Abstract
Transposable elements can affect an organism's fitness through the insertional inactivation of genes and can therefore be used to identify genes that are nonessential for growth in vitro or in animal models. However, these models may not adequately represent the genetic requirements during chains of human infection. We have therefore conducted a genome-wide survey of transposon mutations in Mycobacterium tuberculosis isolates from cases of human infection, identifying the precise, base-specific insertion sites of the naturally occurring transposable element IS6110. Of 294 distinct insertions mapped to the strain H37Rv genome, 180 were intragenic, affecting 100 open reading frames. The number of genes carrying IS6110 in clinical isolates, and hence apparently not essential for infection and transmission, is very much lower than the estimates of nonessential genes derived from in vitro studies. This suggests that most genes in M. tuberculosis play a significant role in human infection chains. IS6110 insertions were underrepresented in genes associated with virulence, information pathways, lipid metabolism, and membrane proteins but overrepresented in multicopy genes of the PPE family, genes of unknown function, and intergenic sequences. Population genomic analysis of isolates recovered from an organism's natural habitat is an important tool for determining the significance of genes or classes of genes in the natural biology of an organism.
The genome sequence of an organism reveals its complete genetic capacity. However, the functions of many open reading frames (ORFs; genes) are uncertain, as is their importance to the organism in its natural habitat. Various approaches, including comparative genomics and saturation mutagenesis, have been used to attempt to identify essential genes (1, 5, 13, 18, 43). Laboratory-based studies of Mycobacterium tuberculosis have suggested that fewer than 1,000 genes are potentially essential and over 3,000 genes are potentially nonessential for infection (22, 27, 39-41). However, more genes are likely to be needed by an organism in its natural environment than laboratory-based studies suggest. For a pathogen, essential or advantageous functions include not only those needed for infection and disease (and for tuberculosis, persistence in the host) but also those required for transmission. These complex requirements are difficult to mimic in vitro.
In a novel approach to this problem, we identified mutations in wild strains of the bacterial pathogen M. tuberculosis. Natural variation of M. tuberculosis is largely due to the mobile insertion sequence IS6110 (26, 47), which causes insertional inactivation and deletion (8, 11, 38). In clinical isolates, genes advantageous for survival, pathogenicity, or transmissibility rarely contain a copy of this element. This enables an assessment of the significance of specific genes in human infection, especially compared with the results of transposon mutagenesis in vitro or in animal models (22, 27, 39-41). To our knowledge, this is the first comprehensive study to utilize transposon-based, natural genomic polymorphisms to assess the genetic requirements of an organism in its natural environment.
MATERIALS AND METHODS
Sources of isolates.
A total of 161 isolates were analyzed, of which 122 were obtained from United Kingdom sources (105 from B. Watt, Edinburgh; 17 low-copy isolates [one to four IS6110 copies detected by restriction fragment length polymorphism] from London [23]; 20 isolates provided by C. Sola [Guadeloupe, France]; and 19 Tanzanian isolates from T. McHugh [Royal Free Hospital, London]). Of the total of 161 patients, 98 were Caucasian, 26 Black African, 5 Indian or Pakistani, 13 other Asian, and 19 other or ethnicity not known. A wide age range was represented (3 to 91 years, mean = 47 years), with different clinical conditions (129 pulmonary, 18 nonpulmonary, 14 unknown). The isolates were heterogeneous by IS6110 restriction fragment length polymorphism typing with a mean IS6110 copy number of 9.2 (range, 1 to 18), and the multicopy isolates had less than 70% relatedness as determined by the Dice coefficient of similarity (BioNumerics, version 2.0; Applied Maths, Kortrijk, Belgium). For further strain information, see Table S4 in the supplemental material (supplemental material will be provided by the corresponding author upon request).
Identification of IS6110 insert sites.
The location of IS6110 copies was determined by heminested inverse PCR as described previously (51), supplemented by ligation-mediated PCR (30) with additional confirmation by specific PCR for the London isolates as described by Dale et al. (8). The sites were mapped by comparison with the published genome sequences of M. tuberculosis strains H37Rv (6) and CDC1551 (12).
Data analysis.
Functional classes of genes are based on M. tuberculosis H37Rv (http://genolist.pasteur.fr/TubercuList). For comparison with other studies (22, 27, 39, 41, 49), the data presented in those papers were reanalyzed using the same classification of gene function. Paralogous gene associations in strain H37Rv were taken from (http://www.tigr.org/tigr-scripts/CMR2/LevelsOfParalogy1.spl?db_data_id=89). Statistical significance was assessed by chi-square and Student t tests. Analysis of the extent of saturation of the genome by IS6110 insertions was by rarefaction analysis (48).
RESULTS AND DISCUSSION
Availability of insertion sites.
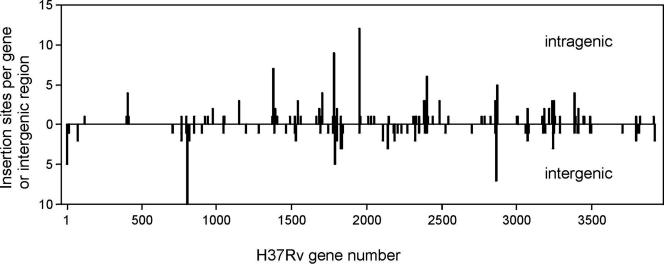
Using 161 diverse clinical isolates, we identified 818 insertions of IS6110 at the sequence level. To exclude the possibility that insertions at certain sites (ancient insertions) occur more frequently because of evolutionary relationships between isolates, we selected insertions at different base positions; there were 340 of these distinct sites (see Table S5 in the supplemental material, available upon request). Thirteen of these were in repetitive regions which could not be unambiguously located and so these were excluded; nine of these were in insertion sequences (six within or immediately adjacent to IS1547 [10]; one site each in IS1081, IS1557, and IS1558), one site occurs twice in a PPE gene (Rv1753c), and three sites are within a pair of very similar genes (Rv1765c and Rv2015c). Comparison with the genome sequence of M. tuberculosis H37Rv (6) enabled the unambiguous mapping of 294 insertion sites (Fig. 1), with a further 32 sites mapped to the genome sequence of M. tuberculosis strain CDC1551 (12). Forty percent of the distinct IS6110 insertion sites were intergenic, which is much greater than the proportion of intergenic regions in the genome (9%, P < 0.001), reflecting the generally deleterious effect of insertions into coding sequences (7, 38). In contrast, in a study of in vitro transposon mutagenesis of M. tuberculosis, using Tn5370, only 19% of the inserts were intergenic (27), consistent with greater selection against intragenic insertion in M. tuberculosis from its natural habitat than when grown in vitro. Two intergenic regions had a larger number of insertions: Rv0794 to Rv0798 (10 sites) is adjacent to or contains IS1547, which we have previously identified as a preferential locus for IS6110 insertion and where it is involved with deletions (10), and Rv2813 to Rv2816 (7 sites), which is the DR region and which is again a preferential locus for IS6110 insertion and IS6110-mediated deletion (11).
FIG. 1.
Genomic distribution of IS6110 insertions. The number of independent insertion sites in each gene (above the line) or intergenic region (below the line), ordered by gene number in the M. tuberculosis H37Rv genome, is shown.
Of the 294 distinct insertions mapped to the strain H37Rv genome, 180 were intragenic, affecting 100 ORFs. Of these, insertions in 90 ORFs were identified in the Edinburgh isolates; the remaining isolates added only 10 ORFs out of the 37 identified (27%) in these strains, which suggests that a broader distribution of sources would not increase the number of affected ORFs dramatically. However, the non-Edinburgh isolates contributed more to the diversity of the insert sites at the sequence position level, with 47 out of 74 sites (63%) in these isolates not being identified in the Edinburgh strains. This is consistent with the existence of constraints on the number of ORFs that can accept an IS6110 insert.
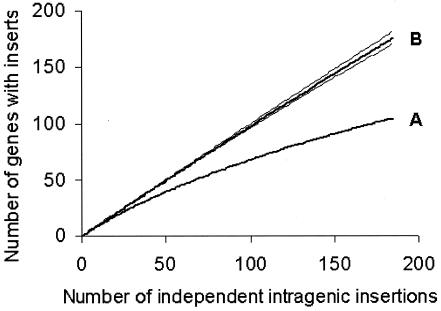
If the genomic distribution of IS6110 inserts was completely random, analysis (19) of a sample of this size indicates that insertions would be expected in 178 ORFs, which also suggests that there is a limitation on the number of ORFs that can accept an IS6110 insertion in clinical isolates. This is supported by examination of the number of independent insertions in each ORF (Fig. 1), which shows that one-third of the affected ORFs have more than one independent insertion. In addition, Fig. 2 shows that the accumulation of genes in which IS6110 inserts were detected was nonlinear with respect to the total number of independent inserts; in contrast, repeated resampling of in vitro transposon mutagenesis data (22) showed a linear response with a similar sample size. We can therefore conclude that relatively few genes (possibly fewer than 300, excluding repetitive elements) can readily accept an IS6110 insert, in bacteria from clinical infections.
FIG. 2.
Degree of saturation of insertions. (A) Number of genes affected compared to the total number of independent intragenic IS6110 insertions, taken in random order, in clinical isolates. (B) Accumulated genes with inserts in random samples of an in vitro transposon mutagenesis library (22). Broken lines indicate 95% confidence limits from repeated resampling by rarefaction analysis (48).
The number of truly nonessential genes may be lower than this, as not all genes with IS6110 inserts are necessarily inactivated. On the other hand, polar effects may result in a nonessential gene being unable to accept an IS6110 insertion due to possible deleterious effects on essential genes downstream in the same operon. Knowledge of the operon structure of M. tuberculosis is not at present adequate for thorough assessment of the likely extent of these polar effects. Nevertheless, it is instructive to compare the number of genes in clinical isolates that can readily accept an IS6110 insert with published estimates of the numbers of essential and nonessential genes in in vitro studies. Previous estimates using transposon mutagenesis indicated that 15 to 35% of the genes were essential in vitro (22, 39) and a further 177 genes, in addition to those required in vitro, were essential in a mouse model of infection (41). These estimates leave over 3,000 genes as potentially nonessential for infection, which is an order of magnitude higher than the number of genes that appear to be able to accept IS6110 in clinical isolates.
The two most likely hypotheses to account for this are (i) that there are relatively few genes into which IS6110 can be inserted without a significant effect on the ability of the organism to propagate human disease and (ii) that the availability of IS6110 insertion sites is limited by the transpositional specificity of the element. On the latter point, examination of all of the insertion sites revealed no detectable sequence specificity. There was some preference for a lower GC content of the region 50 bp to either side of the insertion (60% compared to 65.6% for the whole genome). The GC content for all determined sites compared to that of random 100-bp fragments from the genome was significantly different (P < 0.001); however, the preference was far from absolute, with IS6110 insertion sites ranging from 43 to 73% GC content, suggesting that although transpositional specificity may influence the number of available sites, it is unlikely to account for the effect completely and that the number of genes that can be affected by IS6110 in clinical isolates is much lower than the number of genes identified as essential in in vitro or laboratory animal studies. Clinical isolates are the outcome of repeated passage from person to person, which is likely to provide substantial selective pressure against the inactivation of any genes which have even a minor effect on fitness, as well as maintaining those genes that are needed for transmission and persistence, rather than infection. Such genes may not be identified as essential in short-term in vitro culture or in single infections of laboratory animals. Many genes may in fact play a more important role in the biology of an organism than in vitro transposon saturation studies suggest. For example, data from studies of Saccharomyces cerevisiae show that while 80% of the genes are apparently not essential under laboratory conditions (14), an in silico metabolic network analysis of enzyme dispensability (31) suggests that up to two-thirds of these genes are probably only dispensable under the particular laboratory growth conditions tested. The incidence of paralogues of genes harboring IS6110 insertions was assessed as a measurement of whether the inactivation of these genes might be complemented by an alternative gene. In the strain H37Rv genome, 47% (1,849/3,924) of the genes have at least one paralogue, while of the genes with an IS6110 insertion, 60% (60/100) have at least one paralogue. Thus, while paralogous gene complementation may play a role in mitigating the effects of gene disruption, it is not a generally applicable explanation.
Nature of the affected genes.
The identities of the ORFs affected in clinical isolates in this study were compared with those in previous studies, which were of three types. McAdam et al. (27) and Lamichhane et al. (22) used in vitro transposon mutagenesis with Tn5370 and Himar1, respectively, thus identifying nonessential genes, which can be compared directly with this study. Tsolaki et al. (49) also identified nonessential genes by an analysis of the deletions in a collection of clinical isolates. In contrast, Sassetti and coworkers (39-41) used transposon site hybridization (TraSH) to identify genes that were essential in vitro or for infection of laboratory mice. There was substantial agreement between these studies and the data reported here; of 100 genes containing an IS6110 insertion, only 5 were identified by Sassetti et al. (39, 41) as essential in vivo (Table 1). One of these (Rv2808) also showed a deletion in some clinical isolates (49). Another, ctpD (Rv1469), has been identified as harboring IS6110 in the globally widespread strain W (2) but in that case may be transcribed from a promoter in IS6110. A further four genes with IS6110 inserts in this study were identified by Sassetti et al. (39, 41) as essential in vitro, one of which (Rv2817c) has also been reported as deleted in some clinical isolates (49). Some of these exceptions may be due to complementation; a gene may appear to be essential in studies using one strain, such as H37Rv, if its complementary partner is already inactivated but nonessential in a clinical isolate that contains a complementing gene. Furthermore, in some cases, an IS6110 insertion may not inactivate the gene concerned, either where transcription can be reinitiated from the insertion sequence itself (2, 36, 44) or where the insertion is in the C-terminal region of the protein.
TABLE 1.
Genes previously identified as essential but showing IS6110 insertions in clinical isolates
| Gene no. | Gene name | Product |
|---|---|---|
| Identified as essential in vivoa | ||
| Rv1371 | Rv1371 | Probable membrane protein |
| Rv1469 | ctpD | Probable cation transporter |
| Rv2388c | hemN | Probable oxygen-independent coproporphyrinogen iii oxidase |
| Rv2437 | Rv2437 | Conserved hypothetical protein |
| Rv2808 | Rv2808 | Conserved hypothetical protein |
| Identified as essential in vitrob | ||
| Rv2817cc | Rv2817c | Conserved hypothetical protein |
| Rv3018c | PPE46 | PPE family protein |
| Rv3113 | Rv3113 | Possible phosphatase |
| Rv3343c | PPE54 | PPE family protein |
Analysis of the locations of the IS6110 inserts in the clinical isolates, within the different functional classes of genes in M. tuberculosis (6) (Table 2; see Table S5 of the supplemental material [available upon request] for a detailed list), showed a complete absence of inserts in putative virulence genes, which supports the identification of this class of genes as necessary for infection. Mutations of information pathway genes were also virtually absent from clinical isolates, as has also been found in studies of natural deletions in M. tuberculosis clinical isolates (3, 4, 15, 20, 24, 49); transposon mutagenesis also indicates that many genes of this class are essential in vitro (22, 27, 39). The central role of these genes in the biology of organisms and the general absence of multiple copies of these genes both preclude their inactivation. The one exception in this study was the detection of an IS6110 insertion in sigH; this gene is believed to be needed for virulence, as a sigH knockout mutant was shown to be attenuated in a mouse model (21). It is possible that this IS6110 mutation arose during the infection of this patient, and the organism may not be transmissible to other patients.
TABLE 2.
Distribution of inserts among functional classes of genes
| Function code | Functional classa | No. of genes in genome | % of genes in genome | No. of genes with inserts | % of genes with inserts |
|---|---|---|---|---|---|
| 0 | Virulence, detoxification, adaptation | 102 | 2.6 | 0 | 0.0 |
| 1 | Lipid metabolism | 237 | 6.2 | 4 | 4.1 |
| 2 | Information pathways | 231 | 6.0 | 1 | 1.0b |
| 3 | Cell wall and cell processes | 746 | 19.4 | 11 | 11.2 |
| 6 | PE/PPE | 168 | 4.4 | 13 | 13.3c |
| PE | 100 | 2.6 | 0 | 0 | |
| PPE | 68 | 1.8 | 13 | 13.3c | |
| 7 | Intermediary metabolism and respiration | 893 | 23.2 | 22 | 22.4 |
| 8 | Unknown | 15 | 0.4 | 1 | 1.0 |
| 9 | Regulatory proteins | 191 | 5.0 | 7 | 7.1 |
| 10 | Conserved hypotheticals | 998 | 25.9 | 22 | 22.4 |
| 16 | Conserved hypotheticals with an orthologue in M. bovis | 270 | 7.0 | 17 | 17.3c |
| Total | 3,851 | 100 | 98 | 100 |
Functional classes are based on M. tuberculosis H37Rv (http://genolist.pasteur.fr/TubercuList). Stable RNAs, IS elements, and phages (classes 4 and 5) have been excluded. Thirty-two insertions, affecting 13 ORFs, which mapped only to the M. tuberculosis CDC1551 genome are not included.
Significant difference from expected value. P < 0.05.
Significant difference from expected value. P < 0.01.
Genes involved in cell wall synthesis and lipid metabolism are believed to be important in infection. Cell wall synthesis genes were significantly (P < 0.05) less affected by mutations in clinical isolates than in the in vitro transposition libraries (22, 27). These genes were also significantly (P < 0.05) overrepresented in the list of in vivo essential genes (41). This supports the concept that cell wall synthesis genes are specifically important for infection. The results obtained with lipid metabolism genes as a class were less clear. Although these genes were less affected than expected by insertions of IS6110 in clinical isolates and in other studies overrepresented among genes identified as essential in vivo (41) and underrepresented among genes inactivated by deletion in clinical isolates (49), none of these results were statistically significant. This should not be interpreted as showing that none of these genes are needed for infection but indicates that the class as a whole is not significantly more important than other types of genes.
IS6110 is also found in other M. tuberculosis genes that may be involved in pathogenesis: the ESAT-6 gene esxJ (Rv1038c) (16) and PPE46-esxR-esxS region (25); Rv1265, which is upregulated in macrophages (17); Rv2819, which is upregulated in H37Rv compared to H37Ra (34); and bacterioferritin-encoding Rv3841,which is upregulated under low-oxygen conditions (35).
There is evidence for IS6110 transposition occurring in the bacilli of infected patients at the end of the metabolically dormant latency period (9). Similarly, in Escherichia coli archival cultures that had been stored for up to 3 decades, it was found that there had been extensive transpositional activity of several different transposable elements and that the resultant mutants had a diversity of fitness coefficients. The authors proposed that this genetic plasticity might allow an organism to be more adaptive in a hostile environment (28, 29). Recent evidence of IS6110-mediated transcription of adjacent genes from rightward promoters (36, 44) and possibly leftward promoters (2) also adds to the repertoire of mechanisms by which IS6110 might alter the phenotype of M. tuberculosis. However, in this study there was no obvious bias in the orientation of IS6110 in intergenic regions with respect to the direction of transcription of the adjacent genes, suggesting that activation of adjacent genes was not a major factor in the distribution of intergenic insertions.
Two classes of genes were more affected by IS6110 insertions in clinical isolates than predicted: the PE/PPE gene family and genes of unknown function (including conserved hypothetical genes). Members of the PE/PPE family of genes have been postulated to be involved in host immunity (6) and show a higher degree of sequence polymorphism than the genome as a whole (12), as well as apparently extensive codon volatility (32), which would be consistent with a role as targets for the host immune response. The high number of independent insertions in these genes may indicate that IS6110 also plays a mutagenic role in generating antigenic diversity or may reflect a considerable degree of redundancy among these genes. It is especially notable that subdivision of this family into PE and PPE subfamilies showed that all of the affected genes belonged to the latter category; no inserts were detected in members of the larger PE subfamily (Table 2). This contrasts markedly with the report (41) that few genes of either category are essential for experimental infection of animals. The IS6110 insertion preference for the PPE subfamily rather than the PE subfamily may be influenced by the high GC content of PE genes (75%) compared to PPE genes (64%,), but this is unlikely to fully account for the marked asymmetry detected. It seems likely, therefore, that IS6110 insertion into members of the PE subfamily of genes has some deleterious effect on the specific fitness of the organism in human infection chains.
The overrepresentation of genes of unknown function (including conserved hypothetical genes) among those affected by IS6110 insertion could be taken to indicate that some may have arisen from incorrect identification of ORFs and should be regarded as noncoding. However, subdivision of this category to separate out conserved genes with an orthologue in Mycobacterium bovis (category 16) shows that the overrepresentation is confined to this category (Table 2), which strengthens the belief that these insertions are in genuine coding sequences. The high level of insertions in category 16 genes suggests that the role of these genes is not critical, which is in agreement with the finding (41) that relatively few genes in this category are essential either in vitro or in vivo.
Frequently affected genes.
Substantial numbers of independent insertion sites (up to 12) were found in some genes (Table 3). In contrast, Lamichhane et al. (22), with a larger in vitro transposition library, found no gene with more than eight inserts. This raises the possibility that IS6110 insertion into certain genes may potentiate virulence, or progression from latent to active infection, which is necessary both for transmission and for isolation of the organism (45). Prominent are the genes coding for phospholipase C, which may play a role early in M. tuberculosis infection (33). M. tuberculosis has four plc genes, and multiple IS6110 inserts were detected in all four genes. The prevalence of insertions in these genes may reflect complementation between them, but the level of multiple insertions could also indicate a more complex role in the pathology of tuberculosis, especially during the later stages of the disease (33, 42, 50). Other genes with multiple independent insertions of IS6110 included several members of the PPE family and two major membrane protein genes, Rv0402c (mmpL1) and Rv1522c (mmpL12). PPE34 had 12 independent IS6110 inserts, the highest abundance detected in this study. Sampson et al. (37) noted extensive polymorphisms in this gene, but in variable numbers of tandem repeats, and with their observation that recombinant PPE34 was surface exposed when expressed in M. smegmatis and in M. bovis BCG, this protein may well have a role in the immunological interaction of the pathogen with the host. The hypothetical protein Rv2336 had multiple IS6110 insert sites and is not expressed in the avirulent H37Ra variant of H37Rv (34). Two probable transcription regulatory proteins (Rv1358 and Rv1359) both have several independent insert sites, but like many genes of M. tuberculosis, nothing is known of their biological role.
TABLE 3.
Genes with multiple insertion sites
| Gene no. | Gene name | No. of sites/gene | Gene product |
|---|---|---|---|
| Rv1917c | PPE34 | 12 | PPE family protein |
| Rv1755c | plcD | 9 | Probable phospholipase C (fragment) |
| Rv1358 | Rv1358 | 7 | Probable transcriptional regulatory protein |
| Rv2352c | PPE38 | 6 | PPE family protein |
| Rv2818c | Rv2818c | 5 | Hypothetical protein |
| Rv0402c | mmpL1 | 4 | Probable conserved transmembrane transport protein |
| Rv1359 | Rv1359 | 4 | Probable transcriptional regulatory protein |
| Rv1682 | Rv1682 | 4 | Probable coiled-coil structural protein |
| Rv3323c | moaX | 4 | Probable moaD-moaE fusion protein |
| Rv1135c | PPE16 | 3 | PPE family protein |
| Rv1522c | mmpL12 | 3 | Probable conserved transmembrane transport protein |
| Rv2336 | Rv2336 | 3 | Hypothetical protein |
| Rv2349c | plcC | 3 | Probable phospholipase C |
| Rv2351c | plcA | 3 | Probable phospholipase C (Mtp40 antigen) |
| Rv2435c | Rv2435c | 3 | Probable cyclase (adenylyl or guanylyl) |
| Rv2807 | Rv2807 | 3 | Conserved hypothetical protein |
| Rv2817c | Rv2817c | 3 | Conserved hypothetical protein |
| Rv3175 | Rv3175 | 3 | Possible amidase (aminohydrolase) |
| Rv3189 | Rv3189 | 3 | Conserved hypothetical protein |
Concluding remarks.
The genomic distribution of a transposable element is influenced by its target site preference, by its stability at different sites, and by the consequences of insertions on viability—which, for M. tuberculosis in human disease, is dependent on its pathogenicity and its transmissibility. The absence of IS6110 insertions in many genes, and the relative distributions of insertions in different classes of genes, suggests that M. tuberculosis requires a large repertoire of functional genes and that there are few genes that can be inactivated without a significant deleterious effect during chains of human infection. This is in contrast to the bioinformatic perspective, where there is evidence of duplication of many genes and therefore the possibility of functional redundancy (46). The relative abundance of intergenic IS6110 insertions over intragenic insertions in clinical strains suggests that most of M. tuberculosis’ complement of genes play a role in human infection and transmission and therefore that many of these paralogues are not functionally interchangeable. Although a majority of M. tuberculosis genes may confer an evolutionary benefit on the organism, the benefits of some, perhaps, may only be slight. This is suggested by several studies mimicking the infection process in the laboratory, where comparatively few genes were identified as playing a significant and detectable role. Plotkin et al. (32) claimed that these seemingly essential genes have less apparent codon volatility than genes nonessential in vitro. However, we were unable to detect any relationship between codon volatility and the occurrence of IS6110 insertion sites (data not shown), indicating that the requirements for clinical infection are wider and more subtle than those for in vitro growth.
More generally, definitions of the essential genes of an organism must take into account the environment in which the measurement is made. The essentiality of genes to an organism in its natural habitat reflects the organism's needs, not only over the different stages of its life cycle but also over evolutionary timeframes: slight advantages on infrequent occasions in a competitive environment will, over longer timescales, provide sufficient selective pressure to show up the importance of those genes.
Acknowledgments
We thank I. Wilson, Engineering and Physical Sciences School, University of Aberdeen, for statistical analysis; A. Thomson for technical assistance; and B. Watt, Scottish Mycobacteria Reference Laboratory, Edinburgh, United Kingdom, T. McHugh, Royal Free and University College Medical School, London, United Kingdom, and C. Sola, Institut Pasteur, Guadeloupe, France, for the provision of DNA extracted from clinical isolates.
This work was supported by the Wellcome Trust (grant 005791).
REFERENCES
- 1.Akerley, B. J., E. J. Rubin, V. L. Novick, K. Amaya, N. Judson, and J. J. Mekalanos. 2002. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 99:966-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beggs, M. L., K. D. Eisenach, and M. D. Cave. 2000. Mapping of IS6110 insertion sites in two epidemic strains of Mycobacterium tuberculosis. J. Clin. Microbiol. 38:2923-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 4.Brosch, R., S. V. Gordon, K. Eiglmeier, T. Garnier, F. Tekaia, E. Yeramian, and S. T. Cole. 2000. Genomics, biology, and evolution of the Mycobacterium tuberculosis complex, p. 19-36. In G. F. Hatfull and W. R. Jacobs (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, D.C.
- 5.Camacho, L. R., D. Ensergueix, E. Perez, B. Gicquel, and C. Guilhot. 1999. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34:257-267. [DOI] [PubMed] [Google Scholar]
- 6.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 7.Craig, N. L. 1997. Target site selection in transposition. Annu. Rev. Biochem. 66:437-474. [DOI] [PubMed] [Google Scholar]
- 8.Dale, J. W., H. Al-Ghusein, S. Al-Hashmi, P. Butcher, A. Dickens, F. Drobniewski, K. J. Forbes, S. G. Gillespie, D. Lamprecht, T. D. McHugh, R. Pitman, N. Rastogi, A. T. Smith, C. Sola, and H. Yesilkaya. 2003. Evolutionary relationships among strains of Mycobacterium tuberculosis with few copies of IS6110. J. Bacteriol. 185:2555-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eilders, P. H. C., D. van Soolingen, N. T. N. Lan, R. M. Warren, and M. W. Borgdorff. 2004. Transposition rates of Mycobacterium tuberculosis IS6110 restriction fragment length polymorphism patterns. J. Clin. Microbiol. 42:2461-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang, Z., and K. J. Forbes. 1997. A Mycobacterium tuberculosis IS6110 preferential locus (ipl) for insertion into the genome. J. Clin. Microbiol. 35:479-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang, Z., N. Morrison, B. Watt, C. Doig, and K. J. Forbes. 1998. IS6110 transposition and evolutionary scenario of the direct repeat locus in a group of closely related Mycobacterium tuberculosis strains. J. Bacteriol. 180:2102-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleischmann, R. D., D. Alland, J. A. Eisen, L. Carpenter, O. White, J. D. Peterson, R. DeBoy, R. Dodson, M. Gwinn, D. H. Haft, E. K. Hickey, J. F. Kolonay, W. C. Nelson, L. A. Umayam, M. Ermolaeva, S. L. Salzberg, A. Delcher, T. R. Utterback, J. F. Weidman, H. Khouri, J. Gill, A. Mikula, W. Bishai, W. R. Jacobs, J. C. Venter, and C. M. Fraser. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184:5479-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsyth, R. A., R. J. Haselbeck, K. L. Ohlsen, R. T. Yamamoto, H. Xu, J. D. Trawick, D. Wall, L. Wang, V. Brown-Driver, J. M. Froelich, C. G. Kedar, P. King, M. McCarthy, C. Malone, B. Misiner, D. Robbins, Z. Tan, Z. Y. Zhu Zy, G. Carr, D. A. Mosca, C. Zamudio, J. G. Foulkes, and J. W. Zyskind. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43:1387-1400. [DOI] [PubMed] [Google Scholar]
- 14.Giaver, G., A. M. Chu, L. Ni, C. Connelly, et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 15.Gordon, S. V., B. Heym, J. Parkhill, B. Barrell, and S. T. Cole. 1999. New insertion sequences and a novel repeated sequence in the genome of Mycobacterium tuberculosis H37Rv. Microbiology 145:881-892. [DOI] [PubMed] [Google Scholar]
- 16.He, X. Y., Y. H. Zhuang, X. G. Zhang, and G. L. Li. 2003. Comparative proteome analysis of culture supernatant proteins of Mycobacterium tuberculosis H37Rv and H37Ra. Microbes Infect. 5:851-856. [DOI] [PubMed] [Google Scholar]
- 17.Hobson, R. J., A. J. McBride, K. E. Kempsell, and J. W. Dale. 2002. Use of an arrayed promoter-probe library for the identification of macrophage-regulated genes in Mycobacterium tuberculosis. Microbiology 148:1571-1579. [DOI] [PubMed] [Google Scholar]
- 18.Hutchison, C. A., S. N. Peterson, S. R. Gill, R. T. Cline, O. White, C. M. Fraser, H. O. Smith, and J. C. Venter. 1999. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286:2165-2169. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs, M. A., A. Alwood, I. Thaipisuttikul, D. Spencer, E. Haugen, S. Ernst, O. Will, R. Kaul, C. Raymond, R. Levy, L. Chun-Rong, D. Guenthner, D. Bovee, M. V. Olson, and C. Manoil. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:14339-14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato-Maeda, M., J. T. Rhee, T. R. Gingeras, H. Salamon, J. Drenkow, N. Smittipat, and P. M. Small. 2001. Comparing genomes within the species Mycobacterium tuberculosis. Genome Res. 11:547-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaushal, D., B. G. Schroeder, S. Tyagi, T. Yoshimatsu, C. Scott, C. Ko, L. Carpenter, J. Mehrotra, Y. C. Manabe, R. D. Fleischmann, and W. R. Bishai. 2002. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc. Natl. Acad. Sci. USA 99:8330-8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamichhane, G., M. Zignol, N. J. Blades, D. E. Geiman, A. Dougherty, J. Grosset, K. W. Broman, and W. R. Bishai. 2003. A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 100:7213-7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maguire, H., J. W. Dale, T. D. McHugh, P. D. Butcher, S. H. Gillespie, A. Costetsos, H. Al Ghusein, R. Holland, A. Dickens, L. Marston, P. Wilson, R. Pitman, D. Strachan, F. A. Drobniewski, and D. K. Banerjee. 2002. Molecular epidemiology of tuberculosis in London 1995-7 showing low rate of active transmission. Thorax 57:617-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahairas, G. G., P. J. Sabo, M. J. Hickey, D. C. Singh, and C. K. Stover. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 178:1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marmiesse, M., P. Brodin, C. Buchrieser, C. Gutierrez, N. Simoes, V. Vincent, P. Glaser, S. T. Cole, and R. Brosch. 2004. Macro-array and bioinformatic analyses reveal mycobacterial ‘core’ genes, variation in the ESAT-6 gene family and new phylogenetic markers for the Mycobacterium tuberculosis complex. Microbiology 150:483-496. [DOI] [PubMed] [Google Scholar]
- 26.McAdam, R. A., P. W. Hermans, D. van Soolingen, Z. F. Zainuddin, D. Catty, J. D. A. van Embden, and J. W. Dale. 1990. Characterization of a Mycobacterium tuberculosis insertion sequence belonging to the IS3 family. Mol. Microbiol. 4:1607-1613. [DOI] [PubMed] [Google Scholar]
- 27.McAdam, R. A., S. Quan, D. A. Smith, S. Bardarov, J. C. Betts, F. C. Cook, E. U. Hooker, A. P. Lewis, P. Wollard, M. J. Everett, P. T. Lukey, G. J. Bancroft, W. R. Jacobs, Jr., and K. Duncan. 2002. Characterization of a Mycobacterium tuberculosis H37Rv transposon library reveals insertions in 351 ORFs and mutants with altered virulence. Microbiology 148:2975-2986. [DOI] [PubMed] [Google Scholar]
- 28.Naas, T., M. Blot, W. M. Fitch, and W. Arber. 1994. Insertion sequence-related genetic variation in resting Escherichia coli K-12. Genetics 136:721-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naas, T., M. Blot, W. M. Fitch, and W. Arber. 1995. Dynamics of IS-related genetic rearrangements in resting Escherichia coli K-12. Mol. Biol. Evol. 12:198-207. [DOI] [PubMed] [Google Scholar]
- 30.Palittapongarnpim, P., S. Chomyc, A. Fanning, and D. Kunimoto. 1993. DNA fingerprinting of Mycobacterium tuberculosis isolates by ligation-mediated polymerase chain reaction. Nucleic Acids Res. 21:761-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papp, B., C. Pal, and L. D. Hurst. 2004. Metabolic network analysis of the causes and evolution of enzyme dispensability in yeast. Nature 429:661-664. [DOI] [PubMed] [Google Scholar]
- 32.Plotkin, J. B., J. Dushoff, and H. B. Fraser. 2004. Detecting selection using a single genome sequence of M. tuberculosis and P. falciparum. Nature 428:942-945. [DOI] [PubMed] [Google Scholar]
- 33.Raynaud, C., C. Guilhot, J. Rauzier, Y. Bordat, V. Pelicic, R. Manganelli, I. Smith, B. Gicquel, and M. Jackson. 2002. Phospholipases C are involved in the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 45:203-217. [DOI] [PubMed] [Google Scholar]
- 34.Rindi, L., N. Lari, and C. Garzelli. 1999. Search for genes potentially involved in Mycobacterium tuberculosis virulence by mRNA differential display. Biochem. Biophys. Res. Commun. 258:94-101. [DOI] [PubMed] [Google Scholar]
- 35.Rosenkrands, I., R. A. Slayden, J. Crawford, C. Aagaard, C. E. Barry III, and P. Andersen. 2002. Hypoxic response of Mycobacterium tuberculosis studied by metabolic labeling and proteome analysis of cellular and extracellular proteins. J. Bacteriol. 184:3485-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Safi, H., P. F. Barnes, D. L. Lakey, H. Shams, B. Samten, R. Vankayalapati, and S. T. Howard. 2004. IS6110 functions as a mobile, monocyte-activated promoter in Mycobacterium tuberculosis. Mol. Microbiol. 52:999-1012. [DOI] [PubMed] [Google Scholar]
- 37.Sampson, S. L., P. Lukey, R. M. Warren, P. D. van Helden, M. Richardson, and M. J. Everett. 2001. Expression, characterization and subcellular localization of the Mycobacterium tuberculosis PPE gene Rv1917c. Tuberculosis 81:305-317. [DOI] [PubMed] [Google Scholar]
- 38.Sampson, S. L., R. M. Warren, M. Richardson, G. D. Van Der Spuy, and P. D. van Helden. 1999. Disruption of coding regions by IS6110 insertion in Mycobacterium tuberculosis. Tuber. Lung Dis. 79:349-359. [DOI] [PubMed] [Google Scholar]
- 39.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77-84. [DOI] [PubMed] [Google Scholar]
- 40.Sassetti, C. M., D. H. Boyd, and E. R. Rubin. 2001. Comprehensive identification of conditionally essential genes in mycobacteria. Proc. Natl. Acad. Sci. USA 98:12712-12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sassetti, C. M., and E. R. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 100:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith, D. A., T. Parish, S. M. Smith, H. M. Dockrell, N. G. Stoker, and G. J. Bancroft. 2002. Deletion of mycobacterial phospholipases C and haemolysin alters virulence and inhibits T cell recognition of Mycobacterium tuberculosis H37Rv, p. 1. In Fifth International Conference on the Pathogenesis of Mycobacterial Infections. Congrex, Stockholm, Sweden.
- 43.Smith, V., K. N. Chou, D. Lashkari, D. Botstein, and P. O. Brown. 1996. Functional analysis of the genes of yeast chromosome V by genetic footprinting. Science 274:2069-2074. [DOI] [PubMed] [Google Scholar]
- 44.Soto, C. Y., M. C. Menendez, E. Pérez, S. Samper, A. B. Gómez, M. J. García, and C. Martin. 2004. IS6110 mediates increased transcription of the phoP virulence gene in a multidrug-resistant clinical isolate responsible for tuberculosis outbreaks. J. Clin. Microbiol. 42:212-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka, M. M. 2004. Evidence for positive selection on Mycobacterium tuberculosis within patients. BMC Evol. Biol. http://www.biomedcentral.com/1471-2148/4/31. [DOI] [PMC free article] [PubMed]
- 46.Tekaia, F., S. V. Gordon, T. Garnier, R. Brosch, B. G. Barrell, and S. T. Cole. 1999. Analysis of the proteome of Mycobacterium tuberculosis in silico. Tuber. Lung Dis. 79:329-342. [DOI] [PubMed] [Google Scholar]
- 47.Thierry, D., A. Brisson-Noel, V. Vincent-Levy-Frebault, S. Nguyen, J. L. Guesdon, and B. Gicquel. 1990. Characterization of a Mycobacterium tuberculosis insertion sequence, IS6110, and its application in diagnosis. J. Clin. Microbiol. 28:2668-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Topp, E., M. Welsh, Y. C. Tien, A. Dang, G. Lazarovits, K. Conn, and H. Zhu. 2003. Strain-dependent variability in growth and survival of Escherichia coli in agricultural soil. FEMS Microbiol. Ecol. 44:303-308. [DOI] [PubMed] [Google Scholar]
- 49.Tsolaki, A. G., A. E. Hirsh, K. DeRiemer, J. A. Enciso, M. Z. Wong, M. Hannan, Y. O. Goguet de la Salmoniere, K. Aman, M. Kato-Maeda, and P. M. Small. 2004. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc. Natl. Acad. Sci. USA 101:4865-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viana-Niero, C., P. E. de Haas, D. van Soolingen, and S. C. Leao. 2004. Analysis of genetic polymorphisms affecting the four phospholipase C (plc) genes in Mycobacterium tuberculosis complex clinical isolates. Microbiology 150:967-978. [DOI] [PubMed] [Google Scholar]
- 51.Yesilkaya, H., A. Thompson, C. Doig, B. Watt, J. W. Dale, and K. J. Forbes. 2003. Locating transposable element polymorphisms in bacterial genomes. J. Microbiol. Methods 53:355-363. [DOI] [PubMed] [Google Scholar]