAT CCG TTA CGC CGG GAG AGC AG-3′ |
| PpiAKpn | 5′-GGG GTA CCT GAT CGT CAG GTT ACA TAT ATT TCA GA-3′ |
| Q-55-P1 | 5′-AGT TTA ACC GAA CAT CCG CGT CAG CAG ATG CG-3′ |
| Q-55-P2 | 5′-CGC ATC TGC TGA CGC GGA TGT TCG GTT AAA CT-3′ |
| spy3′Bam | 5′-CGG GAT CCC GCA AGG TAG TGG ACA AGA CCG-3′ |
| spy5′Eco | 5′-GGA ATT CCA ATA ACT GAA AGG AAG GAT ATA G-3′ |
Media, antibiotics, and growth conditions.
All strains were grown on Luria-Bertani (LB) agar (36) or lactose-MacConkey agar (Difco), which is an indicator medium that permits measurement of β-galactosidase levels, at either 37 or 30°C. Liquid cultures were grown in LB broth with aeration. Strains were maintained with the appropriate selection, 100 μg/ml ampicillin, 100 μg/ml chloramphenicol, 25 μg/ml kanamycin, or 25 μg/ml tetracycline. All antibiotics were purchased from Sigma.
Mutagenesis of pPB.
Random cpxP point mutants were isolated by passaging pPB through DNA repair-deficient strain XL-1 Red (Stratagene), according to the manufacturer's specifications. Twenty independently derived samples of mutagenized pPB were collected after passaging through XL-1 Red and were transformed into TR50. Transformants were grown at 37°C for a maximum of 20 h on lactose-MacConkey medium containing ampicillin. Lac+ Ampr derivatives were isolated and characterized further.
DNA sequence analysis.
Mutated pPB plasmid DNA was extracted using a QIAprep Spin mini prep kit (QIAGEN), according to the manufacturer's protocol. The mutated cpxP genes were amplified using the CpxPBla and CpxP5′Kpn primers (Table 2). The PCR products were purified using a QIAGEN PCR purification kit (QIAGEN), according to the manufacturer's directions. Sequencing was carried out using the protocol for the DYEnamic ET terminator cycle sequencing premix kit (Amersham) provided by the Molecular Biology Service Unit (University of Alberta).
β-Galactosidase assay.
Single colonies of each bacterial strain to be assayed were inoculated into 2-ml overnight cultures of LB broth containing the appropriate antibiotics at 37 or 30°C with aeration. The next day the cultures were diluted 1:50 into fresh medium and grown at 37°C to late log phase (optical density at 600 nm = 0.6). Induction of the Cpx response by elevated pH was accomplished by adding 1 M sodium phosphate buffer of the appropriate pH (35) to the medium to a final concentration of 100 mM. β-Galactosidase activity was measured using the microtiter plate assay (37) with slight variation to the protocol; 5 μl of cell mixture was added to 195 μl of Z-buffer due to high expression of β-galactosidase in the strains. The final calculation was then multiplied by 10 to account for the dilution. Each assay was performed in triplicate. Error bars represent the standard deviation.
Western blot analysis.
Overnight cultures of the appropriate strains grown in LB plus antibiotics at 30°C or 37°C with aeration were subcultured 1:50 and grown at 37°C for 4 h. pH induction of the Cpx response was accomplished using the same procedure described for the β-galactosidase assays above. The optical density at 600 nm of the cultures was measured and whole-cell extracts were prepared by pelleting volumes that were adjusted to correspond to 1 ml of the culture that reached the lowest optical density at 600 nm. Cell extracts were resuspended in 50 μl of 2X sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) loading dye (35). Lysates were boiled for 5 min and 10 μL was electrophoresed on a 10% SDS-PAGE minigel system (Bio-Rad) (24). Proteins were transferred to nitrocellulose membrane at 10 V overnight (41). The membrane was labeled as previously described by Raivio et al. (32). β-Lactamase monoclonal antibody (1:1,000 dilution) (QED BioScience Inc.) or bacterial alkaline phosphatase polyclonal antibody (1:20,000 dilution) (Research Diagnostics, Inc.) were used as primary antibodies. A secondary anti-mouse or anti-rabbit antibody conjugated to alkaline phosphatase (Sigma) was used at a 1:10,000 dilution. Proteins were visualized using the Immun-Star alkaline phosphatase substrate pack chemiluminescent kit (Bio-Rad).
Site-directed mutagenesis of pCpxPU to make pCpxPD61EU and pCpxPQ55PU.
The D61E and Q55P mutations were transferred to the pCpxP vector as follows. First, the genomic region surrounding cpxP was PCR amplified using the CpxPBam and CpxPKpn primers and cloned into the BamHI and KpnI sites of pUC18 to construct pCpxPU. Next, the D61E and Q55P mutations were introduced into pCpxPU using the primers D-61-E1 and D-61-E2 (Table 2) for pCpxPD61EU and the primers Q-55-P1 and Q-55-P2 (Table 2) for pCpxPQ55PU using the Quickchange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions with slight variations to the protocol as follows; 2 μL of 10 mM deoxynucleoside triphosphates was used in the extension reaction for 6.5 min at 68°C, 1 μL of DpnI (5 U/μL) was used to digest unmutated template DNA for 2.5 h, One Shot Top10 chemical competent cells (Invitrogen) were used in place of the XL-1 Blue competent cells, 5 μL of DpnI-treated DNA was added to the Top10 cells, and LB was used in place of NZY+ broth.
RESULTS
Inhibition of the Cpx pathway is specific to CpxP overexpression.
Raivio et al. (31, 32) have shown that overproduction of CpxP represses the Cpx regulon in a CpxA-dependent manner. From these findings it was proposed that CpxP acts as a periplasmic inhibitor by interaction with the sensing domain of CpxA. Alternatively it is possible that CpxP overexpression may lead to pathway inhibition by aiding in envelope protein folding, thus relieving envelope stress and causing down-regulation of the Cpx response in this fashion (14). If this is true, then overproduction of other Cpx-regulated members may also down-regulate the pathway.
To test this hypothesis, we overexpressed numerous members of the Cpx regulon: a periplasmic endoprotease, DegP (21, 25), a major disulfide oxidase, DsbA (3, 20), a peptidyl-prolyl-isomerase, PpiA (26), a small periplasmic protein of unknown function, Spy (17), and CpxP (9). Overexpression of CpxP, PpiA, and Spy was confirmed by SDS-PAGE analysis of isopropylthiogalactopyranoside (IPTG)-induced periplasmic extracts (data not shown). DegP and DsbA overexpression was confirmed by complementation of temperature sensitivity (40) and dithiothreitol sensitivity (10), respectively, of degP and dsbA null mutants transformed with pDegP or pDsbA (data not shown).
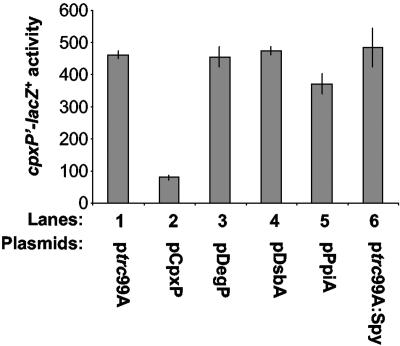
To see what effect overexpression of these factors may have on Cpx-regulated gene expression we used a cpxP′-lacZ+ fusion in single copy on the chromosome. When CpxP is overexpressed there is a fivefold reduction in Cpx-mediated gene expression, as expected (Fig. 1, compare lanes 1 and 2). In contrast, overexpression of DegP, DsbA, PpiA, or Spy had no effect on Cpx-mediated gene expression (Fig. 1, compare lanes 1 with 3 to 6). We conclude that CpxP functions uniquely as an inhibitor of the Cpx response.
FIG. 1.
Inhibition of the Cpx response is specific to CpxP overexpression. β-Galactosidase levels were used to measure the effect of overproduction of specific Cpx-regulated genes on Cpx pathway activity, using strains carrying a cpxP′-lacZ+ fusion. The strains used were DB12 (lane 1), DB8 (lane 2), DB58 (lane 3), DB59 (lane 4), DB60 (lane 5), and DB61 (lane 6).
Isolation of cpxP mutants.
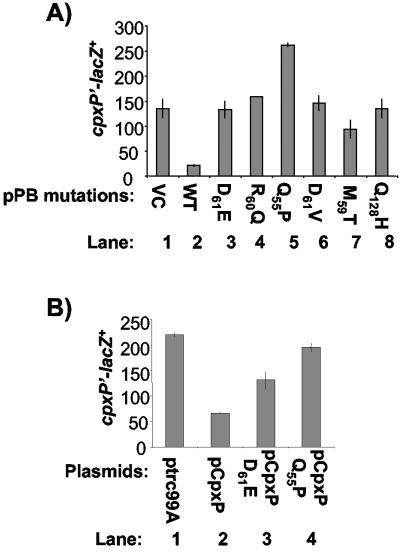
Since CpxP has no informative homologues, we sought to identify functional domains in CpxP that are important for signal transduction. The strongest phenotype available to monitor CpxP function is the inhibition of Cpx-regulated gene expression upon CpxP overexpression (14, 31, 32). Thus, we looked for cpxP loss-of-function mutations that prevented Cpx pathway inhibition upon overexpression of a CpxP-β-lactamase (CpxP′-′Bla) translational fusion protein encoded on plasmid pPB. Overexpression of the CpxP′-′Bla fusion from an exogenous promoter confers ampicillin resistance (Ampr) and down-regulates Cpx-mediated gene expression three- to fivefold, the same as overexpression of the wild-type CpxP protein does (31, 32) (Fig. 2A, compare lanes 1 and 2). Thus, the CpxP′-′Bla protein possesses wild-type activity.
FIG. 2.
(A) Effect of overexpression of wild-type and mutant CpxP′-′Bla fusions on Cpx-mediated gene expression. β-Galactosidase produced from a cpxP′-lacZ+ fusion was measured as a reporter of Cpx-mediated gene expression in TR930 (lane 1), TR932 (lane 2), DB10 (lane 3), DB30 (lane 4), DB31 (lane 5), DB32 (lane 6), DB33 (lane 7), and DB40 (lane 8). VC, vector control; WT, wild-type CpxP′-′Bla protein. (B) Effect of overexpression of wild-type and mutant CpxP on cpxP′-lacZ+ activity. β-Galactosidase production was measured from strain TR50 carrying a chromosomal cpxP′-lacZ+ reporter and either ptrc99A (lane 1), pCpxP (lane 2), pCpxPD61E (lane 3), or pCpxPQ55P (lane 4).
On lactose-MacConkey indicator medium, this inhibition phenotype is manifested as white or Lac− colonies. We used pPB in combination with a chromosomally encoded Cpx-regulated cpxP′-lacZ+ fusion to screen for Lac+ and Ampr phenotypes, thereby eliminating nonsense mutations that would destroy CpxP function by truncating the protein. pPB was subjected to random mutagenesis by passaging through the E. coli mutator strain XL1-Red (Stratagene). Over 10,000 Ampr colonies, resulting from 20 mutated plasmid pools, were screened on lactose-MacConkey indicator plates, resulting in the isolation of 6 different cpxP mutations that yielded Lac+ and Ampr transformants upon plasmid isolation and retransformation into the reporter strain. Several of these mutations were isolated more than once, indicating that the screen was performed to saturation.
cpxP loss-of-function mutations localize to two distinct regions.
Both strands of the mutated pPB plasmids encoding potential loss-of-function CpxP′-′Bla mutants were sequenced to identify the precise mutations. Of the six loss-of-function mutations that we isolated, four of them were transversions and two were transitions in keeping with the broad mutator phenotype of the XL1-Red strain. Five of these mutations altered amino acids between residues 55 and 61 of the unprocessed CpxP and were localized to a single predicted α-helix in the N-terminal region of the protein (Q55P, M59T, R60Q, D61E, D61V), while the sixth mutation is located at position 128 in the last predicted α-helix in the C-terminal region of CpxP (Q128H). Sequence analysis and recloning of the mutant cpxP genes showed that no mutations occurred in the bla gene, the tac promoter, or the plasmid backbone indicating that the observed affects on Cpx-mediated gene expression were due solely to the CpxP amino acid changes.
Loss-of-function CpxP mutants fail to inhibit the Cpx pathway.
In order to determine the extent to which our mutants had lost the ability to inhibit the Cpx response, we transformed the vector control, wild-type and mutant pPB plasmids into a strain that contained a chromosomal cpxP′-lacZ+ fusion, which allowed for detection of Cpx pathway activity. Activity of the Cpx pathway in the presence of the parent vector, pPB, carrying the wild-type CpxP′-′Bla was approximately three to five fold lower than in the presence of either the empty control vector or the six vectors carrying the mutated CpxP′-′Bla fusions (Fig. 2A, compare lane 2 to lanes 1 and 3 through 8). These results show that all of the mutations impair the ability of CpxP′-′Bla overexpression to inhibit cpxP′-lacZ+ expression. To determine if these mutations affected activity of the wild-type CpxP protein in the same fashion and rule out the possibility that our mutations exerted their affects in a manner specific to the CpxP′-′Bla fusion protein, we transferred two of the mutations, D61E and Q55P, to the previously characterized pCpxP vector, which inhibits the Cpx response via overexpression of the native CpxP protein (31, 32). Introduction of the D61E or Q55P mutations into pCpxP resulted in a diminished ability of these plasmids to inhibit cpxP′-lacZ+ activity relative to the wild-type pCpxP control (Fig. 2B, compare lanes 3 and 4 to lane 2). Thus, the mutations we isolated exert their effects by altering the inhibitory capacity of the native CpxP protein.
Mutant CpxP′-′Bla proteins are expressed at varied levels.
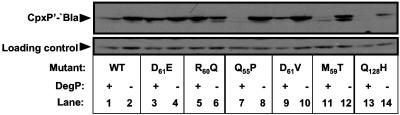
To examine the expression levels of the mutant compared to the wild-type CpxP′-′Bla proteins, whole-cell lysates were prepared and immunoblotting was carried out using a monoclonal antibody to β-lactamase. Of the six loss-of-function mutants, R60Q, D61V, and D61E, produced similar or slightly elevated levels of protein of the same size as the wild-type CpxP′-′Bla protein (Fig. 3, compare lane1 with lanes 3, 5, and 9). The other three mutants, Q55P, M59T, and Q128H, exhibited decreased protein expression compared to the wild-type CpxP′-′Bla fusion (Fig. 3, compare lane 1 to lanes 7, 11, and 13). Since sequence analysis and recloning of the mutant cpxP genes showed that no mutations in the bla gene, tac promoter, or plasmid backbone were responsible for the mutant phenotypes (data not shown), these data indicate that the decreased protein levels were due to effects on translation or protein stability.
FIG. 3.
Expression levels of wild-type and mutant CpxP′-′Bla fusion proteins in wild-type and degP null strain backgrounds. Western blots were performed on whole-cell lysates of wild-type (odd-numbered lanes) and degP null strains (even-numbered lanes) transformed with the CpxP′-′Bla encoding plasmid pPB (TR932 and DB4) (lanes 1 and 2) or the mutated pPB plasmids CpxPD61E′-′Bla (DB10 and DB70) (lanes 3 and 4), CpxPR60Q′-′Bla (DB30 and DB72) (lanes 5 and 6), CpxPQ55P′-′Bla (DB31 and DB71) (lanes 7 and 8), CpxPD61V′-′Bla (DB32 and DB73) (lanes 9 and 10), CpxPM59T′-′Bla (DB33 and DB74) (lanes 11 and 12), and CpxPQ128H′-′Bla (DB40 and DB75) (lanes 13 and 14) using a monoclonal β-lactamase antibody to label CpxP′-′Bla proteins. WT refers to the wild-type CpxP′-′Bla protein. The experiment was performed three times and a representative blot is shown.
Diminished mutant CpxP′-′Bla protein levels are restored by degP mutation.
Since the levels of three of the six CpxP′-′Bla loss-of-function mutants were affected (Fig. 3), we wondered if CpxP might be controlled by proteolysis. To test this idea, we sought to determine if DegP, a Cpx-regulated protease, affected CpxP′-′Bla levels. The cpxP′-′bla overexpression plasmid pPB and mutated plasmids pPB1 to pPB6 were transformed into a strain carrying a degP::Tn10 mutation. Mutation of degP resulted in an increase in the level of the wild-type CpxP′-′Bla protein (Fig. 3, compare lanes 1 and 2). The levels of the three loss-of-function mutant proteins (CpxPQ55P′-′Bla, CpxPM59T′-′Bla, and CpxPQ128H′-′Bla) shown to be present at lower levels than the wild-type control (Fig. 4, lanes 7, 11 and 13), were returned to close to wild-type levels in the degP null strain (Fig. 3, compare lane 2 to lanes 8, 12, and 14). In addition, the levels of the CpxPD61E′-′Bla and CpxPD61V′-′Bla proteins appeared to be slightly increased in the degP strain background (Fig. 4, compare lanes 3 and 4 and 9 and 10). The levels of a cross-reactive protein that served as a convenient loading control were unchanged in either background, as were levels of the periplasmic protein alkaline phosphatase (Fig. 3, bottom, and data not shown). The data suggest that the Q55P, M59T, and Q128H mutant proteins are present at reduced levels due to diminished stability and that DegP affects the stability of both the wild-type and mutant CpxP′-′Bla fusion proteins.
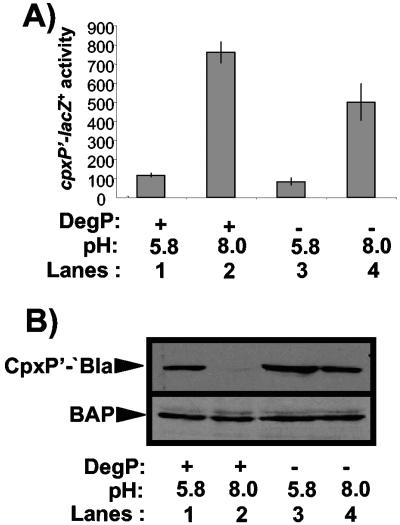
FIG. 4.
(A) Induction of the Cpx pathway is reduced in a degP null strain. A chromosomal cpxP′-lacZ+ fusion was used to determine the effect of pH on Cpx pathway activity in wild-type and degP null strains. β-Galactosidase levels were measured from strains grown in LB buffered with sodium phosphate buffer at a pH of 5.8 (odd-numbered lanes) or 8.0 (even-numbered lanes). The strains used in this experiment were TR50 (lanes 1 and 2) and TR757 (lanes 3 and 4). (B) CpxP′-′Bla levels are diminished in the presence of Cpx-inducing cues in a DegP-dependent fashion. Western blots were carried out on whole-cell lysates of wild-type (TR932) and degP null (DB4) strains overexpressing a CpxP′-′Bla fusion protein that had been grown in LB buffered with sodium phosphate buffer at a pH of 5.8 (lanes 1 and 3) or 8.0 (lanes 2 and 4). The experiment was performed three times and a representative blot is shown.
Cpx pathway activity and CpxP′-′Bla levels are affected by DegP in response to elevated pH.
The above observations suggest that one reason some of the mutant CpxP′-′Bla fusion proteins have lost inhibitory activity is due to a loss of stability that is mediated by DegP-dependent proteolysis. Since we showed that our mutations affect the inhibitory activity of the native CpxP protein in the same fashion (Fig. 2), we speculated that DegP might influence Cpx signaling through proteolysis of CpxP under inducing conditions. To investigate whether DegP influences Cpx signaling, we examined the effects of mutating degP on Cpx pathway activity and CpxP′-′Bla levels in the presence and absence of inducing signals (Fig. 4). To induce the Cpx response, we used alkaline pH, a growth condition shown to induce the Cpx response in both E. coli and Shigella flexneri (9, 27).
We grew either wild-type or degP mutant strains of E. coli bearing a cpxP′-lacZ+ reporter at pH 5.8, where the Cpx response has been shown to be uninduced, or pH 8.0, where the Cpx response is induced (9), and measured β-galactosidase activity. As observed previously, the Cpx response is strongly induced by growth at pH 8.0 relative to pH 5.8 (Fig. 4A, compare lanes 1 and 2). In a degP mutant strain, while the pathway was still induced, the level of cpxP′-lacZ+ expression was diminished almost twofold (Fig. 4, compare lanes 1 and 2 to 3 and 4). When we examined the levels of CpxP′-′Bla present in wild-type strains at pHs of 5.8 and 8.0, we were unable to detect the fusion protein at a pH of 8.0 (Fig. 4B, compare lanes 1 and 2). Conversely, in the absence of DegP, the fusion protein was present at approximately equivalent levels at both pHs of 5.8 and 8.0 (Fig. 4B, compare lanes 3 and 4). The levels of a periplasmic control protein, alkaline phosphatase, were unaffected at either pH in wild-type or degP mutant strains (Fig. 4B, bottom). Thus, the ability to induce the Cpx response in a degP null mutant strain is compromised, and this correlates with increased levels of the CpxP′-′Bla fusion protein observed under the same conditions.
DISCUSSION
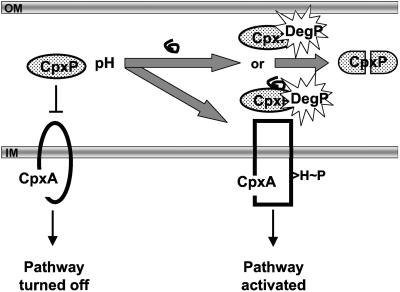
In this study, we have taken advantage of a CpxP′-′Bla fusion construct that mediates Cpx pathway inhibition in the same fashion as the native CpxP protein (Fig. 2) to identify the first essential inhibitory domain in CpxP. We screened over 10,000 mutagenized plasmids and isolated five mutations that localize to a single, highly conserved, predicted α-helix in the N terminus of CpxP (Fig. 5). The mutations alter the activity of the native CpxP protein in the same fashion (Fig. 2) and affect either or both inhibitory function and stability (Fig. 2 and 3). The reduced levels of some mutant proteins can be restored by mutation of DegP and this led us to examine the effect of DegP on Cpx signal transduction and CpxP′-′Bla levels. Strikingly, we found that induction of the Cpx response by elevated pH is correlated with disappearance of the CpxP′-′Bla fusion protein and that mutation of degP abrogates both these effects (Fig. 4). Together, the data support a model in which DegP-mediated proteolysis of CpxP relieves inhibition of the Cpx response in the presence of inducing cues (Fig. 6).
FIG. 5.
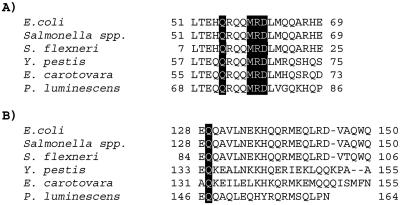
Squence alignment of selected regions of CpxP orthologues. The amino acid sequence of E. coli CpxP was aligned with those of Salmonella spp., Shigella flexneri, Yersinia pestis, Erwinia carotovora, and Photorhabdus luminescens using PepTool (Biotools Inc.). The locations of the six loss-of-function mutations are indicated by the dark boxes. A) The first predicted α-helix of CpxP, where five of the six mutations fall. B) The predicted C-terminal α-helix of CpxP, where the sixth mutation falls.
FIG. 6.
Model for activation of the Cpx pathway. In the absence of envelope stress, CpxP (shaded oval) interacts with the sensing domain of CpxA (central region of the periplasmic domain of CpxA), maintaining the pathway in an off state. Inducing cues, such as elevated pH, lead to either a conformational change in CpxP or its association with misfolded envelope proteins (squiggly line) or both. CpxP or the complex of CpxP with misfolded protein is subsequently degraded by DegP (starburst) and can no longer inhibit CpxA. This leaves the CpxA sensing domain available for maximal induction by cues such as pH. OM, outer membrane; IM, inner membrane; H, histidine; P, phosphate.
Despite repeated attempts using fusion proteins and immunogenic epitopes as antigens, we have been unsuccessful in raising CpxP-specific antisera. Thus, we cannot be certain at this point that the chromosomally expressed CpxP is affected in a similar fashion by DegP in response to inducing cues. However, several lines of evidence suggest that this is the case. Firstly, the mutations we isolated in the CpxP′-′Bla construct affect the activity of the native, overexpressed CpxP in the same manner (Fig. 2). Second, the CpxP′-′Bla construct is dramatically destabilized by the Cpx-specific inducing cue of elevated pH (Fig. 4B) (9), and this destabilization is DegP dependent. Finally, mutation of degP, which would be expected to stabilize CpxP, leads to diminished Cpx pathway activity in the presence of inducing cues (Fig. 4A). Thus, although an analysis of the chromosomally encoded CpxP will be a necessary topic of future studies, these experiments provide strong support for a role of the N terminus of CpxP as well as DegP in a Cpx signaling cascade (Fig. 6).
Cpx envelope stress response is uniquely inhibited by CpxP overexpression.
DiGiuseppe and Silhavy (14) have recently set forth the hypothesis that CpxP has dual functions as an inhibitory protein and a chaperone, suggesting that CpxP inhibition of the Cpx response upon overexpression may be a by-product of combating extracytoplasmic protein-mediated toxicity. If this is true, then overexpression of other protein folding and degrading factors that belong to the Cpx pathway should also cause a decrease in pathway activity. We overexpressed numerous Cpx-regulated genes, and only when CpxP was overexpressed was there a decrease in pathway activity (Fig. 1). This supports previous experiments (31, 32) and shows that simply overexpressing Cpx-regulated genes does not confer inhibition. These results do not allow us to support or refute the idea that CpxP has dual functions, however, they do allow us to speculate that CpxP has a unique function in inhibition of the Cpx response, which none of the other Cpx-regulated protein folding or degrading factors possess.
Predicted N-terminal α-helix plays an essential role in CpxP-mediated inhibition and stability.
In this study we identified two classes of mutations that affect CpxP function and/or stability. Some mutations conferred a loss of inhibition but were otherwise present at comparable levels to the wild-type fusion protein (CpxPR60Q′-′Bla, CpxPD61E′-′Bla, and CpxPD61V′-′Bla). Another class of mutations not only conferred loss of function but also clearly altered protein levels (CpxPQ55P′-′Bla, CpxPM59T′-′Bla, and CpxPQ128H′-′Bla) (Fig. 3). Since most mutations fall in the N terminus, these observations suggest that the N terminus of CpxP contains a domain that is important both for the inhibition of CpxA via its sensing domain, as well as stabilization of CpxP.
Interestingly, five of the six cpxP mutations we identified localized to a span of seven amino acids in a predicted N-terminal α-helix. Since other CpxP homologues show some divergence in the N-terminal region, it has been suggested that the C terminus is responsible for inhibition and possibly interaction with the highly conserved CpxA (12). However, the sequence identities of the N-terminal domains in question compared to those of the entire cpxP open reading frame suggest that this predicted α-helix is highly conserved (Fig. 5). The identities of the predicted N-terminal α-helices relative to the entire cpxP open reading frame compared to those of E. coli are 100 compared to 88 for Salmonella spp., 100 compared to 87.7 for Shigella flexneri, 65 compared to 53 for Yersinia pestis, 70 compared to 46 for Erwinia carotovora, and 60 compared to 44 for Photorhabdus luminescens. Even more strikingly, the five mutations that localize to this putative α-helix affect amino acids that are completely conserved between the seven species identified above (Fig. 5A). This is strong evidence that our mutations are in a critical area of the protein and that this N-terminal α-helix plays an important role in CpxP function.
The C-terminal mutation CpxPQ128H′-′Bla also falls on a completely conserved amino acid (Fig. 5B), once again pointing to the importance of these amino acids in the function and stability of CpxP. Helical wheel analysis (Peptool, BioTools Inc.) shows that the N-terminal α-helix where five of our six loss-of-function mutations localized is polar except for CpxPL51, CpxPL62, and CpxPA66. This suggests that this helix is most likely located on the outside of the protein and thus one possibility is that our mutations alter a possible protein-protein interaction, such as that which is proposed to occur with the sensing domain of CpxA. It has been suggested that CpxP forms a dimer (D. Issac and T. J. Silhavy, personal communication). Thus, another possibility is that this N-terminal α-helix plays a role in the formation of dimers, and that dimerization is essential for inhibition of the Cpx pathway. Experiments are in progress to differentiate among these hypotheses.
DegP influences both Cpx pathway activity and CpxP′-′Bla levels in response to inducing cues.
Since some mutant CpxP′-′Bla proteins exhibited reduced protein levels (Fig. 3), we hypothesized that CpxP might be regulated by proteolysis. We reasoned that DegP might be involved in CpxP proteolysis since it is Cpx regulated (6) and has a broad substrate specificity (19, 21). We found that, in a degP knockout, we were able to recover full-length mutant CpxP′-′Bla proteins that were normally undetectable in a wild-type strain (Fig. 3), supporting the idea that CpxP is regulated by proteolysis and that the protease responsible for this degradation is DegP.
In order to determine if DegP-dependent proteolysis of CpxP plays a role in activation of the Cpx pathway, we analyzed Cpx pathway activity and CpxP′-′Bla levels in wild-type and degP null strains after exposure to the known Cpx-inducing signal of alkaline pH (9, 27). We found that in alkaline growth medium CpxP′-′Bla was undetectable, while at a pH of 5.8 CpxP′-′Bla seems to be stabilized (Fig. 4). CpxP′-′Bla instability at pH 8 was completely abolished in a degP null background (Fig. 4B), suggesting that DegP is responsible for CpxP′-′Bla degradation in response to elevated pH.
The levels of CpxP′-′Bla correlated inversely with induction of the Cpx response. In a wild-type strain, growth at pH 8.0 relative to pH 5.8 leads to a seven- to eightfold increase in cpxP′-lacZ+ reporter activity and this increase was diminished by approximately twofold in the absence of DegP (Fig. 4A). Together, these data support a model for Cpx signal transduction in which CpxP is rapidly degraded by DegP in the presence of inducing cues, fostering increased pathway activation (Fig. 6). Interestingly, when DegP was overexpressed in the wild-type background we saw no evidence of increased pathway activity (Fig. 1). Thus, we hypothesize that inducing cues must be present in order for DegP-mediated proteolysis of CpxP to occur.
It is not surprising that pathway induction is not obliterated in a degP null strain, since it has previously been shown that the Cpx response can still be induced in the presence of an overexpressed CpxP fusion protein, albeit to a lower level (14). Further, CpxP is not required for Cpx signaling (14, 31, 32). Thus, these results support previous reports that suggest an accessory role for CpxP in the signal transduction scheme. Interestingly, the RseB and TcpH regulators of E. coli and Vibrio cholerae, respectively, appear to play similar roles. The periplasmic RseB, although not essential for signaling, has been shown to protect the RseA anti-sigma factor from signal-independent proteolysis that would lead to inappropriate induction of the σE envelope stress response (16). Similarly, the periplasmic domain of TcpH appears to be involved in signaling by protecting the membrane-bound regulatory protein TcpP from untimely proteolysis (4). We propose that CpxP may serve a similar safeguarding function, protecting the sensing domain of CpxA from unwarranted activation or signaling noise in the absence of veritable envelope stresses.
At this time, we cannot say what event would trigger CpxP proteolysis by DegP in the presence of inducing cues. DegP possesses two PDZ domains (23) that are proposed to bind to substrates and direct them to the proteolytic central cavity of the hexameric protease, although the DegP substrate recognition motif remains undefined. Other PDZ-like domains recognize substrates by virtue of C-terminal recognition motifs baring hydrophobic determinants (39). Since the C terminus of CpxP is predominantly polar/charged, perhaps inducing cues lead to the exposure of an internal domain baring a hydrophobic recognition motif. Alternatively, CpxP may interact with denatured proteins generated by envelope stress, which then target the complex to DegP (Fig. 6).
Whatever the mechanism, the current work identifies the first conserved inhibitory domain in the novel regulator CpxP, implicates DegP-dependent proteolysis in the Cpx signaling cascade, and places CpxP among a new class of periplasmic regulatory proteins involved in shielding membrane-bound regulatory proteins from superfluous signaling events. Future studies will be aimed at working out the molecular details of these new facets of Cpx signal transduction.
Acknowledgments
This study was supported by operating grants from the Natural Sciences and Engineering Research Council of Canada and the Canadian Institutes of Health Research. T.L.R. is supported by a Scholar Award from the Alberta Heritage Foundation for Medical Research.
REFERENCES
- 1.Ades, S. E. 2004. Control of the alternative sigma factor σE in Escherichia coli. Curr. Opin. Microbiol. 7:157-162. [DOI] [PubMed] [Google Scholar]
- 2.Alba, B. M., and C. A. Gross. 2004. Regulation of the Escherichia coli σE-dependent envelope stress response. Mol. Microbiol. 52:613-619. [DOI] [PubMed] [Google Scholar]
- 3.Bardwell, J. C., J. O. Lee, G. Jander, N. Martin, D. Belin, and J. Beckwith. 1993. A pathway for disulfide bond formation in vivo. Mol. Microbiol. 90:1038-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck, N. A, E. S. Krukonis, and V. J. DiRita. 2004. TcpH influences virulence gene expression in Vibrio cholerae by inhibiting degradation of the transcription activator TcpP. J. Bacteriol. 186:8309-8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casadaban, M. J. 1976. Transposition and fusion of lac genes to selected promoters in Escherichia coli using bacteriophages lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 6.Danese, P. N., W. B. Snyder, C. L. Cosma, L. Davis, and T. J. Silhavy. 1995. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease DegP. Genes Dev. 9:387-398. [DOI] [PubMed] [Google Scholar]
- 7.Danese, P. N., G. Oliver, K. Barr, G. Bowman, P. Rick, and T. J. Silhavy. 1998. Accumulation of the enterobacterial common antigen lipid II biosynthetic intermediate stimulates degP transcription in Escherichia coli. J. Bacteriol. 180:5875-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danese, P. N., and T. J. Silhavy. 1997. The σE and Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes Dev. 11:1183-1193. [DOI] [PubMed] [Google Scholar]
- 9.Danese, P. N., and T. J. Silhavy. 1998. CpxP, a stress-combative member of the Cpx regulon. J. Bacteriol. 180:831-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dartigalongue, C., H. Nikaido, and S. Raina. 2000. Protein folding in the periplasm in the absence of primary oxidant DsbA: modulation of redox potential in periplasmic space via OmpL porin. EMBO J. 19:5980-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dartigalongue, C., and S. Raina. 1998. A new heat-shock gene, ppiD, encodes a peptidyl-prolyl isomerase required for folding of outer membrane proteins in Escherichia coli. EMBO J. 17:3968-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeWulf, P., B. Akerley, and E. Lin. 2000. Presence of the Cpx system in bacteria. Microbiology 146:247-248. [DOI] [PubMed] [Google Scholar]
- 13.DeWulf, P., A. McGuire, X. Liu, and E. Lin. 2002. Genome-wide profiling of promoter recognition by the two-component response regulator CpxR∼P in Escherichia coli. J. Biol. Chem. 182:1423-1426. [DOI] [PubMed] [Google Scholar]
- 14.DiGiuseppe, P. A., and T. J. Silhavy. 2003. Signal detection and target gene induction by the CpxRA two-component system. J. Bacteriol. 185:2432-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong, J., S. Iuchi, H. Kwan, Z. Lu, and E. C. C. Lin. 1993. The deduced amino-acid sequence of the cloned cpxR gene suggests the protein is the cognate regulator for the membrane sensor, CpxA, in a two-component signal transduction system of Escherichia coli. Gene 136:277-280. [DOI] [PubMed] [Google Scholar]
- 16.Grigorova, I. L., R. C. Chaba, H. J. Zhong, B. M. Alba, V. Rhodius, C. Herman, and C. A. Gross. 2004. Fine-tuning of the Escherichia coli σE envelope stress response relies on multiple mechanisms to inhibit signal-independent proteolysis of the transmembrane anti-sigma factor, RseA. Genes Dev. 18:2686-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagenmaier, S., Y. D. Stierhof, and U. Henning. 1997. A new periplasmic protein in Escherichia coli which is synthesized in spheroplasts but not in intact cells. J. Bacteriol. 179:2073-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, C. H., P. N. Danese, J. S. Pinkner, T. J. Silhavy, and S. J. Hultgren. 1997. The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J. 21:6394-6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, C. H., P. Dexter, A. K. Evans, C. Liu, S. J. Hultgren, and D. E. Hurby. 2002. Escherichia coli DegP protease cleaves between paired hydrophobic residues in a natural substrate: the PapA pilin. J. Bacteriol. 184:5762-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamitani, S., Y. Akiyama, and K. Ito. 1992. Identification and characterization of an Escherichia coli gene required for the formation of correctly folded alkaline phosphatase, a periplasmic enzyme. EMBO J. 11:57-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolmar, H., P. R. Waller, and R. T. Sauer. 1996. The DegP and DegQ periplasmic endoproteases of Escherichia coli: specificity for cleavage sites and substrate conformation. J. Bacteriol. 178:5925-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:801-802. [PubMed] [Google Scholar]
- 23.Krojer, T., M. Garrido-Franco, R. Huber, M. Ehrmann, and T. Clausen. 2002. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 416:455-459. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 25.Lipinska, B., M. Zylicz, and C. Georgopoulos. 1990. Identification, characterization and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J. Bacteriol. 172:1791-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, J., and C. T. Walsh. 1990. Peptidyl-prolyl cis-trans-isomerase from Escherichia coli: a periplasmic homolog of cyclophilin that is not inhibited by cyclosporine A. Mol. Microbiol. 87:4028-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakayama, S. I., and H. Watanabe. 1995. Involvement of cpxA, a sensor of a two-component regulatory system, in the pH-dependent regulation of expression of Shigella sonnei virF gene. J. Bacteriol. 177:5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nevesinjac, A. Z., and T. L. Raivio. 2005. The Cpx envelope stress response affects expression of the type IV bundle-forming pilus of enteropathogenic Escherichia coli. J. Bacteriol. 187:672-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pogliano, J., S. Lynch, D. Belin, E. Lin, and J. Beckwith. 1997. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 11:1169-1182. [DOI] [PubMed] [Google Scholar]
- 30.Raffa, R. G., and T. L. Raivio. 2002. A third envelope stress signal transduction pathway in Escherichia coli. Mol. Microbiol. 45:1599-1611. [DOI] [PubMed] [Google Scholar]
- 31.Raivio, T. L., M. W. Laird, J. C. Joly, and T. J. Silhavy. 2000. Tethering of CpxP to the inner membrane prevents spheroplast induction of the Cpx envelope stress response. Mol. Microbiol. 37:1186-1197. [DOI] [PubMed] [Google Scholar]
- 32.Raivio, T. L., D. L. Popkin, and T. J. Silhavy. 1999. The Cpx envelope stress response is controlled by amplification and feedback inhibition. J. Bacteriol. 181:5263-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raivio, T. L., and T. J. Silhavy. 1997. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J. Bacteriol. 179:7724-7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raivio, T. L., and T. J. Silhavy. 1999. The σE and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr. Opin. Microbiol. 2:159-165. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments in gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Slauch, J. M., and T. J. Silhavy. 1991. cis-Acting ompF mutations that result in OmpR-dependent constitutive expression. J. Bacteriol. 173:7039-7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snyder, W. B., L. Davis, P. N. Danese, C. L. Cosma, and T. J. Silhavy. 1995. Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J. Bacteriol. 177:4216-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sonyang, Z., A. S. Fanning, C. Fu, J. Xu, S. M. Marfatia, A. H. Chishti, A. Crompton, A. C. Chan, J. M. Anderson, and L. C. Cantley. 1997. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science 275:73-77. [DOI] [PubMed] [Google Scholar]
- 40.Strauch, K. L., K. Johnson, and J. Beckwith. 1989. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperatures. J. Bacteriol. 171:2689-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Mol. Microbiol. 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber, R. F., and P. M. Silverman. 1988. The Cpx proteins of Escherichia coli K12: structure of the CpxA polypeptide as an inner membrane component. J. Mol. Biol. 203:467-478. [DOI] [PubMed] [Google Scholar]