Abstract
The rcs phosphorelay pathway components were originally identified as regulators of capsule synthesis. In addition to the transmembrane sensor kinase RcsC, the RcsA coregulator, and the response regulator RcsB, two new components have been characterized, RcsD and RcsF. RcsD, the product of the yojN gene, now renamed rcsD, acts as a phosphorelay between RcsC and RcsB. Transcription of genes for capsule synthesis (cps) requires both RcsA and RcsB; transcription of other promoters, including that for the small RNA RprA, requires only RcsB. RcsF was described as an alternative sensor kinase for RcsB. We have examined the role of RcsF in the activation of both the rprA and cps promoters. We find that a number of signals that lead to activation of the phosphorelay require both RcsF and RcsC; epistasis experiments place RcsF upstream of RcsC. The RcsF sequence is characteristic of lipoproteins, consistent with a role in sensing cell surface perturbation and transmitting this signal to RcsC. Activation of RcsF does not require increased transcription of the gene, suggesting that modification of the RcsF protein may act as an activating signal. Signals from RcsC require RcsD to activate RcsB. Sequencing of an rcsC allele, rcsC137, that leads to high-level constitutive expression of both cps and rprA suggests that the response regulator domain of RcsC plays a role in negatively regulating the kinase activity of RcsC. The phosphorelay and the variation in the activation mechanism (dependent upon or independent of RcsA) provide multiple steps for modulating the output from this system.
Expression of colanic acid, the major exopolysaccharide of Escherichia coli K-12, is a highly regulated process. Initial screens for activators of this pathway identified RcsC, RcsB, and RcsA as the major components of the pathway (17). RcsC and RcsB are members of a two-component signal transduction pathway; RcsC is the transmembrane sensor kinase, and RcsB is the cytoplasmic response regulator (38). However, unlike standard two-component systems, the Rcs pathway also involves an additional cytoplasmic coregulator, RcsA. The RcsA/RcsB heterodimer is necessary for activation of capsule gene expression (cps); under conditions in which either RcsA accumulates or RcsC is activated, colonies overproduce colanic acid, obvious as a mucoid phenotype on plates (23, 39). RcsA is itself subject to rapid degradation by the Lon protease (41).
Recent work has demonstrated that a large number of operons in addition to cps are also subject to regulation by the RcsC/RcsB system. RcsB/RcsA stimulates both cps and transcription of rcsA itself; it also inhibits the transcription of the flhDC flagellar master switch (10, 14, 43, 44). Furthermore, other promoters have been identified which are dependent upon RcsB but do not require RcsA. These include ftsZ, osmC, osmB, bdm, and the small RNA RprA (2, 8, 13, 16, 24). Array studies suggest that the set of genes regulated may be considerably larger (12, 20, 29).
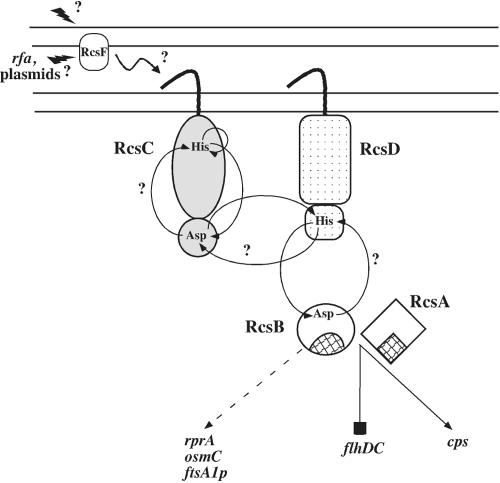
Sequence comparisons of RcsC to other sensor kinases reveal that it is a transmembrane hybrid sensor kinase. At the N terminus are two transmembrane segments, separated by a 273-amino-acid periplasmic domain. After the second transmembrane domain comes an N-terminal kinase domain that includes a conserved histidine residue (His 479) that is the predicted phosphorylation site and is known to be essential for function (5). In addition to this domain, there is a C-terminal domain with homology to response regulators; this contains a conserved aspartate residue (Asp 875), also essential for function (5). RcsB is a traditional response regulator. The N-terminal response regulator domain contains a conserved aspartate that is likely to be the site of phosphorylation; the C-terminal domain contains a helix-turn-helix motif. The presence of the extra response regulator domain on RcsC posed a problem in understanding how this system works. Phosphotransfer in all other systems has been observed to proceed from His→Asp→His→Asp. In some cases, hybrid sensor kinases carry an additional histidine phosphotransmitter domain not present in RcsC (for instance, in ArcB); in other cases, a separate protein acts as the phosphotransmitter (21, 27). Takeda et al. suggested that there was a missing component in this Rcs system that acts as a phosphorelay between the response regulator domain of RcsC and RcsB (40). Their search led them to the discovery of RcsD (formerly YojN), a putative transmembrane protein with a histidine phosphotransmitter domain (HPt) similar to that found in ArcB (40). The rcsD gene is in an operon with and immediately upstream of rcsB and encodes an 890-amino-acid protein with a predicted molecular mass of 100 kDa (Fig. 1A). Takeda et al. established that RcsD is essential in the signaling from RcsC to RcsB and demonstrated phosphate transfer in vitro between RcsD and RcsB (40).
FIG. 1.
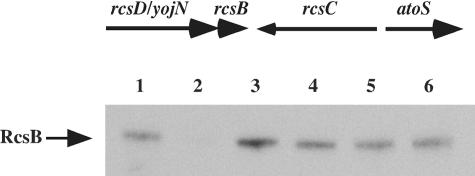
Genetic organization of the rcsB rcsD rcsC region and test of polarity of rcsD mutations on rcsB. The top panel is a schematic representation of the rcs gene organization, with arrows indicating the direction of transcription. The bottom panel is a Western blot analysis with an anti-RcsB antibody showing steady-state amounts of RcsB at mid-exponential growth. Lane 1, MC4100; lane 2, DH311 (rcsB); lane 3, DJ480 (parental); lane 4, DH339 (rcsD542); lane 5, DH351 (rcsD541); lane 6, DH368 (rcsD543).
Another component of the rcs pathway is RcsF. It was initially isolated as a multicopy suppressor of an ftsZ temperature-sensitive phenotype, leading to the demonstration that the ftsA1 promoter is regulated by RcsB (15, 16). RcsF was recently independently identified as a multicopy activator of the promoter of the small regulatory RNA, RprA, and its RcsB-dependent promoter (24). RcsF is a small protein (14 kDa) believed to contain an N-terminal transmembrane domain and no homology to sequences in the database. Gervais and Drapeau originally proposed that RcsF represents an RcsC-independent alternative for phosphorylation of RcsB (15).
In this work, we establish the epistasis through the rcs phosphorelay pathway, and in particular we address the role of RcsF and confirm and extend previous findings on the role of RcsD in this pathway. To this end, we have used the RcsA-dependent cps promoter and the RcsA-independent rprA promoter. In both cases, the data indicate that signaling proceeds through an ordered cascade, RcsF→RcsC→RcsD→RcsB, and that RcsF, rather than playing a role in direct phosphorylation of RcsB, plays a critical role in signal transduction from the cell surface to RcsC.
MATERIALS AND METHODS
Media and chemicals.
Luria-Bertani (LB) Lennox from KD Medical (Gaithersburg, MD) and tryptone broth (TB) media were used as indicated. Antibiotics and o-nitrophenyl-β-d-galactopyranoside were all from Sigma (St. Louis, MO). Oligonucleotides for the DH strain constructions and pNM plasmids are from Operon (Huntsville, AL).
Strain construction.
The strains used are listed in Table 1 or described below. DH324 (ΔrcsF) was constructed by first deleting the rcsF open reading frame (ORF), replacing it with a PCR-amplified chloramphenicol resistance plus sucrase cassette (cat-sacB) using recombineering (45). The cassette was then removed by this same technique with a single-stranded oligonucleotide, counterselecting for sucrose resistance for loss of the cat-sacB cassette as described in supplemental materials (supplemental materials and methods). The unmarked rcsF deletion was verified by sequencing.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant genotype | Comment(s) or referencea |
|---|---|---|
| DJ480 | MG1655 Δ(argF-lac)U169 | Parental to all DH strains |
| DH300 | rprA-lacZ | 24 |
| DH311 | rprA-lacZ rcsB::Kanr | 24 |
| DH312 | rprA-lacZ rcsC::Kanr | 24 |
| DH324 | rprA-lacZ ΔrcsF1 | See Materials and Methods |
| DH333 | rprA-lacZ ΔrcsF1 rcsC137 zei-10::Cmr | This study |
| DH339 | rprA-lacZ ΔrcsD542::Kanr | This study |
| DH351 | rprA-lacZ ΔrcsD541 | See Materials and Methods |
| DH354 | rprA-lacZ Δrfa-1::Cmr | This study |
| DH355 | rprA-lacZ Δrfa-1::CmrrcsC::Tetr | This study |
| DH356 | rprA-lacZ Δrfa-1::Cmr ΔrcsF1 | This study |
| DH357 | rprA-lacZ Δrfa-1::Cmr ΔrcsF1 rcsC::Tetr | This study |
| DH358 | rprA-lacZ ΔrcsD541 Δrfa-1::Cmr | This study |
| DH366 | rprA-lacZ rcsC137 zei-10::Cmr | This study |
| DH368 | rprA-lacZ ΔrcsD543::Kanr | This study |
| DH369 | rprA-lacZ rcsC137 zei-10::Cmr ΔrcsD543::Kanr | See Materials and Methods |
| DH371 | rprA-lacZ rcsC137 zei-10::Cmr ΔrcsD543::Kanr | See Materials and Methods |
| SG20781 | cpsB-lacZ lon+ | Parental to all VS and MH1-3 strains (3) |
| SG20803 | cpsB-lacZ rcsC137 ompC::Tn5 | 3; parental to MH4 |
| MH1 | cpsB-lacZ rfa-1::CmrrcsF118::Kanr | This study |
| MH2 | cpsB-lacZ rcsD339::Kanr | This study |
| MH3 | cpsB-lacZ rcsD339::Kanrrfa-1::Cmr | This study |
| MH4 | cpsB-lacZ rcsC137 ompC::Tn5 rcsF118::Kanr | This study |
| VS20299 | cpsB-lacZ rcsC10::Kanr | 18 |
| VS20324 | cpsB-lacZ rcsF118::Kanr | 18; rcsF allele from reference 15 |
| VS20302 | cpsB-lacZ rfa-1::Cmr | This study |
Strains made by P1 transduction for this work are indicated by “This study”; strains constructed in more complicated ways are indicated by “See Materials and Methods,”where the specifics of the construction are described.
To move the unmarked rcsF deletion into other strains, the cat-sacB cassette was transduced by phage P1 into CAG18447, a proAB::Tn10 strain (36), to link ΔrcsF to the proAB marked mutation, selecting for Cmr and screening for Tetr (strain NM110). P1 grown on NM110 was then used to transduce the ΔrcsF::cat-sacB and proAB::Tn10 mutations into DH300 (rprA-lacZ) selecting for Tetr and screening for Cmr (DH303). P1 grown on NM12 (described in the supplemental materials) was used to transduce the unmarked rcsF deletion into DH303, selecting for growth on minimal glucose plates (Pro+) and screening for loss of Cmr and loss of Tetr to create DH324. Three rcsD alleles are used in this work. All three delete the same 541 nucleotides at the 5′ end of rcsD (yojN) but have somewhat different sequences inserted. Strain DH351 (ΔrcsD541) was constructed according to the method of Datsenko and Wanner (7), using pKD13 as template and the ΔyojN.F and ΔyojN.R primers listed in supplementary table S1. The strain containing the Kanr cassette is DH339 (rcsD542::Kanr); the Kanr cassette was then eliminated by the Flp recombinase (7) to create DH351, and polarity on RcsB was tested in a Western blot (Fig. 1B). Only a modest decrease in RcsB amounts was found.
Strain DH369 (ΔrcsD543 rcsC137) was constructed by sequential P1 transductions (see supplemental materials). Starting with DH300 (rprA-lacZ), the rcsC137 mutation was introduced by selecting for the closely linked zei-10::Cmr marker to yield a very mucoid strain (DH366). This strain was then transduced with P1 grown on the rcsD543::Kanr mutant strain, selecting for kanamycin resistance. Because of the proximity of rcsD and rcsC on the chromosome and to make sure we retained the rcsC137 mutation, several colonies were cross-streaked on a background of phage SY7 (rcsD+) (3). Three of 12 colonies became mucoid in the presence of the phage; these were assumed to still carry the rcsC137 allele. Complementation was confirmed by transforming these candidates with a pBAD-rcsD plasmid, which also restored mucoidy, and by sequencing the rcsC137 mutation; one was designated DH369.
Strain DH371 was made in a multistep procedure (see supplemental materials). First, recombineering was used to insert a Cmr cassette to delete and replace a 4.5-kb region extending from the 5′ end of rcsD to the region at the 3′ of rcsC where the rcsC137 mutation is located. Second, from an rcsD::Kanr strain, a PCR product spanning this same deleted region was generated by using a primer that introduces the rcsC137 mutation into the product. This PCR product now carries the rcsD543::Kanr deletion-insertion and incorporates the rcsC137 point mutation as well as flanking regions of homology upstream and downstream of the deletion. Third, recombineering was used to replace the Cmr cassette in the first strain by the PCR product carrying the Kanr cassette and screening for the loss of Cmr. This mutation was transduced by P1 into a DH300 (rprA-lacZ) background selecting for Kanr and complemented to check for mucoidy with the pBAD-rcsD plasmid, and the rcsC137 mutation was sequenced. All the other DH, MH, and VS strains listed in Table 1 were constructed by P1 transduction into the appropriate wild-type or mutant background.
Cloning of rcsF.
Amplification of rcsF from the chromosome of E. coli was done using forward primer 5′-GGAATTCCTCGAGATGCGTGCTTTACCG and reverse primer 5′-CGCGGATCCGGAAACTGCTCATTTCGCCGT (IDT, Coralville, IA). AB1157 (E. coli Genetic Stock Center no. 1157) cells were grown overnight in LB medium at 37°C and diluted 1:100 in water, and 1 μl was used for a 50-μl PCR using Taq polymerase (Promega, Madison, WI). The PCR product was cloned into Invitrogen's TOPO2.1 plasmid (Carlsbad, CA) according to the manufacturer's specifications. Plasmids containing rcsF were digested and subcloned into pTrc99a from Amersham Pharmacia (Piscataway, NJ) using EcoRI and BamHI restriction enzymes (Promega, Madison, WI). The resulting plasmid construct is designated pMH300.
Cloning of rcsD.
The pNM501 arabinose-inducible rcsD plasmid was constructed by restriction-less cloning or recombineering (6, 45). The Expand High Fidelity PCR system from Roche (Basel, Switzerland) was used for inverse PCR according to the manufacturer's specifications. The pBAD24 plasmid (19) was used as template for inverse PCR along with forward primer 5′-GGTAGGAGTGAAAAGCGGGTCGTGGCCGTTGTCTCTTTCTGACGCATGAATTCCTCCTGCTAGCCCAAAAAAAC-3′ and reverse primer 5′-GAAAAATACATCAGCGACATTGACAGTTATGTCAAGAGCTTGCTGTAGGTACCCGGGGATCCTCTAGAGTCG-3′) to generate a linear pBAD24 vector with homologies to the start and end of the rcsD ORF. The PCR product was digested with 2 U of NcoI enzyme (NEB, Beverly, MA) for 2 h at 37°C to linearize the template and reduce the background in subsequent steps. The mix was purified with a Wizard PCR purification kit from Promega (Madison, WI) and checked on a gel for a single band of 4.5 kb. One-hundred nanograms of this DNA was electroporated into NM300 (MG1655, mini-λ-Tetr) after induction of the λ functions (6), selecting for Ampr transformants. Plasmids from 6 of 16 Ampr clones contained an insert of the appropriate size. All six were used to transform DH351, an rprA-lacZ rcsD strain, to test for complementation on lactose MacConkey plates with ampicillin. The rcsD mutant strain is somewhat Lac+ compared to the Lac− rcsD+ strain (see Table 4); all of the clones were complemented without any arabinose, returning the strain to Lac−. However, when the plasmid was introduced into either the rcsD mutant host or a (Lac−) rcsD+ host, DH300, the level of expression was increased in the presence of arabinose (data not shown), suggesting that higher levels of RcsD may allow signaling to RcsB, either via RcsC or via cross-talk, even in the absence of an inducing signal. The junction borders were confirmed by sequencing for two of these plasmids. They were identical, and one was designated pNM501.
TABLE 4.
Effect of rcsD mutations on rprA-lacZ expression
| Strain | Relevant genotype | β-Galactosidase expression (sp act)a |
|---|---|---|
| DH300 | Wild type | 1.3 ± 0.3 |
| DH351 | rcsD541 | 4.9 ± 0.6 |
| DH354 | rfa-1 | 9.2 ± 0.4 |
| DH358 | rfa-1 rcsD541 | 6.4 ± 0.4 |
| DH366 | rcsC137 | 37 ± 2.6 |
| DH369 | rcsC137 rcsD543 | 3.1 ± 0.5 |
| DH371 | rcsC137 rcsD543 | 1.9 ± 0.1 |
| DH371/pBAD-rcsD+ | rcsC137 rcsD543/pBAD-rcsD+ | 25 ± 3.0 (without arabinose) |
| 18.5 ± 3.7 (with arabinose) |
Average of at least two independent kinetic β-galactosidase experiments.
β-Galactosidase assays.
For the rprA-lacZ fusion, β-galactosidase kinetic assays were performed in a microtiter plate format and read in a SpectraMax250 plate reader as described previously (24). These specific activity units are about 1/25th of a Miller unit. For the cps-lacZ fusion, the standard Miller assay was used (26).
RNA extraction and fluorescent RT-PCR.
RNA extraction was performed with the hot-phenol procedure described previously (25). Prior to reverse transcription-PCR (RT-PCR), to reduce or eliminate residual genomic DNA, RNA samples were treated with Ambion's Turbo DNase (Austin, TX) according to the manufacturer's specifications. The RNA samples were extracted once with phenol, precipitated, and resuspended in diethyl pyrocarbonate water. First-strand cDNA synthesis was performed on 1 μg of this RNA in a 20-μl volume as follows: 4 μl Superscript 5× RT buffer, 2 μl deoxynucleoside triphosphates (10 mM stock), 2 μl dithiothreitol (100 mM stock), 0.2 μl bovine serum albumin (10 mg/ml stock), and 0.2 μl random hexamer primer (2 pmol stock). The mix was heated to 70°C for 5 min to denature the RNA and transferred to 50°C for 30 min to allow the primer to hybridize to the RNA. One microliter of Superscript III enzyme was added (Invitrogen, Carlsbad, CA) and incubated at 50°C for 30 min. The PCRs were performed in triplicates using the SYBR Green PCR kit from QIAGEN (Valencia, CA) according to the manufacturer's specifications and run in the Opticon II real-time cycler from MJ Research (Waltham, MA). The ompA mRNA was used as an endogenous control in all samples. Specific primers for rcsF and ompA (control) were designed using Oligo 6.8 software (MBI, Cascade, CO) to generate a 120-nucleotide fragment and have similar annealing temperatures (supplemental materials, Table S1). Relative quantification of the RT-PCR results is described by Pfaffl (33) and by R. Soong, J. Ruschoff, and K. Tabiti (2000) in a Roche Diagnostics internal publication. The threshold cycle parameter (CT) values for ompA were averaged and came out to 14.05, a number that was used in subsequent calculations. The comparative expression level (CEL) is 2−ΔΔCT and is derived as follows: first, the equation ΔCT = CT (target) − CT (ompA normalizer 14.05) is calculated for each sample. Second, from among those samples, the negative control DH324 is chosen as the baseline signal. The next step is to calculate the equation ΔΔCT = ΔCT (sample) − ΔCT (baseline DH324). Finally, that number is used for calculating the CEL.
Western blot of RcsB.
For Western blots of RcsB, cells were grown to an optical density at 600 nm of ∼0.5, and 1 ml of culture was immediately transferred to a tube containing trichloroacetic acid (TCA), 5% final concentration. Tubes were incubated on ice for 10 min and centrifuged for 10 min. Supernatants were removed, and pellets were washed once with cold acetone, resuspended in sodium dodecyl sulfate sample buffer, boiled for 5 min, and loaded proportionally to their respective optical densities. The 10% NuPAGE gels (Invitrogen, CA) were run and transferred onto a nitrocellulose membrane, blocked with 5% dry milk (Bio-Rad, CA), and probed with anti-RcsB antibody (Animal Research Center, Arizona State University) (Fig. 1).
Sequencing the rcsC137 mutation.
The rcsC gene from the genome of strain SG20803 was amplified in fragments of 750 bp using Promega's Taq polymerase (Madison, WI) according to the manufacturer's specifications. Both strands of each fragment were sequenced and assembled to cover the 2.8-kb rcsC gene in its entirety. Note that the originally published sequence for rcsC showed an initiation site that is 16 amino acids after that now currently defined by comparison with the sequence of rcsC in other organisms. Numbering of RcsC amino acids in this work uses the longer 949-amino-acid protein.
RESULTS
RcsF is required to sense signals.
Previously, we had isolated rcsF in a screen of multicopy plasmids that stimulated the activity of the rprA-lacZ fusion. In that screen, several other plasmids were also isolated that activated rprA-lacZ activity; all were dependent upon RcsB (24). We tested the dependence of these plasmids on RcsF and RcsC for activation of the rprA-lacZ fusion. We had previously found that an insertion in rcsC led to a modest increase in expression of the rprA-lacZ fusion (24), presumably a reflection of cross-talk to RcsB and loss of the phosphatase activity of RcsC. None of the plasmids were able to further stimulate the fusion in the rcsC deletion, suggesting that they were all dependent upon RcsC (data not shown). This is consistent with previous observations of RcsC dependence of signaling by osmotic shock, mutations in the cell surface, and overexpression of some proteins (4, 11, 31, 37). We investigated whether RcsF was also required for these signals.
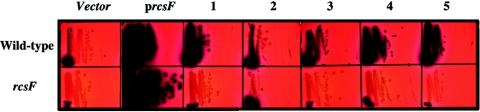
An rcsF mutant was constructed and introduced into both the rprA-lacZ fusion strain and the cps-lacZ fusion. This mutation had a modest effect in reducing the basal level of expression of the fusions (Fig. 2, vector panels, compare top and bottom, and Table 2, compare VS20324 to SG20781). As expected, the plasmid carrying rcsF stimulated expression of the fusion and complemented the rcsF mutant (Fig. 2). All other plasmids failed to activate the rprA-lacZ fusion in the rcsF mutant background (Fig. 2). These results demonstrate that signaling from these plasmids is dependent upon RcsF as well as RcsC.
FIG. 2.
Multicopy inducers of rprA-lacZ fusion require RcsF. A set of previously isolated plasmids (24) was used to transform DH300 (wild-type) and DH324 (rcsF) strains carrying an rprA-lacZ fusion. Plasmid-bearing cells were streaked on MacConkey lactose-ampicillin plates and incubated at 37°C. A representative gene is listed, and plasmids are described fully in Table 1 of reference 24. Lane 1, pwzxC; lane 2, pdnaK; lane 3, ptolB; lane 4, pspot42; and lane 5, pyeiU.
TABLE 2.
Regulation of expression of cps-lacZ fusion
| Strain | Relevant genotype
|
Units of β-galactosidasea in tryptone broth | |||
|---|---|---|---|---|---|
| rfa | rcsF | rcsC | rcsD | ||
| SG20781 | + | + | + | + | 1 ± 0.2 |
| VS20299 | + | + | − | + | 0.6 ± 0.1 |
| VS20324 | + | − | + | + | 0.4 ± 0.2 |
| MH2 | + | + | + | − | 0.7 ± 0.3 |
| VS20302 | − | + | + | + | 54 ± 4 |
| MH1 | − | − | + | + | 0.3 ± 0.3 |
| MH3 | − | + | + | − | 0.4 ± 0.1 |
| SG20803 | + | + | C137 | + | 455 ± 113 |
| MH4 | + | − | C137 | + | 447 ± 54 |
| SG20781(prcsF+) | + | +++ | + | + | 79 ± 10 |
| VS20299(prcsF+) | + | +++ | − | + | 0.5 ± 0.1 |
| MH2(prcsF+) | + | +++ | + | − | 0.6 ± 0.3 |
Average of three independent Miller assays.
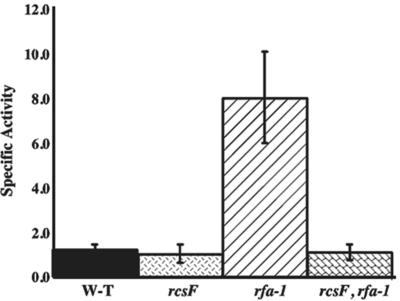
Deep rough mutants (rfa) carry mutations in the lipopolysaccharide biosynthesis pathway that result in constitutive cps expression and mucoid colonies on plates (31), affording us another way to induce the Rcs pathway. We introduced an rfa-1 mutation into a wild-type or an rcsF null strain of E. coli carrying a single-copy chromosomal cps-lacZ or rprA-lacZ fusion and measured the β-galactosidase activity. As shown in Table 2 (VS20302 and MH1), activity of the cps-lacZ fusion increased significantly in the rfa-1 mutant; all of this increase was abolished in an rcsF mutant. Similarly for rprA-lacZ, in the rfa-1 strain activity of the fusion is high, but in the rcsF rfa-1 mutant the fusion was not activated above basal levels (Fig. 3). As previously shown for cps synthesis (31), signaling to both rprA and cps also required RcsC; an rfa-1 rcsC double mutant expressed the fusions at the level of the rcsC mutant (data not shown). Therefore, as for the plasmids, RcsF is necessary for signal transmission in the rfa-1 mutant, both for cps-lacZ and for rprA-lacZ.
FIG. 3.
Signaling through RcsF in an rfa mutant. Strains DH300 (wild-type), DH324 (rcsF), DH354 (rfa-1), and DH356 (rfa-1 rcsF) were grown overnight and diluted into LB media at 37°C. From an optical density at 600 nm of 0.1 and onward, samples were taken at 25-min time intervals and sampled in a kinetic microtiter plate assay as described in Materials and Methods. Bars represent the average specific activity over the length of sampling and are an average of three separate experiments. W-T, wild type.
rfa mutations activate the phosphorelay without affecting the transcription of rcsF.
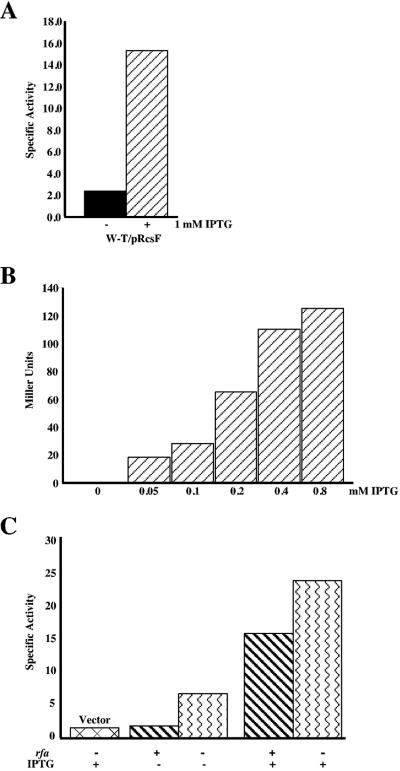
How do rfa-1 mutants lead to activation of the RcsC/RcsB phosphorelay via RcsF? Do they increase the synthesis of RcsF or somehow change its activity? The isolation of plasmids containing rcsF as a multicopy activator suggested that increased synthesis was sufficient for activation (15, 24). We confirmed this using a multicopy isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible rcsF plasmid. As expected, induction of rcsF expression leads to an increase in activity from both the rprA-lacZ fusion and from the cps-lacZ fusion (Fig. 4A and B).
FIG. 4.
Dose-dependent stimulation by RcsF and rfa acts independently of rcsF transcription. A) DH300 (rprA-lacZ) cells carrying pMH300 (ptac-rcsF) were grown without or with IPTG as indicated. A kinetic β-galactosidase assay was performed, and bars represent an average of three assays. B) SG20781 (cps-lacZ) cells carrying pMH300 were grown in TB medium with increasing concentrations of IPTG and assayed in a standard Miller assay. Each bar represents the average of three independent assays. C) DH324 (rcsF) and DH356 (rcsF rfa-1) strains carrying pMH300 were grown in the absence or presence of 1 mM IPTG as indicated. The first bar is DH356 with a vector control grown in 1 mM IPTG shown for basal levels. Cultures were assayed three times in a kinetic β-galactosidase assay and averaged. W-T, wild type.
If the rfa-1 mutant activates the Rcs relay by increasing transcription from the rcsF promoter, we would not expect it to lead to further activation of the phosphorelay when rcsF is made from a foreign promoter. We tested this using the multicopy IPTG-inducible rcsF plasmid to transform an rcsF and an rcsF rfa-1 strain carrying the rprA-lacZ fusion. We measured the lacZ activity in both of these strains using either no IPTG or 1 mM IPTG (Fig. 4C). In the double rcsF rfa-1 mutant strain, rcsF activation of rprA-lacZ was still enhanced by the rfa-1 mutation both in the absence and presence of IPTG (Fig. 4C, compare column 2 to 3 and column 4 to 5). Therefore, the effect of rfa-1 is independent of the rcsF promoter, and we conclude that the rfa mutant is not activating expression of the Rcs system by affecting the transcription of rcsF. It may act to affect its translation or, more likely, its activity.
To confirm this result with rcsF in its normal context, we used RT-PCR to measure the levels of rcsF transcription from the chromosome in a wild-type strain compared to an rfa-1 strain. While we could detect the increased rcsF transcription from a multicopy plasmid, the rfa-1 mutant showed no increase in rcsF transcripts over an rfa+ host (Table 3).
TABLE 3.
Relative mRNA levels of rcsF by RT-PCR
| Variable | DH300 (wild type) | DH324 (ΔrcsF) | DH354 (rfa-1::Cmr) | DH300/prcsF plus IPTG (wild type/prcsF) |
|---|---|---|---|---|
| CT | 21.8 | 33.8 | 22.3 | 15.9 |
| ΔCTa | 7.75 | 19.75 | 8.25 | 1.85 |
| ΔΔCTb | −12.0 | 0 | −11.5 | −17.9 |
| CELc | 4096 | 1 | 2896 | 244589 |
ΔCT = CT (target) − CT (ompA normalizer 14.05).
ΔΔCT = ΔCT (sample) − ΔCT (baseline DH324).
The comparative expression level is 2−ΔΔCT.
RcsF is upstream of RcsC and RcsD in the signaling pathway.
RcsC is the sensor kinase for the Rcs phosphorelay and has been shown to be necessary for activation of the system by osmotic shock, some mutants, and other signals (4, 11, 31, 37). RcsD (formerly YojN) has been shown to be essential for RcsC→ RcsB signaling (40). As indicated in Fig. 1, rcsD is the upstream gene in an operon with rcsB with convergent orientation to rcsC. There is a high similarity in domain organization between RcsD and the histidine kinase RcsC with two crucial differences: RcsD lacks the His autophosphorylation site, and the C-terminal receiver domain is replaced by a predicted phosphotransfer (HPt) domain (40). We asked whether RcsF is upstream or downstream of RcsC and RcsD in the signaling pathway.
The relative positions of RcsF, RcsC, and RcsD in the signaling pathway were tested using epistasis experiments, activating the system initially by RcsF overproduction from a plasmid in cells carrying rcsC or rcsD mutants. An rcsC::Kanr insertion mutant was used to inactivate rcsC. Two rcsD insertion-deletion derivatives were created and tested for polarity on rcsB as described in Materials and Methods. About a twofold decrease in RcsB levels was seen in the presence of the rcsD mutations (Fig. 1), confirming previous results suggesting that RcsB can be expressed from sequences within rcsD (38). Generally, the rcsD541::Kan allele was used in the experiments described below.
The results of such experiments for the cps-lacZ fusion are shown in Table 2 and for the rprA-lacZ fusion in Fig. 5A. At a concentration of 1 mM IPTG, both fusions are expressed at a significant level, as shown above. In contrast, in both the rcsC and rcsD mutants, induction of rcsF expression from the plasmid fails to stimulate either fusion above basal level (Table 2, compare SG20781 prcsF to VS20299 and MH2, each carrying prcsF; Fig. 5A, compare column 2 to columns 4 and 6). Thus, RcsF is dependent upon both RcsC and RcsD for signaling to rprA and cps promoters.
FIG. 5.
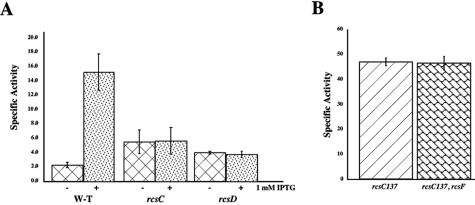
Epistasis of rcsD and rcsC to rcsF. A) DH300 (wild-type), DH312 (rcsC), and DH351 (rcsD) cells harboring the pMH300 plasmid were grown in LB media in the absence or presence of IPTG as indicated. Kinetic β-galactosidase assays were performed, and the average of three assays is graphed. B) DH366 (rcsC137) and DH369 (rcsC137 rcsF) were grown in LB media and assayed in a kinetic β-galactosidase assay. Bars represent the averages of two independent experiments. W-T, wild type.
In a second test, the epistasis between rfa-1 and rcsD was determined. If the effect of an rfa-1 mutant is upstream of rcsF, and rcsD is downstream of rcsF, rfa-1 signaling should be abolished in an rcsD mutant. This is what was observed (Table 2). The high-level expression of cps-lacZ in an rfa-1 mutant (VS20302) was abolished when an rcsD mutation was introduced (MH3). A similar effect was seen for rprA-lacZ (Table 4, compare DH354 to DH358), although the high basal level of expression of the rprA-lacZ fusion in the absence of rcsD (DH351) masks some of the effect.
If RcsF acts upstream of RcsC to activate the RcsC histidine kinase, a mutant form of rcsC that is already active should be independent of RcsF. We used the rcsC137 mutation to test this prediction. Previously isolated as an rcsC recessive mutation that strongly activated the cps-lacZ fusion (17), rcsC137 was sequenced and found to carry a single-nucleotide mutation in the codon GCT, changing it to GTT, which results in changing Ala904 to Val in the response regulator domain of RcsC. The mutation leads to constitutive and unregulated phosphorylation of RcsB (data not shown). Previously, we had noted in Northern blots that the amounts of RprA RNA increased by about 30-fold in an rcsC137 mutant over the wild type (24), consistent with activation of the Rcs phosphorelay for rprA as well as for cps. Isogenic strains carrying rcsC137, with and without an rcsF mutant, and either the rprA-lacZ fusion (Fig. 5B) or the cps-lacZ fusion (Table 2) were assayed. As expected, introduction of the rcsC137 allele significantly increased expression of both the rprA-lacZ fusion (Fig. 5B, column 1) and the cps-lacZ fusion (Table 2, SG20803), consistent with previous experiments. The presence of the rcsF null mutation had no effect on expression, confirming that rcsC is epistatic to rcsF (Fig. 5B, column 2; Table 2, MH4).
Testing epistasis between rcsC137 and rcsD was complicated by their tight linkage. Therefore, strains were constructed both by P1 transduction and by linear transformation, as described in Materials and Methods (also see supplemental materials). In both cases, the rcsC137 rcsD double mutant lost the mucoid characteristic of the rcsC137 parent, consistent with epistasis of the rcsD mutant.
We further quantitated this by assay of the rprA-lacZ fusion in the double mutant. The increased expression of rprA-lacZ in the rcsC137 strain was totally abolished by the rcsD mutant, reducing fusion expression to that found for the rcsD mutant alone (Table 4). Introduction of a plasmid expressing RcsD from an arabinose-inducible promoter complemented the rcsD defect, restoring activity to about half of that seen in the rcsC137 rcsD+ case in the absence or presence of arabinose (Table 4). The twofold decrease in activity may reflect some polarity of the rcsD insertion mutation on rcsB (Fig. 1).
Possible cross-talk in the Rcs phosphorelay.
The phenotype of an rcsC or rcsD mutant is very close to that of a wild-type strain for the cps-lacZ fusion in the absence of any inducing treatment (Table 2). However, we noted previously that the wild-type basal level for the rprA-lacZ fusion is significantly elevated in an rcsC mutant (24); we find a similar, not quite as marked elevation in the rcsD mutant strain (Table 4). In the experiments reported here, the normal basal level of the rprA-lacZ fusion in a wild-type strain is about 1.5 U, while it is about 6 and 4 U for the rcsC and rcsD mutant strains, respectively. One interpretation of these results is that there is phosphorylation of RcsB from other sources (cross-talk); in the absence of the specific phosphotransfer and phosphatase proteins (RcsD and RcsC, respectively), this phosphate cannot be removed, leading to higher expression of the fusion. This would also suggest that low levels of RcsB-phosphate give a detectable signal for rprA-lacZ but not for cps-lacZ. Whether this is a characteristic of RcsA-independent promoters or is specific to rprA is not known. Deletion of two genes necessary for synthesis of acetyl-phosphate, a small molecule known to be able to phosphorylate various response regulators, ackA and pta, did not affect the elevated basal level of the fusion in the rcsC mutant (data not shown), suggesting that cross-talk must come from another, as yet unidentified, source.
DISCUSSION
While many two-component systems and phosphorelays have been studied, in many cases we know relatively little about how signals are transmitted to the sensor kinase. The data we present here establish RcsF as an important component of the Rcs phosphorelay and place it upstream of RcsC for signaling both to the cps promoter and the rprA promoter. The dependence on RcsF was true in rfa mutants (Fig. 3) and in cells carrying a number of multicopy plasmids that activate the Rcs system (Fig. 2). In studies by other labs, mutations in pgsA, necessary for synthesis of acidic phospholipids, induced the Rcs system, as did growth of wild-type cells at low temperature in the presence of excess Zn2+; in both cases, induction required both RcsF and RcsC (20, 35). Therefore, a variety of perturbations to the cell surface and/or environmental signals act via RcsF to activate RcsC.
Our experiments also show that the induction of the signal via RcsF is independent of the normal transcriptional signals for RcsF (Fig. 4C) and does not result in increased transcription of rcsF (Table 3). Nonetheless, overproduction of RcsF in the absence of another inducing signal is sufficient to activate the phosphorelay (Fig. 4A and B). These data are most consistent with a model in which RcsF interacts, directly or indirectly, with RcsC to activate the phosphorelay (Fig. 6). Under the usual inducing conditions, changes in the RcsF structure or modifications to the protein may increase this interaction; overproduction may drive the interaction in the absence of this proposed structural change. Alternatively, overproduced RcsF may otherwise perturb the cell to provide a signal, which then acts through RcsC, much as the other plasmids in Fig. 2 do. Some, but not all, of the plasmids in Fig. 2 encode membrane proteins, and we do not currently know how directly any of them act on RcsF to induce the Rcs signaling cascade.
FIG. 6.
Model of the Rcs signaling pathway. The schematic diagram is not to scale but shows the signaling flow through the Rcs cascade. Question marks indicate experimentally unresolved issues. RcsF localization to the outer membrane is according to in silico predictions based on the amino acid sequence and A. Jacq (personal communication; see Discussion). Amino acids involved in the phosphorelay are indicated. The shaded areas of RcsA and RcsB indicate the DNA-binding domains of these proteins. A dashed arrow points to genes that are RcsB dependent but RcsA independent. A blocked line indicates a gene repressed by an RcsA-RcsB heterodimer, and a solid arrow points to genes that require RcsA and RcsB for activation (see the text for details).
RcsF has the sequence characteristics of an outer membrane lipoprotein and includes disulfide bonds that need to be maintained by interactions with DsbA (22). Palmitate labeling and density fractionation confirm that RcsF is an outer membrane lipoprotein (A. Jacq, personal communication). As an outer membrane protein, it may be poised to sense perturbations in the cell surface caused by osmotic shock or mutation of lipopolysaccharide synthesis, although the mechanism of this sensing remains unknown. If it interacts directly with RcsC, that interaction is most likely to take place in the periplasm (Fig. 6). We note that another lipoprotein, NlpE, has been implicated in the signaling pathway to Cpx (9, 30). The Cpx sensor kinase and RcsC have another characteristic in common: both are activated by interaction of the bacteria with surfaces, although possibly at different steps in the attachment process (12, 30). Finally, it is intriguing that overproduction of another putative lipoprotein, YpdI, has been found to activate the Rcs phosphorelay independently of RcsF (34); overproduction of a lipoprotein-specific chaperone, LolA, has also been found to activate the Rcs phosphorelay (4), although its dependence on RcsF or YpdI is not known. These findings suggest that lipoproteins may play important roles in the upstream sensing of signals for membrane sensor kinases; exactly what those signals are and how they are sensed is not yet understood.
RcsF was first identified by Gervais and Drapeau by its ability, in multicopy, to activate transcription from the RcsB-dependent ftsA1p promoter (15, 16). They suggested that RcsF is capable of phosphorylating RcsB directly or indirectly but in an RcsC-independent manner, based on measurements of colanic acid in an rcsC mutant carrying an rcsF plasmid. In our tests of epistasis between rcsF and rcsC, we unequivocally demonstrate that RcsF is upstream of and dependent upon RcsC. One possible explanation for the results of Gervais and Drapeau comes from our observation that an rcsC null mutant does increase activation of the very sensitive rprA-lacZ fusion. Possibly the increased colanic acid they measured reflected this same modest increase in activation of cps that the cps-lacZ fusion is not sensitive enough to detect.
The elevated basal level of the rprA-lacZ fusion observed in an rcsC or rcsD strain suggests that the Rcs system is capable of integrating signals from other sources. We are assuming that, in the absence of RcsD and RcsC, phosphorylation of RcsB by other means cannot be reversed, as it presumably is in the wild-type case. Therefore, dephosphorylation of RcsB may proceed through RcsD to RcsC. This is in agreement with studies on the dephosphorylation of response regulators ArcA and TorR, both of which have complex upstream signaling sensor kinases; dephosphorylation in these cases was shown to require the phosphotransfer domains as well as receiver domains in the sensor kinase (1, 32). We do not currently know which systems might provide the cross-talk phosphorylation of RcsB. ArcB kinase was able to phosphorylate RcsD in vitro (40), and we have also observed EnvZ and FixL phosphorylation of RcsB in vitro (M. Heck, G. Gupte, and V. Stout, unpublished data). An in vivo genome-wide analysis by Hagiwara et al. found evidence of cross-talk between the PhoQ/PhoP and Rcs systems (20). Other mechanisms for cooperation between a response regulator like PhoP and RcsB are also possible, however. For instance, PhoP appears to promote activation of an RcsB-dependent promoter of ugd under some conditions by binding to a site upstream of the RcsB site (28). Similarly, interactions between RcsA and RcsB can activate cps transcription under conditions that are not known to activate the RcsC cascade (stabilization of RcsA in a lon mutant) (39). Thus, any mechanism for improving RcsB binding to its target might bypass the requirement for phosphorylation. However, it is not clear why rcsD or rcsC mutants would improve the effect of cooperative binding by another protein, for instance, in activating rprA transcription. Therefore, we currently favor phosphorylation of RcsB from an unknown source as the explanation for increased expression of rprA in rcsC and rcsD null mutants.
rcsC137 leads to high-level activation of both capsule synthesis and RprA synthesis. We found that this mutation changes Ala904 to Val. Alanine is absolutely conserved in RcsC proteins from different species; Ala or Gly is found at this position in 80% of response regulators (42). rcsC137 is recessive to wild-type rcsC and can also be complemented by the response regulator domain of rcsC, consistent with the sequence localization and suggesting that rcsC137 is a loss-of-function mutant (3, 5). However, it seems unlikely that rcsC137 is solely a mutant that has lost phosphatase activity. If so, we would have expected the same low level of activity of cps and rprA seen with an rcsC null mutation (Fig. 5), which must certainly have also lost phosphatase activity. Therefore, this domain of RcsC must also act negatively in some other way; we suggest that it may negatively regulate the activity of the histidine kinase as well as play a necessary role in dephosphorylation. A number of other mutations isolated for increased cps-lacZ expression also map to this domain but had less dramatic effects than rcsC137 (M. Heck and V. Stout, unpublished results). Deletion of the domain will not itself lead to activation of RcsB, since it also plays an essential role in the phosphorelay. Under conditions where no signal is activating the Rcs phosphorelay, any phosphorylation of RcsB from other sources will have little effect, due to reversal of the phosphorylation through RcsD to RcsC. Under activating conditions, the flow of phosphate is reversed. Whether activation of the kinase activity of RcsC mimics the effect of an rcsC137 mutation, relieving the negative regulation by the D1 domain, or works by another pathway that overcomes this negative effect is not yet known. Also not known is whether there are environmental conditions under which cross-talk is not efficiently reversed and becomes physiologically important.
In summary, our results show that regulation of the RcsA-dependent cps promoter and the RcsA-independent rprA promoter are completely parallel in their dependence on the components of the Rcs phosphorelay, although the sensitivities of the promoters may be somewhat different. All activating signals we have tested here are dependent upon both RcsF and RcsC. RcsF thus plays an important upstream role in transmitting many signals to RcsC, and the activity, rather than the transcription, of RcsF appears to be controlled by a far-upstream signal, such as that created by an rfa-1 mutant. This paradigm for upstream signaling to a sensor kinase by a lipoprotein may be preserved for other phosphorelays as well.
Supplementary Material
Acknowledgments
We thank Don Court for strains and advice on strain construction and members of our laboratories for comments and suggestions on this work. We thank Annick Jacq for allowing us to cite her unpublished results.
This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute Center for Cancer Research.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ansaldi, M., C. Jourlin-Castelli, M. Lepelletier, L. Theraulaz, and V. Mejean. 2001. Rapid dephosphorylation of the TorR response regulator by the TorS unorthodox sensor in Escherichia coli. J. Bacteriol. 183:2691-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boulanger, A., A. Francez-Charlot, A. Conter, M.-P. Castanie-Cornet, K. Cam, and C. Gutierrez. 2005. Multistress regulation in Escherichia coli: expression of osmB involves two independent promoters responding either to σS or to the RcsCDB His-Asp phosphorelay. J. Bacteriol. 187:3282-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brill, J. A., C. Quinlan-Walshe, and S. Gottesman. 1988. Fine-structure mapping and identification of two regulators of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 170:2599-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, M. H., S. Takeda, H. Yamada, Y. Ishii, T. Yamashino, and T. Mizuno. 2001. Characterization of the RcsC->YojN->RcsB phosphorelay signaling pathway involved in capsular synthesis in Escherichia coli. Biosci. Biotechnol. Biochem. 65:2364-2367. [DOI] [PubMed] [Google Scholar]
- 5.Clarke, D. J., S. A. Joyce, C. M. Toutain, A. Jacq, and I. B. Holland. 2002. Genetic analysis of the RcsC sensor kinase from Escherichia coli K-12. J. Bacteriol. 184:1204-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Court, D. L., S. Swaminathan, D. Yu, H. Wilson, T. Baker, M. Bubunenko, J. Sawitzke, and S. K. Sharan. 2003. Mini-lambda: a tractable system for chromosome and BAC engineering. Gene 315:63-69. [DOI] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davalos-Garcia, M., A. Conter, I. Toesca, C. Gutierrez, and K. Cam. 2001. Regulation of osmC gene expression by the two-component system rcsB-rcsC in Escherichia coli. J. Bacteriol. 183:5870-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiGiuseppe, P. A., and T. J. Silhavy. 2003. Signal detection and target gene induction by the CpxRA two-component system. J. Bacteriol. 185:2432-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebel, W., and J. E. Trempy. 1999. Escherichia coli RcsA, a positive activator of colanic acid capsular polysaccharide synthesis, functions to activate its own expression. J. Bacteriol. 181:577-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebel, W., G. J. Vaughn, H. K. Peters III, and J. E. Trempy. 1997. Inactivation of mdoH leads to increased expression of colanic acid capsular polysaccharide in Escherichia coli. J. Bacteriol. 179:6858-6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrieres, L., and D. J. Clarke. 2003. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. Mol. Microbiol. 50:1665-1682. [DOI] [PubMed] [Google Scholar]
- 13.Francez-Charlot, A., M. P. Castanie-Cornet, C. Gutierrez, and K. Cam. 2005. Osmotic regulation of the Escherichia coli bdm (biofilm-dependent modulation) gene by the RcsCDB His-Asp phosphorelay. J. Bacteriol. 187:3873-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francez-Charlot, A., B. Laugel, A. Van Gemert, N. Dubarry, F. Wiorowski, M. P. Castanie-Cornet, C. Gutierrez, and K. Cam. 2003. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol. Microbiol. 49:823-832. [DOI] [PubMed] [Google Scholar]
- 15.Gervais, F. G., and G. R. Drapeau. 1992. Identification, cloning, and characterization of rcsF, a new regulator gene for exopolysaccharide synthesis that suppresses the division mutation ftsZ84 in Escherichia coli K-12. J. Bacteriol. 174:8016-8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gervais, F. G., P. Phoenix, and G. R. Drapeau. 1992. The rcsB gene, a positive regulator of colanic acid biosynthesis in Escherichia coli, is also an activator of ftsZ expression. J. Bacteriol. 174:3964-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottesman, S., P. Trisler, and A. S. Torres-Cabassa. 1985. Regulation of capsular polysaccharide synthesis in Escherichia coli K12: characterization of three regulatory genes. J. Bacteriol. 162:1111-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupte, G., C. Woodward, and V. Stout. 1997. Isolation and characterization of rcsB mutations that affect colanic acid capsule synthesis in Escherichia coli K-12. J. Bacteriol. 179:4328-4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagiwara, D., M. Sugiura, T. Oshima, H. Mori, H. Aiba, T. Yamashino, and T. Mizuno. 2003. Genome-wide analyses revealing a signaling network of the RcsC-YojN-RcsB phosphorelay system in Escherichia coli. J. Bacteriol. 185:5735-5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoch, J. A., and K. I. Varughese. 2001. Keeping signals straight in phosphorelay signal transduction. J. Bacteriol. 183:4941-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadokura, H., H. Tian, T. Zander, J. C. A. Bardwell, and J. Beckwith. 2004. Snapshots of DsbA in action: detection of proteins in the process of oxidative folding. Science 303:534-537. [DOI] [PubMed] [Google Scholar]
- 23.Majdalani, N., and S. Gottesman. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59:379-405. [DOI] [PubMed] [Google Scholar]
- 24.Majdalani, N., D. Hernandez, and S. Gottesman. 2002. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 46:813-826. [DOI] [PubMed] [Google Scholar]
- 25.Massé, E., F. E. Escorcia, and S. Gottesman. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17:2374-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Mizuno, T. 1997. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res. 4:161-168. [DOI] [PubMed] [Google Scholar]
- 28.Mouslim, C., T. Latifi, and E. A. Groisman. 2003. Signal-dependent requirement for the co-activator protein RcsA in transcription of the RcsB-regulated ugd gene. J. Biol. Chem. 278:50588-50595. [DOI] [PubMed] [Google Scholar]
- 29.Oshima, T., H. Aiba, Y. Masuda, S. Kanaya, M. Sugiura, B. L. Wanner, H. Mori, and T. Mizuno. 2002. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol. Microbiol. 46:281-291. [DOI] [PubMed] [Google Scholar]
- 30.Otto, K., and T. J. Silhavy. 2002. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. USA 99:2287-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker, C. T., A. W. Kloser, C. A. Schnaitman, M. A. Stein, S. Gottesman, and B. W. Gibson. 1992. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J. Bacteriol. 174:2525-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pena-Sandoval, G. R., O. Kwon, and D. Georgellis. 2005. Requirement of the receiver and phosphotransfer domains of ArcB for efficient dephosphorylation of phosphorylated ArcA in vivo. J. Bacteriol. 187:3267-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Potrykus, J., and G. Wegrzyn. 2004. The ypdI gene codes for a putative lipoprotein involved in the synthesis of colanic acid in Escherichia coli. FEMS Microbiol. Lett. 235:265-271. [DOI] [PubMed] [Google Scholar]
- 35.Shiba, Y., Y. Yokoyama, Y. Aono, T. Kiuchi, J. Kusaka, K. Matsumoto, and H. Hara. 2004. Activation of the Rcs signal transduction system is responsible for the thermosensitive growth defect of an Escherichia coli mutant lacking phosphatidylglycerol and cardiolipin. J. Bacteriol. 186:6526-6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating resistance elements for genetic mapping. Microbiol. Rev. 53:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sledjeski, D. D., and S. Gottesman. 1996. Osmotic shock induction of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 178:1204-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stout, V., and S. Gottesman. 1990. RcsB and RcsC: a two-component regulator of capsule synthesis in Escherichia coli. J. Bacteriol. 172:659-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stout, V., and S. Gottesman. 1991. Regulation of capsular polysaccharide synthesis in Escherichia coli K12. Mol. Microbiol. 5:1599-1606. [DOI] [PubMed] [Google Scholar]
- 40.Takeda, S., Y. Fujisawa, M. Matsubara, H. Aiba, and T. Mizuno. 2001. A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC->YojN->RcsB signalling pathway implicated in capsular synthesis and swarming behavior. Mol. Microbiol. 40:440-450. [DOI] [PubMed] [Google Scholar]
- 41.Torres-Cabassa, A. S., and S. Gottesman. 1987. Capsule synthesis in Escherichia coli K-12 is regulated by proteolysis. J. Bacteriol. 169:981-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Volz, K. 1995. Structural and functional conservation in response regulators, p. 53-64. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, D.C.
- 43.Wehland, M., and F. Bernhard. 2000. The RcsAB box. Characterization of a new operator essential for the regulation of exopolysaccharide biosynthesis in enteric bacteria. J. Biol. Chem. 275:7013-7020. [DOI] [PubMed] [Google Scholar]
- 44.Wehland, M., C. Kiecker, D. L. Coplin, O. Kelm, W. Saenger, and F. Bernhard. 1999. Identification of the RcsA/RcsB recognition motif in the promoters of exopolysaccharide biosynthetic operons from Erwinia amylovora and Pantoea stewartii subspecies stewartii. J. Biol. Chem. 274:3300-3307. [DOI] [PubMed] [Google Scholar]
- 45.Yu, D. G., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.