Abstract
Bacteriophage T4 effects host lysis with a holin, T, and an endolysin, E. T and E accumulate in the membrane and cytoplasm, respectively, throughout the period of late gene expression. At an allele-specific time, T triggers to disrupt the membrane, allowing E to enter the periplasm and attack the peptidoglycan. T triggering can be blocked by secondary infections, leading to the state of lysis inhibition (LIN). LIN requires the T4 antiholin, RI, and is sensitive to the addition of energy poisons. T is unusual among holins in having a large C-terminal periplasmic domain. The rI gene encodes a polypeptide of 97 residues, of which 72 are predicted to be a periplasmic domain. Here, we show that the periplasmic domain of RI is necessary and sufficient to block T-mediated lysis. Moreover, when overexpressed, the periplasmic domain of T (TCTD) was found to abolish LIN in T4 infections and to convert wild-type (wt) T4 plaques from small and fuzzy edged to the classic “r” large, sharp-edged plaque morphology. Although RI could be detected in whole cells, attempts to monitor it during subcellular fractionation were unsuccessful, presumably because RI is a highly unstable protein. However, fusing green fluorescence protein (GFP) to the N terminus of RI created a more stable chimera that could be demonstrated to form complexes with wild-type TCTD and also with its LIN-defective T75I variant. These results suggest that the function of the unusual periplasmic domain of T is to transduce environmental information for the real-time control of lysis timing.
An Escherichia coli cell infected at 37°C by a wild-type (wt) T4 phage undergoes lysis at about 25 min and releases ∼200 progeny virions. Lysis requires the muralytic activity of the T4 lysozyme, E, one of the best characterized soluble enzymes in terms of its structure, enzymatic mechanism, and thermodynamic stability (26). The precise timing of lysis, however, is not determined by E, which accumulates fully folded and active in the cytoplasm throughout the morphogenesis period. Instead, like all double-stranded DNA phages, the timing of T4 lysis is controlled by its holin, T, an integral membrane protein that suddenly triggers to disrupt the bilayer at an allele-specific time (35, 39). Membrane disruption allows the T4 lysozyme to attack the cell wall, after which the infected cell bursts and releases the progeny virions. T4 t, like the λ holin gene S, is genetically malleable, in that many missense alleles have been isolated, with lysis times either advanced or delayed relative to the wt allele (13, 28, 31, 35, 37). This malleability is significant, because it is thought that investing lysis timing exclusively in the holin gene allows double-stranded DNA phages to evolve rapidly in response to changed conditions. For example, an environment with reduced host numbers should favor phages with an extended latent period, allowing the intracellular accumulation of more progeny virions before they are released into the host-poor medium (34).
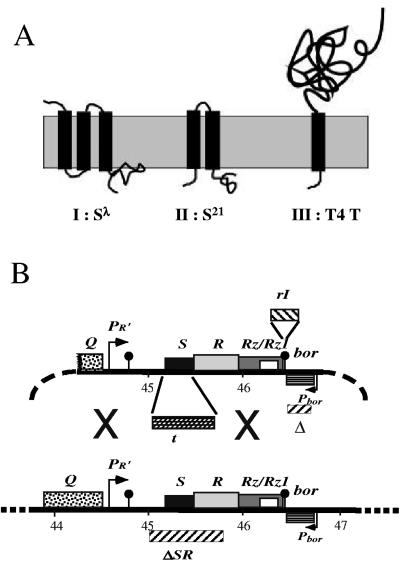
Holin genes have enormous diversity, with more than 50 unrelated gene families having been described (35). However, compared to other known holins, the T4 holin has an unusual topology. All other characterized holins have either three (class I; e.g., the S105 product of phage λ gene S) or two (class II; e.g., the S2168 product of S21, the holin gene of lambdoid phage 21) transmembrane domains (TMDs) (Fig. 1A). In contrast, T and its orthologs in T4-like phages constitute a single protein family with bitopic topology (Fig. 1A and 2). T, at 218 residues, is substantially larger than other holins (e.g., the λ holin S105 is only 105 residues and the S2168 is only 68 residues). The extra mass comes principally from its large C-terminal periplasmic domain of 163 residues (Fig. 2A).
FIG. 1.
(A) Topology of class I, II, and III holins (35, 38). (B) Features of plasmid and phage constructs. All plasmids used for inductions are based on the structure of pS105 (14) (upper structure), with the λ late promoter and the promoter-proximal genes of the late transcriptional unit (lower structure), except that in the plasmid pT4T, the S gene in the λ SRRzRz1 lysis cassette of pER157 (30) is replaced by the phage T4 t gene. In plasmid pT4TRI, the rI gene is inserted 13 nucleotides downstream of the Rz gene of pT4T. “X” designates areas where homologous recombination can occur for generation of λt recombinants.
FIG. 2.
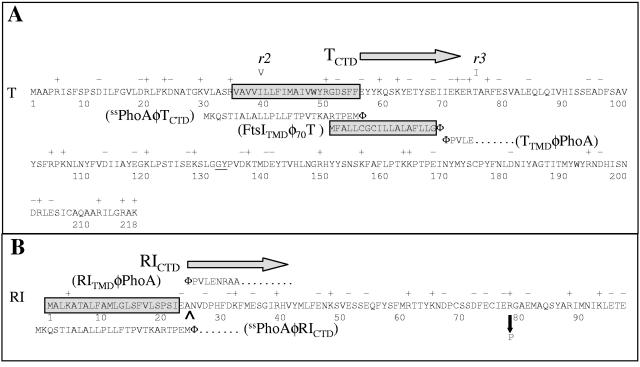
(A) Amino acid sequence of T and its derivatives. The predicted TMD of T is boxed and shaded. The rV mutants r2(I39V) and r3(T75I) are indicated above the sequence. In This, a hexahistidine tag (HHHHHHGG) was inserted between the residues G132 and Y133 (underlined). For the ssPhoAΦTCTD construct, the signal sequence of PhoA, shown below the T sequence, was substituted for residues 1 to 55 of T. For the FtsITMDΦT construct, the transmembrane domain of FtsI, shown in a shaded box below the T sequence, was substituted for residues 2 to 69 of T. For the TTMDφPhoA construct, residues 1 to 70 of T were substituted for residues 1 to 26 in the full-length primary gene product of phoA; the PhoA sequence begins with PVLE (from PhoA residue 27 on, as indicated). (B) Amino acid sequence of RI and its derivatives. The predicted signal sequence of RI is boxed, with the leader peptidase I cleavage site predicted by the SignalP program (http://www.cbs.dtu.dk/services/SignalP/) (2) indicated by a carat. In the c-myc-tagged version of RI, the c-myc tag (QKLISEEDL) was inserted after the terminal glutamate at position 97, while in the GFPΦRIcmyc constructs, the entire GFP sequence was inserted after the initial Met residue. For the ssPhoAφRICTD construct, the signal sequence of PhoA, shown below the RI sequence, was substituted for residues 1 to 24 of RI. For the RINTDφPhoA construct, the predicted signal sequence of RI replaced residues 1 to 26 in the uncleaved precursor to mature PhoA. The sole known rI missense mutation, R78P, is shown by a down arrow.
In addition to its unusual topology and size, the function of the T4 holin is subject to a type of control not seen with the prototypical class I and II holins. Almost 60 years ago, in publications now considered landmarks in the history of molecular genetics, it was reported that T4-infected cells are subject to “lysis inhibition,” or LIN (8, 16). The LIN state, in which the normal lysis timing of the holin is overridden, is established if a T4-infected cell undergoes superinfection by another T4 particle. The LIN state is unstable, requires continued superinfection to be maintained, and can be subverted by addition of energy poisons that collapse the membrane potential (1). In infected cultures at visible optical densities, the lysis of a small fraction of the cells generates sufficient free virions in the medium to establish and maintain LIN throughout the bulk culture, allowing progeny to accumulate to >103 virions per cell over a period of hours. On agar lawns, LIN causes T4 to make small, indistinct plaques, and it is easy to isolate T4 mutants defective in LIN by virtue of their large, clearly defined plaque morphology (15). These mutants were called r mutants (for “rapid lysis”) and were mapped to multiple T4 loci, including rI, rIIAB, rIII, and rV, depending on the host used (37). The r genetic system was extensively exploited to establish many of the fundamental principles of molecular genetics (7). Ultimately, only two genes, rI (3, 27) and rV (20, 21), later shown to be allelic to t (10), are required to maintain the wild-type plaque phenotype and to establish LIN with E. coli K-12. Nevertheless, despite the central importance of the r genetic system in the history of molecular biology, the molecular basis of LIN has remained obscure.
Recently, we have undertaken a molecular analysis of T4 lysis and the LIN phenomenon as part of our study of the mechanisms of phage lysis and its regulation. We reported evidence that RI (Fig. 2B) is an antiholin that specifically binds to and inhibits the T holin. This clearly distinguishes T4 from bacteriophage λ, whose antiholin, S107, is the product of an alternative translational start in its holin gene, S, which also encodes S105, the λ holin. Given its near identity with S105 and the fact that holins oligomerize in the process of forming membrane lesions, it is not surprising that S107 dimerizes with S105. The formation of these dimers is responsible for the ability of S107 to prevent the spontaneous triggering of S105. In contrast, RI has no sequence similarity to T (Fig. 2) that might support homotypic interactions of the type observed in the λ S105/S107 system. Here we report experiments designed to identify the topological determinants of RI and T that lead to the LIN state. The results are discussed in terms of the unique ability of RI to respond to an environmental signal (i.e., a superinfecting T4 phage).
MATERIALS AND METHODS
Bacterial strains, bacteriophages, plasmids, and culture growth.
The bacterial strains, bacteriophages, and plasmids used in this work are described in Table 1. T4 phage stocks were prepared as described previously (30). Bacterial cultures were grown in standard LB medium supplemented with ampicillin (100 μg/ml), kanamycin (40 μg/ml), and chloramphenicol (10 μg/ml) when appropriate. Growth and lysis of cultures were monitored by A550 as previously described (30). When indicated, isopropyl β-d-thiogalactoside (IPTG), KCN, or CHCl3 was added to give final concentrations of 1 mM, 10 mM, or 1%, respectively.
TABLE 1.
Phages, strains, and plasmids used in this work
| Phage, strain, or plasmid | Genotype/features | Source or reference |
|---|---|---|
| Phages | ||
| T4 wt | T4D | I. Molineux |
| T4rI | rI48, single-base deletion at position 195; N66-E97 replaced with MTRALLILNV | D. Hall |
| λkanΔ(SR) | λb515 b519 att::Tn903 cI857 nin5 Δ(SR) | 28 |
| λkan this | λb515 b519 att::Tn903 cI857 nin5 ΔS::this | This study |
| Strains | ||
| CQ21 | E. coli K-12 ara leu lacIq1purE gal his argG rpsL xyl mtl ilv | 28 |
| CQ21λkanΔ(SR) | Lysogen carrying λkanΔ(SR) prophage | 28 |
| CQ21[λkanΔ(SR)] recA srl::Tn10 | Lysogen carrying λkanΔ(SR) prophage | This study |
| CQ21(λkan this) | Lysogen carrying λkan this prophage | This study |
| MG1655 | F−ilvG rfb50 rph1 | E. coli Genetic Stock Center |
| MDS12 | MG1655 with 12 deletions, totalling 376,180 nt, including cryptic prophages | 19 |
| MDS12 tonA::Tn10 lacIq1 | This study | |
| XL-1Blue | E. coli K-12 recA endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac[F′ proAB lacZΔM15::Tn10] | Stratagene |
| BL21(DE3) | F−ompT hsdSB (rB− mB−) gal dcm (DE3) | Novagen |
| BL21(DE3) tonA::Tn5 slyD::Tetr | This study | |
| Plasmids | ||
| pT4T | pBR322 derivative carrying late promoter and lysis cassette of λ with S gene replaced by T4 t | I. N. Wang |
| pT4This | pT4T with HHHHHHGG inserted between residues 132 and 133 of T | This study |
| pT4TRI | pT4T with rI gene inserted distal to Rz1 gene | This study |
| pZA32-luc | p15A origin, PLlacO-1 promoter, Camr | 25 |
| pZA-RI | pZA32Δluc::rI | This study |
| pZA-RICTD | pZA32Δluc::rICTD, codons 25 to 97 of rI | This study |
| pZA-PhoA | pZA32Δluc::phoA, codons 27-471 of phoA, encoding mature PhoA | This study |
| pZA-ssPhoAΦRICTD | Codons 1-24 of rI in pZA-RI replaced by codons 1-26 of phoA, encoding signal sequence | This study |
| pZA-ssPhoAΦRICTDcmyc | pZA-ssPhoAΦRICTD with 10 codons encoding QKLISEEDL inserted after codon 97 of rI | This study |
| pZA-RITMDΦPhoA | First 24 codons of rI inserted in front of codon 27 of phoA in pZA-PhoA | This study |
| pZA-T | pZA32Δluc::t | This study |
| pZA-TCTD | pZA32Δluc::tCTD, codons 56-218 of t | This study |
| pZA-ssPhoAΦTCTD | Codons 1-55 of t in pZA-T replaced by codons 1-26 codons of phoA, encoding signal sequence | This study |
| pZA-ssPhoAΦTCTDT75I | pZA-ssPhoAΦTCTD carrying T75I rV mutation | This study |
| pZA-TTMDΦPhoA | First 70 codons of t inserted before codon 27 of phoA in pZA-PhoA | This study |
| pZA-T70-218 | pZA32Δluc::t70-218, codons 70-218 of t | This study |
| pZA-FtsITMDΦ70T | Codons 24 to 40 of ftsI, encoding TMD, inserted before codon 70 of t in pZA-T70-218 | This study |
| pZE12-luc | ColE origin, PLlacO-1 promoter, Ampr | 25 |
| pZE12-RI | pZE12 Δluc::rI | This study |
| pZE12-RIcmyc | pZE12 Δluc::rIcmyc with codons encoding QKLISEEDL inserted after codon 97 of rI | This study |
| pZE12-GFPΦRIcmyc | DNA fragment encoding GFP inserted between codons 1 and 2 of rIcmyc in pZE12-luc | This study |
| pDS439 | pBR322 origin, ParaBAD promoter, Ampr, carrying gfpmut2 | 33 |
| pET11a-TCTDhis | pBR322 origin, T7 promoter, Ampr, carrying DNA fragment encoding Met residue, then residues 70-218 of T with HHHHHHGG insertion between residues 132 and 133 | This study |
| pER-t | Carrying lysis cassette of λ except S is replaced by T4 t gene, Ampr, Kanr | 30 |
| pPRI | pBR322 origin, λ pPR′ promoter, Ampr, carrying DNA fragment encoding RI | This study |
| pPRIhis | pPRI with 8 codons encoding GGHHHHHH inserted after codon 97 of rI | This study |
| pET11a-RICTDhis | pBR322 origin, T7 promoter, Ampr, carrying DNA fragment carrying Met codon, then codons 25-97 of RI, followed by codons for sequence GGHHHHHH | This study |
Standard DNA manipulations, PCR, and DNA sequencing.
Isolation of plasmid DNA, DNA amplification by PCR, DNA transformation, and DNA sequencing were performed as previously described (36). Oligonucleotides were obtained from Integrated DNA Technologies, Coralville, IA, and were used without further purification. The sequences of the oligonucleotides used are listed in Table 2. The Rapid DNA ligation kit from Roche Molecular Biochemicals was used for ligation reactions. All other enzymes were purchased from New England Biolabs, except for Pfu polymerase, which was from Stratagene. Automated fluorescent sequencing was performed at the Laboratory for Plant Genome Technology at the Texas Agricultural Experiment Station.
TABLE 2.
Sequences of the oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|---|
| HindIIIpR′for | CATTAAAGCTTGAAGGAAATA |
| CRzNRIrev | TGTATTTACTTTGTGCCGATGATGGGCAACTCTATCTGCACT |
| CRzNRIfor | AGTGCAGATAGAGTTGCCCATCATCGGCACAAAGTAAATACA |
| ClaIRIrev | ATATATATCGATCTAGACCACTTTGTGAAAAGT |
| 132-H6G2− | GAAAAATCACTTGGAGGACACCACCACCACCACCACGGAGGA |
| 133for | TATCCTGTTGATAAAACT |
| 132-H6G2− | AGTTTTATCAACAGGATATCCTCCGTGGTGGTGGTGGTGGTG |
| 133rev | TCCTCCAAGTGATTTTTC |
| Kpn1RIfor | GCGCGCGGTACCATGGCCTTAAAAGCAACAGCA |
| RIAvrIIrev | ATATATCCTAGGCTCTAGACCACTTTGTGA |
| 25RIKpnfor | ATATATGGTACCAATGTCGATCCTCATTTTGAT |
| ss25RIfor | ATTAAAGAGGAGAAAGGTACCATGAAACAAAGCACTATTGCA |
| ss25RIrev | ATCAAAATGAGGATCGACATTCATTTCTGGTGTCCGGGCTTT |
| KpnItfor | ATATATGGTACCATGGCAGCACCTAGAATA |
| AvrIItrev | ATATATCCTAGGAGCAGCGAACAAATAATTATT |
| 56tKpnIfor | ATATATGGTACCGAGTACTATAAGCAATCAAAG |
| PhoAss70tfor | ATTAAAGAGGAGAAAGGTACCATGAAACAAAGCACTATTGCAC |
| 56tssrev | CTTTAGTTGCTTATAGTACTCCATTTCTGGTGTCCGGGCTTT |
| KpnIphoAfor | ATATATGGTACCCCTGTTCTGGAAAACCGGGCT |
| AvrIIphoArev | ATATATCCTAGGTTATTTCAGCCCCAGGGC |
| RItmphoAfor | ATTAAAGAGGAGAAAGGTACCATGGCCTTAAAAGCAACAGCA |
| RItmphoArev | AGCCCGGTTTTCCAGAACAGGCGCTTCAATCGATGGAGATAA |
| KpnI70tfor | ATATATGGTACCATTGAAAAGGAAAGAACTGCA |
| FtsITM70tfor | ATTAAAGAGGAGAAAGGTACCATGTTTGCGTTGTTATGCGGCTGT |
| FtsITM70trev | TGCAGTTCT TTCCTTTTCAATTCCGAGCAGAAAAGCCAGCGC |
| T4TtmphoAfor | ATTAAAGAGGAGAAAGGTACCATGGCAGCACCTAGAATA TCA |
| T4TtmphoArev | AGCCCGGTTTTCCAGAACAGGAATTTCACTGTATGTTTCATA |
| KpnIgfpfor | ATATATGTTACCATGAGTAAAGGAGAAGAACTT |
| N-RIC-gfprev | AAGTGCTGTTGCTTTTAAGGCTTTTAAGGCTTTGTATAGTTCATCCATGCC |
| C-gfpN-RIfor | GGCATGGATGAACTATACAAAGCCTTAAAAGCAACAGCACTT |
| RI-XbaIrev | GTCGACTCTAGACCACTTTGTGAA |
| NdeI70tfor | ATATATCATATGATTGAAAAGGAAAGAACTGCA |
| BamH1trev | ATATATGGATCCAGCAGCGAACAAATAATTATT |
| 25RINdeIfor | ATATATCATATGAATGTCGATCCTCATTTT |
| RIhisBamrev | ATATATGGATCCTCAGTGGTGGTGGTGGTGGTG |
| RIG2H6for | AACATTAAATTGGAGACTGAAGGAGGACACCACCACCACCACCACTGAAATTCAGCGACTTTTCAC |
| RIG2H6rev | GTGAAAAGTCGCTGAATTTCAGTGGTGGTGGTGGTGGTGTCCTCCTTCAGTCTCCAATTTAATGTT |
Single-base changes and small insertions were introduced using commercially synthesized primers in conjunction with the QuikChange kit from Stratagene. Larger insertions, replacements, and gene fusions were generated using a modification of the basic QuikChange site-directed mutagenesis protocol. Here, a donor sequence is PCR amplified using primers that have 5′ ends that anneal to appropriate sequences in a target plasmid. The first PCR product is then used as the primer for a second PCR using the target plasmid as a template. All subsequent steps are identical to those in the basic QuikChange protocol.
Construction of plasmids.
pT4T was derived by removing the aphI (kanamycin resistance) gene from pER-t (30) and was a gift from I.-N. Wang. It carries a hybrid lysis cassette in which the T4 t gene (Fig. 2A, nucleotides [nt] 160204 to 160884 of the T4 genome) replaces the λ S gene (nt 45157 to 45465 of the λ genome) in a DNA segment comprising pR′, the λ late promoter, the downstream genes SRRzRz1, and a deletion of the bor gene (Fig. 1B). This lysis cassette is flanked by unique HindIII and ClaI sites (not shown). The plasmid pT4TRI was constructed by PCR amplification of the lysis cassette from pT4T using the forward and reverse primers HindIIIpR′for and CRzNRIrev. In a separate PCR, the rI gene was amplified using the forward and reverse primers CRzNRIfor and ClaIRIrev. The rI gene in the template used for this reaction had its internal ClaI site destroyed by introduction of the silent mutation G63A by site-directed mutagenesis. Since the primers CRzNRIrev and CRzNRIfor are complementary, it was possible to fuse the rI gene sequence (nt 59540 to 59177 of the T4 genome) to the 3′ end of the hybrid lysis cassette (after the base corresponding to 46437 of the λ genome, beyond the end of the Rz gene; Fig. 1B) by using the two PCR products as templates in a splicing by overlapping extension (SOE) reaction (17) using the HindIIIpR′for and ClaIRIrev primers. The product from this reaction was digested with HindIII and ClaI and ligated into the vector backbone produced by digesting pT4T with the same enzymes. The plasmid pT4This was generated by introducing a hexahistidine tag between codons 132 and 133 of the t gene (Fig. 2A) in pT4T, using a pair of oligonucleotides, 132-H6G2for/rev, encoding His6Gly2.
Two plasmids were constructed for overexpression of the His-tagged C-terminal domains of RI (RICTDhis) and T (TCTDhis). For the TCTD plasmid, the DNA fragment encoding TCTDhis was PCR amplified from pT4This using the primer pair NdeI70tfor and BamHItrev. The doubly digested PCR product was inserted into the multiple cloning site of plasmid pET11a to generate the plasmid pET11a-TCTDhis. To construct the RICTD plasmid, a fragment carrying gene rI was produced by cleaving pZE12-RI with EcoRI and XbaI. The plasmid pER-t was digested using the same enzymes to generate the backbone for the plasmid pPRI, and this backbone fragment was ligated to the EcoRI-XbaI fragment carrying rI to produce pPRI. The plasmid pPRIhis was made by inserting hexahistidine tag after codon 97 of the rI gene (Fig. 2B) in pPRI, using the primer pair, RIG2H6for/rev, encoding Gly2His6. The DNA fragment encoding the RICTDhis PCR was amplified from pPRIhis using 25RINdeIfor and RIhisBamH1rev with NdeI and BamHI restriction sites at their 5′ ends, respectively. The doubly digested PCR product was inserted into the multiple cloning site of pET11a to yield pET11a-RICTDhis.
λthis was generated by homologous recombination between pT4This and the lysis-defective phage λkanΔ(SR) (formerly designated as λΔSR) (28) (Fig. 1B). Recombinants were identified by their plaque-forming ability, and the presence of the hybrid lysis cassette was verified by DNA sequencing. Lysogens were prepared by infecting cells with λthis and plating at 30°C for survivors on media containing kanamycin.
The T4 t and rI genes and their derivatives were also expressed from the lac promoter of the pZA and pZE plasmids from the family of modular pZ vectors (25). To construct pZA-RI and pZA-RIcmyc, the primer pair, Kpn1RIfor and RIAvrIIrev was used to PCR amplify the rI gene from pZE-RI or pZE-RIcmyc, respectively. After digestion with KpnI and AvrII, these PCR products were used to replace the luc gene in pZA32-luc. The plasmid pZA-RICTD, carrying a DNA fragment encoding the C-terminal domain (residues 25 to 97; Fig. 2B) of the RI protein, was similarly constructed using the primers 25RIKpnIfor and RIAvrIIrev. The signal sequence of alkaline phosphatase (PhoA) was fused to the RI fragment in pZA-RICTD by the modified site-directed mutagenesis procedure described above. In the first PCR, the PhoA signal sequence was amplified using the ss25RIfor and ss25RIrev primers. The second PCR used pZA-RICTD as the template yielding pZA-ssPhoAΦRICTD. The identical reactions were used to generate pZA-RICTDcmyc and pZA-ssPhoAΦRICTDcmyc from pZA-RIcmyc. A similar strategy was used to generate the complementary series of plasmids pZA-T, pZA-TCTD, and pZA-ssPhoAΦTCTD, using the primer pairs KpnIfor/AvrIItrev, 56tKpnIfor/AvrIItrev, and ss25RIfor/56tss rev, respectively. Here, the TCTD consists of residues 56 to 218 of T (Fig. 2A).
The sequence encoding residues 27 to 471 of the phoA gene was PCR amplified using the forward and reverse primers KpnIphoAfor and AvrIIphoArev, with KpnI and AvrII restriction sites at their 5′ ends, respectively. The doubly digested PCR product was used to replace the luc gene in pZA32-luc to yield pZA-PhoA. To fuse the N-terminal domain (residues 1 to 24, RINTD) of RI to the mature form of PhoA, the DNA encoding the RINTD was PCR amplified using the primers RItmphoAfor and RItmphoArev. The PCR product was then used to conduct a modified site-directed mutagenesis reaction using pZA-PhoA as the template to generate pZA-RINTDΦPhoA. The plasmid pZA-TTMDΦPhoA, in which residues 1 to 70 of T are fused to the mature sequence of PhoA, was constructed in a similar fashion using the primers T4TtmphoAfor and T4TtmphoArev.
The ftsItmdφt chimera, encoding a protein with the TMD of FtsI (Fig. 2A) replacing the TMD of T, was constructed in two steps. First, a DNA fragment encoding residues 70 to 218 of T was PCR amplified using the forward and reverse primers AvrIItrev and KpnI70tfor, with AvrII and KpnI restriction sites at their 5′ ends, respectively. The doubly digested PCR product was used to replace the luc gene in pZA32-luc, generating the intermediate plasmid, pZA-T70-218. Then, a DNA fragment encoding a methionine codon followed by the transmembrane segment of FtsI (residues 24 to 40) was PCR amplified using the forward and reverse primers FtsITM70tfor and FtsITM70trev. The PCR product was then used to conduct a modified site-directed mutagenesis reaction by using pZA-T70-218 as the template to generate pZA-FtsITMDΦT.
Green fluorescent protein (GFP) was fused to c-myc-tagged RI (Fig. 2B) by another SOE reaction. First, a DNA fragment encoding GFP was PCR amplified from pDS439 (33) using the primers KpnIgfpfor and N-RIC-gfprev. Separately, a DNA fragment encoding residues 2 to 106 of the c-myc-tagged RI protein was PCR amplified from pZE-RIcmyc using the primers C-gfpN-RIfor and RI-Xbarev. The PCR products from the two reactions were combined and amplified using the primers KpnIgfpfor and RI-Xbarev. The fusion product was digested with KpnI and XbaI and ligated into pZE12 digested with the same enzymes, yielding pZE-GFPΦRIcmyc.
Subcellular fractionation.
To prepare total membrane and soluble fractions, cell pellets from 150-ml cultures were resuspended in 1 ml of French press buffer (100 mM Na2HPO4, 100 mM KCl, 5 mM EDTA, pH 8.0, 1 mM phenylmethylsulfonyl fluoride, 25 mM MgCl2, 50 μg/ml DNase, 50 μg/ml RNase) and 2.5 μl of protease inhibitor cocktail [4-(2-aminoethyl)benzenesulfonyl fluoride, bestatin, pepstatin, E-64, and phosphoramidon; Sigma]. The cells were disrupted by passage through a French pressure cell (Spectronic Instruments, Rochester, N.Y.) at 16,000 lb/in2 (1 lb/in2 = 6.89 kPa). The unbroken cells were removed by centrifugation in a Damon-IEC Spinette clinical centrifuge at 1,000 × g for 10 min. The membrane and soluble fractions were separated by centrifugation at 100,000 × g for 60 min at 4°C. To identify periplasmic proteins, cells from 30-ml cultures were collected by centrifugation and the pellets were resuspended in 250 μl of 25% sucrose, 30 mM Tris-HCl, pH 8.0. Then, 10 μl of 0.25 M EDTA, 10 μl of lysozyme (20 mg/ml in water), and 250 μl of distilled water were added sequentially (6). When microscopic examination showed that ∼95% of the cells had formed spheroplasts, the samples were centrifuged at 9,000 × g for 10 min to separate the periplasm from the spheroplasts (membrane and cytosol).
SDS-PAGE and Western blotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting were performed as previously described (13). To generate antibodies against RI and T, BL21(DE3) tonA::Tn5 slyD::Tetr cells harboring either pET11a-TCTDhis or pET11a-RICTDhis were grown in 1 liter of LB-ampicillin to an A550 of ∼ 0.5 and then the cultures were induced with IPTG for 3 h. The cell pellets were resuspended in 20 ml of 20 mM BES, 0.5 M NaCl (pH 7.5) supplemented with 20 μl protease inhibitor cocktail, 700 μl MgCl2 (1 M), 100 μl RNase (10 mg/ml), 100 μl DNase (10 mg/ml), and 20 μl dithiothreitol (1 M). The cells were lysed by passage through a French pressure cell (Spectronic Instruments, Rochester, N.Y.) at 16,000 lb/in2 (1 lb/in2 = 6.89 kPa). Inclusion bodies were collected by centrifugation of the French press lysates at 17,500 × g for 30 min. The pellets were extracted with 20 ml of 20 mM BES (N,N-bis[2-hydroxyethyl]-2-aminoethanesulfonic acid; Sigma), 6 M guanidine hydrochloride (United States Biochemical) (pH 7.5) for 3 h. The extracts were cleared by centrifugation at 17,500 × g for 30 min. The supernatant was used as the starting material for purifying TCTDhis and RICTDhis using Talon metal affinity resin (Clontech). The 5-ml resin bed was equilibrated with 20 mM BES, 6 M guanidine hydrochloride, pH 7.5, and the bound proteins were eluted with 20 mM BES, 6 M guanidine hydrochloride, 0.5 M imidazole (Sigma), pH 7.5.
Antibodies against the purified, C-terminal domains of This and RIhis were prepared in rabbits by ProSci Incorporated, Poway, CA. Antibodies against the c-myc epitope were purchased from Babco (Richmond, CA). Reagents and methods for immunodetection have been described previously (36). Equivalent sample loadings were used whenever multiple fractions from the same culture were analyzed.
Immunoprecipitation of RI-T complexes.
MDS12 tonA::Tn10 lacIq1 cells harboring the indicated allele of pZA-ssPhoAΦTCTD and pZE-GFPΦRIcmyc either alone or in combination were grown to an A550 of ∼0.4 and then induced with 1 mM IPTG for 30 min. A 30-ml volume of each culture was taken through the EDTA-lysozyme treatment used to prepare spheroplasts (6). Instead of centrifuging the samples after the addition of water, the spheroplasts were lysed by adding 5 μl of protease inhibitor cocktail and 500 μl lysis buffer (100 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP40). Next, 100 μl of 1 M MgCl2, 10 μl of 10 mg/ml DNase I, and 10 μl of 10 mg/ml pancreatic RNase were added and the samples were incubated at room temperature for 15 min with occasional mixing.
To collect complexes containing the TCTD and GFPΦRIcmyc proteins, 200 μl of the indicated lysate was diluted with an equal volume of wash buffer (lysis buffer containing 1 mg/ml bovine serum albumin). Next, 2 μl of a preimmune rabbit serum, rabbit anti-T, or mouse anti-GFP (Stressgen) was added and the samples were incubated for 2 h at 4°C with slow agitation. Then, 50 μl of the appropriate iron-conjugated secondary antibody (Pierce) was added and the mixture was incubated for an additional 2 h at 4°C. The immune complexes were collected magnetically and washed three times with 0.5 ml of wash buffer. The complexes were dissociated by boiling in SDS-PAGE sample buffer and analyzed by SDS-PAGE and Western blotting followed by immunodetection using anti-T or anti-c-myc antibodies, as indicated.
Phage accumulation during LIN.
For assessment of the LIN state, CQ21 cells lysogenic for λkan this and harboring either no plasmid, pZA-RI, or pZA-ssPhoAΦRICTD were grown at 30°C and induced both by adding IPTG and simultaneously shifting the growth temperature to 42°C for 15 min and then to 37°C. At 30, 60, 90, 120, and 150 min after induction, 1 ml was taken from each culture and the cells were lysed by the addition of CHCl3. Debris was removed by centrifugation, and the plaque-forming titers in the cleared lysates were determined in triplicate on MDS12 tonA::Tn10 lacIq1.
RESULTS
Domain analysis of the RI antiholin.
Paddison et al. (27) used primary structure analysis algorithms to predict that RI has an N-terminal secretory signal and was thus a secreted periplasmic protein. Although RI has been visualized as a cross-linked complex with T, it had previously escaped detection as an independent polypeptide, which presumably was due to proteolytic lability (29). Thus, whether this N-terminal sequence serves as a signal anchor, leaving its periplasmic C-terminal domain (RICTD) tethered to the cytoplasmic membrane, or a cleavable signal allowing release of the RICTD into the periplasm was unclear. Sequence analysis of the classical rI alleles of Doermann (9) revealed that most were frameshifts distal to the predicted secretory signal. Moreover, the single missense allele (R78P; listed as R78G by Paddison et al. [27]) (Fig. 2B), also mapped there, implicating RICTD as critical to the antiholin function of RI.
To resolve RI into topological components, we constructed two chimeric genes: one, ssphoAφrICTD, with RICTD fused to the signal sequence of alkaline phosphatase (PhoA); the other, rINTDφphoA, with the N-terminal domain of RI (RINTD) fused to the periplasmic domain of PhoA (Fig. 2B). Using antibodies raised against RICTD, the ssPhoAΦRICTDcmyc protein could be detected in whole cells (Fig. 3A). However, efforts to localize it using conventional subcellular fractionation were unsuccessful, again presumably due to rapid proteolysis after cell disruption. To provide evidence that ssPhoAφRICTDcmyc was present in the periplasm, the ssphoAφrICTD gene was expressed in cells grown in the presence or absence of azide to inhibit SecA. A slower-migrating species accumulated in the presence of azide, indicating that, when SecA is not inhibited, ssPhoAΦRICTDcmyc is processed and localized to the periplasm (Fig. 3A). In contrast, the product of the rINTDφphoA construct was stable, unprocessed, and was found in both the membrane and soluble fractions (Fig. 3B). The dual localization of the unprocessed RINTDΦPhoA protein will be considered elsewhere (T. A. T. Tran., D. K. Struck, and R. Young, manuscript in preparation). We next compared the ability of these chimeras to support LIN with that of wt RI. While the rINTDΦphoA fusion had no biological function, the ssphoAφrICTD chimera blocked t-mediated lysis, as assessed by monitoring the turbidity of the induced culture (Fig. 4A). Moreover, in addition to preventing the loss of optical density, both rI and ssphoAφrICTD allowed the extended intracellular accumulation of virions and suppressed their release to the medium (Fig. 4B). Finally, the LIN state supported by both rI and ssphoAφrICTD could be subverted by the addition of energy poisons (Fig. 4A). We conclude that the periplasmic domain of RI is necessary and sufficient for authentic LIN. Indeed, the ssphoAφrICTD allele is more effective than the parental rI gene, as judged by the stability of the LIN phenotype (Fig. 4A).
FIG. 3.
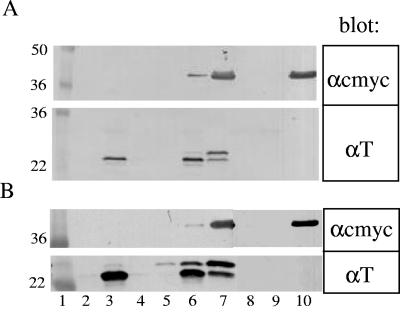
Localization of T and RI chimeras. Subcellular fractions were prepared and analyzed by SDS-PAGE and Western blotting as described in Materials and Methods. (A) Cells carrying pZA-ssPhoAΦRICTDcmyc were grown in the absence (lanes 2 and 3) or the presence (lanes 4 and 5) of 1 mM azide for 10 min in advance of induction. Cells from these cultures were collected by TCA precipitation and centrifugation, resuspended in SDS-PAGE sample buffer, and subjected to SDS-PAGE and Western blotting using anti-RI antisera as the primary antibody. Lane 1, molecular mass standards; lanes 2 and 4, samples from uninduced cultures; lanes 3 and 5, samples from induced cultures. (B) Cells carrying the pZA-RINTDΦPhoA were induced, fractionated, and analyzed by SDS-PAGE and Western blotting using anti-PhoA as the primary antibody. Lane 1, molecular mass standards; lane 2, mature form of PhoA; lane 3, blank; lane 4, cells from an uninduced culture; lane 5, cells from an induced culture; lane 6, total cell lysate; lane 7, 1,000 × g pellet; lane 8, 1,000 × g supernatant; lane 9, 100,000 × g supernatant (soluble fraction); lane 10, 100,000 × g pellet (membrane fraction); lane 11, detergent-extractable (1% NP40) membrane fraction; lane 12, detergent-insoluble fraction. (C) Cells carrying pZA-ssPhoAΦTCTD were grown in the absence (lanes 2 to 6) or the presence (lanes 7 to 11) of 1 mM azide for 10 min in advance of induction, harvested, fractionated, and analyzed by SDS-PAGE and Western blotting using anti-T antisera as the primary antibody. Lane 1, molecular mass standards; lanes 2 and 7, uninduced cells; lanes 3 and 8, induced cells; lanes 4 and 9, cells after spheroplasting; lanes 5 and 10, spheroplasts; lanes 6 and 11, periplasm.
FIG. 4.
The C-terminal domains of T and RI are the determinants of LIN. (A) ssPhoAΦRICTD is necessary and sufficient for LIN. CQ21(λkanΔ(SR)) cells carrying the indicated plasmids were induced at time zero, and culture turbidity was monitored as a function of time: ⊠, pT4T; ▪, □, pT4T and pZA-RI; ○, •, pT4T and pZA-ssPhoAΦRICTD; ▴, pT4T and pZA-RINTDΦPhoA; ⧫, pS105; ⋄, pS105 and pZA-RI; ▿, pS105 and pZA-ssPhoAΦRICTD. To demonstrate premature triggering, KCN was added to two cultures (pT4T and pZA-RI, □; and pT4T and pZA-ssPhoAΦRICTD, ○) at the time indicated by the solid arrow. (B) Phage accumulation during ssPhoAΦRICTD-mediated LIN. CQ21(λ-this) carrying the indicated plasmids was induced at time zero, and culture turbidity (solid symbols) and phage accumulation (open symbols) were monitored as a function of time. ○, •, no plasmid; ⋄, ⧫, pZA-RI; □, ▪, pZA-ssPhoAΦRICTD. (C) Periplasmic TCTD interferes with LIN. CQ21(λkanΔ(SR)) cells carrying the indicated plasmids were induced at time zero, and culture turbidity was monitored as a function of time. •, pT4T; ○, pT4T and pZA-ssPhoAΦTCTD; □, pZA-ssPhoAΦTCTD; ▵, pT4TRI; ▿, pT4TRI and pZA-ssPhoAΦTCTD; ⋄, pT4TRI and pZA-TCTD; ▴, pT4TRI and pZATTCD; ▾, pT4TRI and pZA- FtsITMDΦT70-218; ▪, pT4TRI and pZA-TTMDΦPhoA. (Inset) Detail for growth of cells carrying pT4T alone (•) and pT4T and pZA-ssPhoAΦTCTD (○). (D) Periplasmic TCTD blocks LIN during T4 phage infections. CQ21 cells carrying either pZA32-luc (solid symbols) or pZA-ssPhoAΦTCTD (open symbols) were induced at time zero and were grown without infection (○, •) or infected at a multiplicity of infection of 10 with either T4D (□, ▪) or T4rI (▵,▴). In panels A, C, and D, CHCl3 was added at the time indicated by the arrow.
RI-dependent LIN requires binding to the C-terminal domain of T.
The unusually large C-terminal periplasmic domain of T (TCTD) is a feature that distinguishes it from the numerous class I and class II holins (35). To determine whether this domain is the target for RICTD, we constructed an allele of t in which the sequences encoding the predicted cytoplasmic and transmembrane domains were replaced by the segment of phoA encoding its secretory signal sequence (ssphoAφtCTD; Fig. 2A). Like the RICTD, the TCTD was also efficiently secreted by the PhoA signal sequence (Fig. 3C). This allele was lytically incompetent but exerted a weak dominant-negative phenotype in that it caused a short delay in T-mediated lysis (Fig. 4C, inset), suggesting that homotypic interactions in the TCTD are involved in the lytic function of T. The biological function of this chimera was assessed in a system in which t and rI are both expressed from the λ late promoter, shown in previous work to support physiologically meaningful lysis timing with the λ lysis cassette (13). The results clearly showed that supplying periplasmic TCTD partially blocked the imposition of LIN (Fig. 4C). More dramatic results were obtained when cells producing the ssPhoAΦTCTD were infected with T4 phage; LIN was completely subverted by the presence of the TCTD (Fig. 4D). In a control infection, induction of ssphoAφtCTD had no effect on the lysis kinetics of T4rI (Fig. 4D). When T4 phage were plated on bacteria secreting TCTD to the periplasm, wt T4 (T4D) generated large, distinct plaques which were nearly identical to the plaques produced by rI mutants (Fig. 5). We conclude that interactions between the periplasmic domain of RI and the periplasmic domain of T are required for LIN.
FIG. 5.
Periplasmic TCTD causes T4 to form r-type plaques. Lawns of cells of MDS12 tonA::Tn10 lacIq1 (A), MDS12 tonA::Tn10 lacIq1 harboring uninduced pZA-ssphoAΦTCTD (B), or MDS12 tonA::Tn10 lacIq1 harboring induced ssPhoAΦTCTD (C) were infected with either T4D or T4rI phage.
T and RI form a complex.
To provide further evidence that the T and RI proteins interact, we performed coimmunoprecipitation experiments using lysates prepared from cells expressing ssPhoAΦTCTD and a GFPΦRIcmyc chimera. The GFPΦRIcmyc chimera was used since it was readily visualized by immunoblotting, presumably because of decreased lability compared to either RI or ssPhoAφRICTD, both of which were undetectable in immunoprecipitations. Using either anti-T or anti-GFP as the first antibody, the two proteins were found to coprecipitate (Fig. 6A). These complexes were also formed when detergent-solubilized extracts prepared from cells expressing T or GFPΦRIcmyc separately were mixed and then subjected to immunoprecipitation (not shown). Identical results were obtained when the T75I mutation found in a t allele known to be insensitive to RI-mediated LIN was introduced into the ssPhoAφTCTD protein (Fig. 6B). Since the R78P allele of rI is defective for LIN, we attempted to test the effect of this mutation on the ability of RI to form complexes with T. Unfortunately, the product of the R78P allele of gfpφrIcmyc does not accumulate in whole cells to levels detectable by Western blotting.
FIG. 6.
T and RI form a complex. (A) Immunoprecipitations were performed with samples containing either ssPhoAΦTCTD (lanes 2 to 4), both ssPhoAΦTCTD and GFPΦRIcmyc (lanes 5 to 7), or GFPΦRIcmyc only (lanes 8 to 10), prepared from induced cells carrying either of the plasmids pZA-ssPhoAΦTCTD or pZE-GFPΦRIcmyc or both. Primary antibodies for immunoprecipitations: lanes 2, 5, and 8, rabbit preimmune serum; lanes 3, 6, and 9, anti-T rabbit antibody; lanes 4, 7, and 10, anti-GFP monoclonal antibody. Samples were analyzed by SDS-PAGE and immunoblotted with either anti-T or polyclonal anti-c-myc, as indicated to the right. (B) Same as panel A, except that the rV variant ssPhoAΦTCTDT75I, produced from the plasmid pZA-ssPhoAΦTCTDT75I, was used. The molecular mass appears in lane 1 for each blot.
Interestingly, the ratio of unprocessed ssPhoAΦTCTD to mature periplasmic TCTD was reproducibly higher when GFPΦRIcmyc was present (Fig. 6A and B, lanes 5 to 7). This suggests that the binding of RI to the periplasmic domain can occur while TCTD is nascent and that this binding interferes with leader peptidase cleavage of the signal sequence.
DISCUSSION
Infections of T4 and the other T-even phages are the only examples where lysis timing has been demonstrated to be directly affected by the environmental conditions surrounding the infected cell. The classic LIN phenotype can be thought of as a “no-quorum” signal, in the sense that an effector molecule (i.e., a superinfecting T4 phage) indicating the lack of available hosts binds to an infected cell and initiates an as yet unknown signal transduction pathway leading to activation of an effector, RI. The simplest interpretation of our data is that activated RI directly binds to and inhibits the target molecule, T, so that lysis is blocked. Both RI and T have hydrophobic domains near their N termini, but the results presented here demonstrate conclusively that the interactions essential for the transmission of the signal are entirely between the periplasmic domains of the two proteins. Moreover, we have shown that RI and T are present in a complex that can be recovered by immunoprecipitation without prior covalent cross-linking. Unlike class I and II holins, which consist of two or three transmembrane domains, short interconnecting loops, and a 10- to 25-residue C-terminal cytoplasmic domain, T and its orthologs in T4-like phage have a single transmembrane domain and a relatively large C-terminal periplasmic domain (31). Our findings suggest that this periplasmic domain has evolved to serve as a receptor for the LIN signal provided by “activated” RI. This idea is consistent with the observation that large deletions within the TCTD can be tolerated without entirely abolishing holin function (31). In addition, the coimmunoprecipitation experiments also suggest that RI binds to nascent T, since the presence of GFPΦRIcmyc appears to reduce the efficiency of the maturation of ssPhoAΦTCTD (Fig. 6A, lanes 5 to 7). This is consistent with the finding that in T4 infections, T4 superinfection can confer RI-mediated LIN on a preexisting pool of wt LIN-sensitive T molecules if the incoming phage has a wt, LIN-sensitive t allele, but not if it has an rV, LIN-insensitive t allele (30). The interference with signal sequence processing also suggests that conformational changes derived from binding of RI to the TCTD can be transmitted to the membrane, which is consistent with the fact that one of the classic rV, LIN-defective mutations is a subtle missense change, I39V, in the hole-forming TMD of T.
The classic LIN phenotype appears to involve a direct interaction between the periplasmic domains of RI and T and can be subverted by the collapse of the membrane potential by energy poisons. Thus, at a superficial level, the antiholin function of RI resembles those of S107 and S2171, the antiholins of λ and lambdoid phage 21, respectively. The latter two proteins are identical to their cognate holins except for a short N-terminal extension. In both cases, antiholin-holin interactions are thought to prevent quaternary rearrangements that are required to convert holin oligomers into “holes.” We suggest that in the energized membrane, the single TMD of T is unable to oligomerize into a functional “hole” in the absence of interactions between TCTDs in the periplasm. The antiholin activity of RI would then be due to its ability to bind to the TCTD preventing these interactions. Mutations that alter the periplasmic interactions between TCTD molecules and between TCTD and RICTD would allow for the genetic malleability of lysis timing for T4. For λ bacteriophages (4, 11, 12, 28) and the lambdoid phage 21 (T. Park, D. K. Struck, and R. Young, submitted), collapse of the membrane potential allows premature triggering of the holin by causing topological changes in the antiholin that lead to its inactivation. Moreover, in these cases, representing canonical class I and II holins, respectively, these topological changes in the antiholin effectively convert it into the functional equivalent of its cognate holin. It seems unlikely that T and RI have the same relationship as is seen with the antiholin/holin pairs of phage λ and 21 for several reasons. First, the homotypic interactions that presumably characterize the S105/S107 and S2171/S2168 systems are not possible since T and RI do not share amino acid sequence homology. Second, RINTD is not essential for its antiholin activity, indicating that RI does not directly interact with the hole-forming TMD of the T holin. Finally, in contrast to S107 or S2171, RI appears to be a labile protein whose antiholin activity is only realized physiologically under conditions of superinfection.
While the mechanism of action of RI may be fundamentally different from that of S107 or S2171, a feature common to the antiholin function of all three proteins is its abrogation by collapse of the membrane potential. Although the reason for this behavior is not obvious, we propose that it endows bacteriophage lysis systems with a “sentinel” function. Here, the injection of a heterologous phage DNA into a previously infected cell is detected by the resident holin as a transitory depolarization of the membrane, associated with the channel formed in the bilayer through which the DNA passes. The resident holin is thus triggered prematurely, aborting the new infection and allowing release of progeny from the initial infection.
The results presented here indicate that the large periplasmic domain of the T4 holin is fundamentally involved in real-time regulation by RI. In the T4 infection cycle, there is evidence that rI is transcribed from both early and late promoters (27). Moreover, the RI-dependent LIN phenotype is imposed by superinfections at 3 min after infection and beyond, before the first molecule of T, as a late gene product, is made. Given the temporal relationship between the expression of the rI and t genes, why does RI not inhibit T-mediated lysis in the absence of superinfection? The answer to this question may lie with the stability of RI. LIN is a transient phenomenon which requires continual reinfection to significantly prolong the latent period of the initial T4 infection (1). This, in itself, suggests that the effector molecule that transmits the LIN signal to the T protein is unstable. In fact, we can detect RI in whole cells collected by trichloroacetic acid (TCA) precipitation, but not if cells are fractionated, which suggests that it is extremely labile. This leads to the parsimonious model that LIN is imposed only if RI reaches a certain level, which can be attained either by virtue of a stabilization signal provided by superinfection or by overexpression from induction of a multicopy plasmid. Our hypothesis is thus that RI function is regulated by its proteolytic instability in the periplasm. The nature of the stabilization signal is unknown, but consideration of the molecular events during superinfection may provide a clue. It is thought that Imm, a small cytoplasmic membrane protein produced in quantity early in infection, causes secondary infections to fail, resulting in ectopic periplasmic localization of the capsid contents, which includes the 170-kb T4 chromosome and the more than 1,000 molecules of internal head proteins (1, 23, 24, 32). T4 “ghosts,” emptied of DNA and internal proteins, do not cause LIN, although the ability to undergo tail contraction and induce lethal channels in the cytoplasmic membrane is unaffected (18). The simplest model is that either the T4 DNA or the internal head proteins interfere in proteolysis of RI, thus indirectly activating the r system to block T holin. Recently Los et al. (22) reported that T4 wt, but not T4rI, exhibited delayed lysis in slow-growing cells cultured in chemostats, even when there were insufficient free phage to effect LIN by superinfection. This may reflect a significant stabilization of RI due to cellular responses to slow chemostat growth conditions rather than to the ectopic localization of the contents of superinfecting phage. In any case, experiments to test our model and address other unanswered questions about T4 lysis and LIN that arose decades ago during the classical era of the Delbrück “Phage Church” (5, 7), including the roles of the intensively studied rIIAB genes and also rIII, will be presented elsewhere.
Acknowledgments
We thank the members of the Young laboratory, past and present, for their helpful criticisms and suggestions. The skillful clerical assistance of Daisy Wilbert is gratefully acknowledged.
This work was supported by PHS grant GM27099 to R.Y., the Robert A. Welch Foundation, and the Program for Membrane Structure and Function, a Program of Excellence grant from the Office of the Vice President for Research at Texas A&M University.
REFERENCES
- 1.Abedon, S. T. 1994. Lysis and the interaction between free phages and infected cells, p. 397-405. In J. D. Karam, J. W. Drake, K. N. Kreuzer, G. Mosig, D. H. Hall, F. A. Eiserling, L. W. Black, E. K. Spicer, E. Kutter, K. Carlson, and E. S. Miller (ed.), Molecular biology of bacteriophage T4. American Society for Microbiology, Washington, D.C.
- 2.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol 340:783-795. [DOI] [PubMed] [Google Scholar]
- 3.Benzer, S. 1957. The elementary units of heredity, p. 70-93. In B. Glass (ed.), The chemical basis of heredity. The John Hopkins Press, Baltimore, Md.
- 4.Bläsi, U., C.-Y. Chang, M. T. Zagotta, K. Nam, and R. Young. 1990. The lethal λ S gene encodes its own inhibitor. EMBO J. 9:981-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brock, T. D. 1990. The emergence of bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 6.Broome-Smith, J. K., and B. G. Spratt. 1986. A vector for the construction of translational fusions to TEM beta-lactamase and the analysis of protein export signals and membrane protein topology. Gene 49:341-349. [DOI] [PubMed] [Google Scholar]
- 7.Cairns, J., G. S. Stent, and J. D. Watson. 1966. Phage and the origins of molecular biology. Cold Spring Harbor Laboratory of Quantitative Biology, Cold Spring Harbor, N.Y.
- 8.Doermann, A. H. 1948. Lysis and lysis inhibition with Escherichia coli bacteriophage. J. Bacteriol. 55:257-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doermann, A. H. 1952. The intracellular growth of bacteriophages. I. Liberation of intracellular bacteriophage T4 by premature lysis with another phage or with cyanide. J. Gen. Physiol. 35:645-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dressman, H. K., and J. W. Drake. 1999. Lysis and lysis inhibition in bacteriophage T4: rV mutations reside in the holin t gene. J. Bacteriol. 181:4391-4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graschopf, A., and U. Bläsi. 1999. Functional assembly of the lambda S holin requires periplasmic localization of its N-terminus. Arch. Microbiol. 172:31-39. [DOI] [PubMed] [Google Scholar]
- 12.Graschopf, A., and U. Bläsi. 1999. Molecular function of the dual-start motif in the λ S holin. Mol. Microbiol. 33:569-582. [DOI] [PubMed] [Google Scholar]
- 13.Gründling, A., U. Bläsi, and R. Young. 2000. Biochemical and genetic evidence for three transmembrane domains in the class I holin, λ S. J. Biol. Chem. 275:769-776. [DOI] [PubMed] [Google Scholar]
- 14.Gründling, A., U. Bläsi, and R. Young. 2000. Genetic and biochemical analysis of dimer and oligomer interactions of the λ S holin. J. Bacteriol. 182:6082-6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hershey, A. D. 1946. Mutation of bacteriophage with respect to type of plaque. Genetics 31:620-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hershey, A. D. 1946. Spontaneous mutations in bacterial viruses. Cold Spring Harbor Symp. Quant. Biol. 11:67-77. [Google Scholar]
- 17.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 18.Karam, J. D., J. W. Drake, K. N. Kreuzer, G. Mosig, D. H. Hall, F. A. Eiserling, L. W. Black, E. K. Spicer, E. Kutter, K. Carlson, and E. S. Miller. 1994. Molecular biology of bacteriophage T4. ASM Press, Washington, D.C.
- 19.Kolisnychenko, V., G. Plunkett III, C. D. Herring, T. Feher, J. Posfai, F. R. Blattner, and G. Posfai. 2002. Engineering a reduced Escherichia coli genome. Genome Res. 12:640-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krylov, V. N. 1966. A new r gene of bacteriophage T4B? Microb. Genet. Bull. 24:4-5. [Google Scholar]
- 21.Krylov, V. N., and A. Zapadnaya. 1965. Bacteriophage T4B r mutations sensitive to temperature (rts). Genetika 1:7-11. [Google Scholar]
- 22.Los, M., G. Wegrzyn, and P. Neubauer. 2003. A role for bacteriophage T4 rI gene function in the control of phage development during pseudolysogeny and in slowly growing host cells. Res. Microbiol. 154:547-552. [DOI] [PubMed] [Google Scholar]
- 23.Lu, M., and U. Henning. 1989. The immunity (imm) gene of Escherichia coli bacteriophage T4. J. Virol. 63:3472-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu, M.-J., and U. Henning. 1994. Superinfection exclusion by T-even-type coliphages. Trends Microbiol. 2:137-139. [DOI] [PubMed] [Google Scholar]
- 25.Lutz, R., and H. Bujard. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matthews, B. W. 1996. Structural and genetic analysis of the folding and function of T4 lysozyme. FASEB J. 10:35-41. [DOI] [PubMed] [Google Scholar]
- 27.Paddison, P., S. T. Abedon, H. K. Dressman, K. Gailbreath, J. Tracy, E. Mosser, J. Neitzel, B. Guttman, and E. Kutter. 1998. The roles of the bacteriophage T4 r genes in lysis inhibition and fine-structure genetics: a new perspective. Genetics 148:1539-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raab, R., G. Neal, C. Sohaskey, J. Smith, and R. Young. 1988. Dominance in lambda S mutations and evidence for translational control. J. Mol. Biol. 199:95-105. [DOI] [PubMed] [Google Scholar]
- 29.Ramanculov, E. R., and R. Young. 2001. An ancient player unmasked: T4 rI encodes a t-specific antiholin. Mol. Microbiol. 41:575-583. [DOI] [PubMed] [Google Scholar]
- 30.Ramanculov, E. R., and R. Young. 2001. Functional analysis of the T4 t holin in a lambda context. Mol. Genet. Genomics 265:345-353. [DOI] [PubMed] [Google Scholar]
- 31.Ramanculov, E. R., and R. Young. 2001. Genetic analysis of the T4 holin: timing and topology. Gene 265:25-36. [DOI] [PubMed] [Google Scholar]
- 32.Shapira, A., E. Giberman, and A. Kohn. 1974. Recoverable potassium fluxes variations following adsorption of T4 phage and their ghosts on Escherichia coli B. J. Gen. Virol. 23:159-171. [DOI] [PubMed] [Google Scholar]
- 33.Siegele, D. A., and J. C. Hu. 1997. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc. Natl. Acad. Sci. USA 94:8168-8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, I. N., D. E. Dykhuizen, and L. B. Slobodkin. 1996. The evolution of phage lysis timing. Evol. Ecol. 10:545-558. [Google Scholar]
- 35.Wang, I. N., D. L. Smith, and R. Young. 2000. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54:799-825. [DOI] [PubMed] [Google Scholar]
- 36.Xu, M., D. K. Struck, J. Deaton, I. N. Wang, and R. Young. 2004. The signal arrest-release (SAR) sequence mediates export and control of the phage P1 endolysin. Proc. Nat. Acad. Sci. USA 101:6415-6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young, R. 1992. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 56:430-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young, R. 2002. Bacteriophage holins: deadly diversity. J. Mol. Microbiol. Biotechnol. 4:21-36. [PubMed] [Google Scholar]
- 39.Young, R., I. N. Wang, and W. D. Roof. 2000. Phages will out: strategies of host cell lysis. Trends Microbiol. 8:120-128. [DOI] [PubMed] [Google Scholar]