Abstract
RyhB is a noncoding RNA regulated by the Fur repressor. It has previously been shown to cause the rapid degradation of a number of mRNAs that encode proteins that utilize iron. Here we examine the effect of ectopic RyhB production on global gene expression by microarray analysis. Many of the previously identified targets were found, as well as other mRNAs encoding iron-binding proteins, bringing the total number of regulated operons to at least 18, encoding 56 genes. The two major operons involved in Fe-S cluster assembly showed different behavior; the isc operon appears to be a direct target of RyhB action, while the suf operon does not. This is consistent with previous findings suggesting that the suf genes but not the isc genes are important for Fe-S cluster synthesis under iron-limiting conditions, presumably for essential iron-binding proteins. In addition, we observed repression of Fur-regulated genes upon RyhB expression, interpreted as due to intracellular iron sparing resulting from reduced synthesis of iron-binding proteins. Our results demonstrate the broad effects of a single noncoding RNA on iron homeostasis.
Iron (Fe) is an essential element for virtually all organisms. This metal is an integral part of heme and is used as a cofactor in Fe-S proteins involved in major biological processes such as electron transport, the trichloroacetic acid (TCA) cycle, photosynthesis, N2 fixation, gene regulation, and DNA biosynthesis (2). Iron is one of the most abundant elements on earth; it is readily soluble under anaerobic conditions but becomes extremely insoluble in the presence of oxygen at neutral pH. Although it is essential for the physiology of most organisms, under aerobic conditions free iron is extremely toxic because of its ability to catalyze the formation of reactive oxygen species that can damage a variety of cellular components. To overcome iron toxicity, bacteria like Escherichia coli strictly regulate iron uptake and storage according to the availability of iron in the environment.
Iron can be transported actively from the environment to the cytoplasm via specific iron-binding transporters located in the bacterial membrane. Under conditions of iron limitation, E. coli cells produce the iron-chelating siderophore enterobactin (or enterochelin) (34), as well as cell surface iron transport proteins that recognize iron-loaded siderophores or other sources of iron, such as ferric citrate (12, 6, 13). While binding to surface proteins is energy independent, the transport through the outer membrane of iron-siderophore complexes is driven by energy-transducing proteins TonB-ExbB-ExbD, also called the TonB complex (26, 23). Once inside the cell, the metal is deposited into Fe-S proteins, heme, or iron-storage proteins. Iron-storage proteins called ferritin, encoded by ftnA, and bacterioferritin, encoded by bfr, store excess intracellular iron that is not required for incorporation into cellular enzymes.
When intracellular iron levels become sufficiently high, expression of the iron uptake genes is repressed by the Fur (ferric uptake regulator) protein (9, 19). To repress transcription, Fur binds to the promoter region of iron-regulated genes with ferrous iron (Fe2+) as a cofactor (21). In contrast, when iron concentrations fall below a certain threshold, Fur becomes inactive and repression of iron acquisition genes is relieved. Because Fur senses intracellular iron levels and adjusts gene expression accordingly, it is assumed to be the master regulator of bacterial iron homeostasis.
In addition to its role as a repressor, Fur was characterized as a positive regulator of genes such as acnA, fumA, ftnA, bfr, and sodB (22, 20). acnA and fumA encode iron-binding enzymes of the TCA cycle, and sodB encodes an Fe-superoxide dismutase. However, we have shown that Fur-mediated positive regulation of these mRNAs, and of sdhCDAB, encoding succinate dehydrogenase, is through repression of a small regulatory RNA, RyhB, which causes degradation of these mRNAs when it is expressed (30). The RNA chaperone Hfq is necessary for the activity and stability of the small RNA RyhB (30). We have recently demonstrated that RyhB recruits RNase E and facilitates degradation of mRNA targets (29). Thus, Fur and iron act together to repress expression of RyhB, which itself promotes degradation of mRNAs encoding iron-utilizing proteins, suggesting a role for the small RNA in prioritizing cellular iron use under iron-limiting conditions.
In the present study, we used microarrays to compare global transcription patterns in cells lacking or overproducing RyhB to identify other RyhB mRNA targets. In these experiments, a ryhB mutant host was used, and RyhB expression was restricted to a plasmid containing ryhB under the control of the heterologous pBAD promoter, which is induced only in the presence of arabinose. Thus, RyhB expression was independent of the Fur repressor, allowing analysis of target behavior independently of iron stress and in the presence of a functional Fur repressor. Our results suggest that RyhB directly regulates more than 18 operons in the bacterium E. coli. Moreover, this approach also identified several indirect RyhB targets, suggesting that the small RNA RyhB plays a major role, along with the Fur protein, in the regulation of iron metabolism.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and RNA isolation.
Derivatives of MG1655 carrying either pBAD-ryhB or the control vector pNM12 were used in all experiments. Both plasmids have been previously described (29). pNM12 was specifically designed to clone noncoding RNAs; it is a derivative of pBAD24, a pBR322-based plasmid containing araC and the arabinose-inducible pBAD promoter (29). pBAD-ryhB is a derivative of pNM12 in which the first nucleotide of the transcribed product is the +1 of RyhB. This is an important consideration when cloning noncoding RNAs under the control of heterologous promoters, since altering the natural RNA start site may have unintended consequences for its function. The strains used also contain the Δara714 allele (18) to prevent catabolism of arabinose and the ΔryhB::cat allele (30) to restrict RyhB expression to the inducible pBAD-ryhB vector.
Overnight bacterial cultures were incubated in LB medium with ampicillin at a final concentration of 50 μg/ml at 37°C and diluted 1,000-fold into 50 ml of fresh LB-ampicillin medium at 37°C with agitation. To induce RyhB expression, cultures carrying the pBAD-ryhB construct were grown to an optical density at 600 nm of 0.5 and arabinose was added to the culture at a final concentration of 0.1%. In some experiments, 50 μM FeSO4 was added to the new culture after dilution from an overnight culture. Total RNA was extracted from cells at the indicated time using the hot phenol procedure (1). To remove residual chromosomal DNA in the RNA sample, 30 to 35 μg of RNA was treated with 12 units of Turbo DNase from Ambion (Austin, TX) in a total volume of 150 μl. After incubating at 37°C for 30 min, the RNA was extracted once with phenol-chloroform, precipitated with ethanol, and resuspended in diethyl pyrocarbonate-treated water. RNA samples were stored at −70°C until used.
cDNA synthesis and labeling.
The protocol used was adapted from reference 38. To generate cDNA, 20 μg of RNA was mixed with 1 μg of random hexamer primers in a total volume of 60 μl. The mixture was heated to 70°C for 10 min and then cooled at 25°C for 10 min. The SuperScript reaction buffer from Invitrogen (Carlsbad, CA), dithiothreitol, and deoxynucleoside triphosphates were added according to the manufacturer's specifications. After 10 min at 25°C, the incubation temperature was increased to 50°C for 10 min. The SuperScript III enzyme from Invitrogen was added (2,400 U), and the reaction was incubated for an additional 50 min. The enzyme was heat inactivated at 70°C for 15 min. To remove RNA from the cDNA, 1 μg of RNase A from Epicenter (Madison, WI) and 20 U of RNase H from Invitrogen were added in the reaction mixture and incubated at 37°C for 10 min. The cDNA was purified using a QiaQuick PCR purification column from QIAGEN (Valencia, CA) and quantitated at 260 nm. To fragment the cDNA, a partial digestion with 0.2 U of DNase I per μg of cDNA was performed in 1× One-Phor-All buffer from Amersham Pharmacia (Piscataway, NJ) using 1.5 to 5 μg of cDNA at 37°C for 10 min. DNase I was heat inactivated at 95°C for 10 min, and fragmentation of cDNA to 50 to 100 bp was confirmed on an agarose gel. The fragmented cDNA was then labeled with an Enzo Bio Array Terminal Labeling Kit from Affymetrix (Santa Clara, CA) according to the manufacturer's specifications and used as a probe for Affymetrix E. coli antisense microarrays.
Array hybridization and data analysis.
Hybridization, washing, and staining of microarrays were performed as recommended in the Affymetrix technical manual. Briefly, the cDNA fragments were hybridized for 16 h at 45°C to an Affymetrix GeneChip E. coli antisense microarray. Arrays were washed at 25°C with nonstringent wash buffer, followed by a wash at 50°C with stringent buffer. Then, the arrays were stained with phycoerythrin-conjugated streptavidin (Molecular Probes). The arrays were read at 570 nm with a GeneChip Scanner 3000, and the data were analyzed with the Affymetrix Microarray Suite 5.0 software. Sample variations were standardized by scaling the average of all genes to a constant target intensity of 500 for all arrays used. The statistical significance of the data was analyzed by using rank-based (nonparametric) methods to generate P values for each gene. The data were passed through a first screening to discard absent genes, which have P values > 0.065. Then, from the present genes (P value < 0.05) or genes with a moderate P value (P value between 0.05 and 0.065), only those with a signal ratio (pNM12/pBAD-ryhB) higher than twofold, positive or negative, were kept for further studies. Operons were included in the Tables only when at least one gene within the operon had a ratio of over 2. Usually, when the detection level was low (signal less than 200), the genes were discarded, unless otherwise indicated in the text (e.g., bfr). Array experiments were done in duplicate, and the reproducibility of the experiments is shown on the scatter plot of the values for all present genes available at the following URL: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE3105. Genes repressed by RyhB in all three experiments (wild type, fur, and wild type with added FeSO4) have been listed in Table 1. Table 2 includes genes that are repressed by RyhB only in a fur+ background (unless indicated otherwise in the text). Table 3 includes genes that increased in expression upon RyhB expression.
TABLE 1.
RyhB-regulated genes
| Genea | Blattner no. | Fold change when RyhB is expressed
|
Descriptionb | Reference or source | ||
|---|---|---|---|---|---|---|
| Wild type | fur mutant | + FeSO4 | ||||
| acnB | b0118 | −4.4 | −2.0 | −1.4 | Aconitate hydrase B [4Fe-4S], major, exponential phase | |
| cueOc | b0123 | −2.1 | −1.3 | −1.2 | Multicopper oxidase, phenoloxidase and ferroxidase activities | 36 |
| sdhC | b0721 | −6.1 | −2.5 | −4.5 | Succinate dehydrogenase, cytochrome b556 | |
| sdhD | b0722 | −6.1 | −2.3 | −5.0 | Succinate dehydrogenase, hydrophobic subunit | |
| sdhA | b0723 | −4.0 | −2.6 | −3.0 | Succinate dehydrogenase, flavoprotein subunit | |
| sdhB | b0724 | −5.1 | −2.8 | −1.9 | Succinate dehydrogenase, 2Fe-2S, 3Fe-4S, 4Fe-4S | |
| cydA | b0733 | −1.9 | −2.2 | −2.4 | Cytochrome d terminal oxidase, polypeptide subunit I | |
| cydB | b0734 | −1.7 | −1.8 | −2.2 | Cytochrome d terminal oxidase, polypeptide subunit II | |
| pflA | b0902 | −3.8 | −2.9 | −2.8 | Pyruvate formate lyase activating enzyme 1, 4Fe-4S | |
| acnAd | b1276 | −1.7 | −1.3 | −3.8 | Aconitate hydrase A | |
| ydbK | b1378 | −2.9 | −2.8 | −1.3 | Putative oxidoreductase, 4Fe-4S | |
| fumA | b1612 | −7.6 | −3.8 | −3.2 | Fumarase A = fumarate hydratase Class I, 4Fe-4S | |
| ydhD | b1654 | −3.0 | −3.1 | −1.7 | Similar to some glutaredoxins | |
| sodB | b1656 | −19.3 | −10.6 | −20.9 | Iron superoxide dismutase | |
| yeaC | b1777 | −2.0 | −2.3 | −1.6 | Hypothetical protein | |
| msrB | b1778 | −2.8 | −1.9 | −2.2 | Methionine sulfoxide reductase | 17 |
| mrp/apbC | b2113 | −4.7 | −3.0 | −3.3 | Fe-S cluster formation/repair in Salmonella enterica | 43 |
| nuoN | b2276 | −3.0 | −2.5 | −4.1 | NADH dehydrogenase I chain N | |
| nuoM | b2277 | −4.1 | −3.5 | −4.2 | NADH dehydrogenase I chain M | |
| nuoL | b2278 | −4.3 | −3.2 | −4.8 | NADH dehydrogenase I chain L | |
| nuoK | b2279 | −5.0 | −3.3 | −5.0 | NADH dehydrogenase I chain K | |
| nuoJ | b2280 | −5.5 | −3.9 | −6.6 | NADH dehydrogenase I chain J | |
| nuoI | b2281 | −8.2 | −3.5 | −7.7 | NADH dehydrogenase I chain I, 4Fe-4S | |
| nuoH | b2282 | −6.8 | −3.5 | −5.8 | NADH dehydrogenase I chain H | |
| nuoG | b2283 | −4.9 | −3.7 | −4.3 | NADH dehydrogenase I chain G, 2Fe-2S, 4Fe-4S | |
| nuoF | b2284 | −3.2 | −2.4 | −2.5 | NADH dehydrogenase I chain F, potential 4Fe-4S | |
| nuoE | b2285 | −3.6 | −2.5 | −3.0 | NADH dehydrogenase I chain E, potential 2Fe-2S | |
| nuoC | b2286 | −4.5 | −3.0 | −4.7 | NADH dehydrogenase I chain C, D | |
| nuoB | b2287 | −4.4 | −2.3 | −3.9 | NADH dehydrogenase I chain B, potential 4Fe-4S | |
| nuoA | b2288 | −3.6 | −2.5 | −3.3 | NADH dehydrogenase I chain A | |
| sseB | b2522 | −2.5 | −2.0 | −1.3 | Enhanced serine sensitivity | |
| pepB | b2523 | −2.7 | −2.1 | −2.5 | Peptidase B | |
| iscX | b2524 | −5.6 | −3.5 | −4.7 | Open reading frame, hypothetical protein | |
| fdx | b2525 | −4.3 | −3.6 | −3.1 | Ferredoxin [2FE-2S], electron carrer protein | |
| hscA | b2526 | −2.9 | −2.6 | −2.4 | Heat shock protein, chaperone, member of Hsp70 protein family | |
| hscB | b2527 | −3.7 | −2.6 | −4.4 | Scaffold protein involved in iron-sulfur cluster assembly | |
| iscA | b2528 | −3.5 | −2.3 | −2.5 | Putative regulator | |
| iscU | b2529 | −4.9 | −2.8 | −3.2 | Scaffold protein involved in iron-sulfur cluster assembly | |
| iscS | b2530 | −6.1 | −3.8 | −5.1 | Subunit of cysteine desulfurase | |
| iscR | b2531 | −1.6 | −1.2 | −1.3 | IscR transcriptional regulator | |
| yggG | b2936 | −3.0 | −2.1 | −2.7 | Putative metalloprotease | ExPASy |
| hybG | b2990 | +0.9 | −1.1 | −1.1 | Maturation of large subunit of hydrogenase-2 | |
| hybF | b2991 | +0.9 | −2.8 | −2.3 | May modulate levels of hydrogenase-2 | |
| hybE | b2992 | −1.2 | −1.4 | −1.5 | Member of hyb operon | |
| hybD | b2993 | −1.2 | +0.9 | −1.4 | Probable processing element for hydrogenase-2 | |
| hybC | b2994 | −1.3 | −6.1 | −9.2 | Large subunit, hydrogenase-2 | |
| hybB | b2995 | −2.1 | −1.9 | −3.0 | Probable cytochrome Ni/Fe component of hydrogenase-2 | |
| hybA | b2996 | −2.3 | −1.6 | −2.1 | Hydrogenase-2 small subunit, 4Fe-4S, 3Fe-4S | |
| hybO | b2997 | −2.4 | −1.6 | −8.8 | Small hydrogenase subunit, 4Fe-4S, 3Fe-4S | |
| fdoI | b3892 | −3.8 | −2.6 | −3.4 | Formate dehydrogenase, cytochrome b556 subunit | |
| fdoH | b3893 | −5.6 | −3.1 | −5.2 | Formate dehydrogenase-O, iron-sulfur subunit | |
| fdoG | b3894 | −5.0 | −3.3 | −4.2 | Formate dehydrogenase-O, major subunit | |
| frdD | b4151 | −6.0 | −2.6 | −2.3 | Fumarate reductase, anaerobic, membrane anchor polypeptide | |
| frdC | b4152 | −10.7 | −3.3 | −2.5 | Fumarate reductase, anaerobic, membrane anchor polypeptide | |
| frdB | b4153 | −5.3 | −1.8 | −1.9 | Fumarate reductase, anaerobic, iron-sulfur protein subunit | |
| frdA | b4154 | −4.4 | −2.6 | −2.6 | Fumarate reductase, anaerobic, flavoprotein subunit | |
Individual genes or operons are grouped by lightface or boldface gene names. The gene orientation from top to bottom is clockwise on the chromosome.
Descriptions are mostly from Affymetrix or, when mentioned, from ExPASy (http://au.expasy.org) or cited reference.
While cueO is weakly regulated by RyhB in the fur mutant, it is not repressed by Fur.
acnA is repressed by RyhB as previously shown by Northern blotting (30).
TABLE 2.
Fur-regulated genes
| Genea | Blattner no. | Fold change when RyhB is expressed
|
Fur-regulation reference | Description | Reference and/or source | ||
|---|---|---|---|---|---|---|---|
| Wild type | fur mutant | + FeSO4 | |||||
| fhuA | b0150 | −4.2 | +0.9 | −1.7 | 28 | Outer membrane protein receptor for ferrichrome and colicin M | |
| fhuC | b0151 | −2.0 | +0.9 | −1.4 | ATP-binding component of hydroxymate-dependent iron transport | ||
| fhuD | b0152 | −2.1 | +0.9 | +0.7 | Hydroxamate-dependent iron uptake, cytoplasmic membrane | ||
| fhuB | b0153 | −2.9 | 1.0 | −1.3 | Hydroxamate-dependent iron uptake, cytoplasmic membrane | ||
| ycdNb | b1016 | −5.5 | +0.8 | +0.8 | 31 | Belongs to the FTR1 family (high-affinity iron permease) | ExPASy |
| ycdO | b1018 | −6.5 | +0.9 | −3.0 | Periplasmic or exported protein, similar to Bacillus subtilis ywbM | 44, ExPASy | |
| ycdB | b1019 | −1.3 | 1.0 | −1.2 | Predicted iron-dependent peroxidase | NCBIe | |
| sufE | b1679 | −1.9 | −1.6 | −1.1 | 35 | Stimulates SufS | |
| sufS | b1680 | −7.1 | −1.8 | −1.3 | Cysteine desulfurase | ||
| sufD | b1681 | −2.4 | −1.5 | −8.2 | Stimulates SufS, stabilizes FhuF protein | ||
| sufC | b1682 | −2.3 | −1.7 | +0.9 | Stimulates SufS | ||
| sufB | b1683 | −3.2 | −2.4 | −1.4 | Stimulates SufS | ||
| sufA | b1684 | −1.4 | −1.8 | −2.6 | Scaffold protein for assembly of iron-sulfur cluster | ||
| yojId | b2211 | −14.5 | +0.9 | −3.6 | Pos. cons.d | ATP-dependent transport | 40 |
| exbD | b3005 | −5.1 | −1.1 | −1.6 | 8 | Uptake of enterochelin, forms a complex with tonB | |
| exbB | b3006 | −4.6 | 1.0 | −2.3 | Uptake of enterochelin, forms a complex with tonB | ||
| bfrc | b3336 | −2.4 | −1.3 | −3.4 | 20 | Bacterioferrin, iron storage | |
| bfd | b3337 | −1.3 | −1.1 | −1.6 | 31 | Bacterioferritin-associated ferredoxin | |
| feoA | b3408 | −2.1 | −1.1 | −1.6 | 25 | Ferrous iron transport protein A | |
| feoB | b3409 | −1.2 | −1.1 | −3.2 | Ferrous iron transport protein B | ||
| sodA | b3908 | −7.1 | −1.7 | −4.0 | 11 | Superoxide dismutase, manganese | |
| fecE | b4287 | −2.8 | −1.1 | −4.5 | 3 | ATP-binding component of citrate-dependent iron(III) transport | |
| fecD | b4288 | −3.9 | −1.1 | −2.5 | Citrate-dependent iron transport, membrane-bound protein | ||
| fecC | b4289 | −2.8 | −1.1 | −2.8 | Citrate-dependent iron(III) transport protein, cytosolic | ||
| fecB | b4290 | −2.5 | −1.1 | −2.0 | Citrate-dependent iron transport, periplasmic protein | ||
| fecA | b4291 | −1.6 | +0.7 | −1.4 | Outer membrane receptor; citrate-dependent iron transport | ||
| fecR | b4292 | −4.0 | 1.0 | 1.0 | 5 | Regulation of iron dicitrate transport | |
| fecI | b4293 | −1.6 | 1.0 | 1.0 | Probably regulates fec genes for iron dicitrate transport | ||
| fhuF | b4367 | −14.3 | 1.0 | −1.2 | 45 | Reduction of ferric iron in ferrioxamine B, binds 2Fe-2S cluster | |
Individual genes are grouped by lightface or boldface gene names.
Possible frameshift in the sequence; also named b4490.
Although bfr has been described as regulated by RyhB (30), it corresponds more nearly to a Fur-regulated gene in these arrays.
Possible consensus with Fur-binding site (see text).
NCBI, National Center For Biotechnology Information.
TABLE 3.
Positively regulated genes
| Gene | Blattner no. | Fold change when RyhB is expressed
|
Description | ||
|---|---|---|---|---|---|
| Wild type | fur mutant | + FeSO4 | |||
| ackA | b2296 | +2.0 | +1.4 | +1.5 | Catalyzes acetyl-coenzyme A into acetate and ATP |
| pta | b2297 | +1.8 | +1.2 | +1.3 | Subunit of phosphate acetyltransferase |
| ygdQ | b2832 | +4.0 | +1.9 | +1.9 | Putative transmembrane protein, transport |
| brnQ | b0401 | +2.4 | +1.5 | +1.3 | Branched chain amino acid transporter |
| ftnA | b1905 | +4.2 | +1.0 | +1.7 | Cytoplasmic ferritin, iron storage protein |
| kgtP | b2587 | +2.2 | +1.4 | +1.7 | α-ketoglutarate transporter |
| nhaB | b1186 | +2.2 | +1.3 | +1.5 | Sodium-proton antiporters |
| ompX | b0814 | +2.8 | +1.5 | +2.1 | Integral outer membrane protein X |
| oppA | b1243 | +2.3 | +1.9 | +1.7 | Oligopeptide ABC transport system (Opp) |
| oppB | b1244 | +1.8 | +1.4 | +1.4 | Oligopeptide transport permease protein |
| oppC | b1245 | +1.6 | +1.4 | +1.4 | Transporter of permease protein |
| oppD | b1246 | +1.4 | +1.2 | +1.3 | Oligopeptide ABC transport system |
| oppF | b1247 | +1.4 | +1.2 | +1.2 | Oligopeptide ABC transport system |
| shiA | b1981 | +5.1 | +3.0 | +1.7 | Transporter of shikimate |
| ybaK | b0481 | +3.3 | +1.7 | +2.2 | Haemophilus influenzae homolog, tRNA synthetase |
Northern blot analysis.
Northern blot hybridization was performed as described in reference 30. The biotinylated probes used for detection of sodB and exbBD mRNAs were, respectively, 5′-CCAGTAGAAAGTATGGTTCCAGACCTGAGCTGCGT-3′ (EM33) and 5′-TGGCTTCCAGATCCAGGTCACGGCTTTGCAGCAAC-3′ (EM120). The biotinylated probes used for 5S and 16S rRNA hybridization were, respectively, 5′-GTTTCACTTCTGAGTTCGGCATGGGGTCAGGTGGG-3′ and 5′-TTACGGCGTGGACTACCAGGGTATCTAATCCTGTTTGCT-3′. The probes were detected using a Bright-Star Biodetect kit from Ambion according to the manufacturer's instructions.
RESULTS AND DISCUSSION
Ectopic expression of RyhB expression from a pBAD-inducible promoter.
Our previous work demonstrated that expression of the small regulatory RNA RyhB specifically down-regulated six mRNAs encoding iron-containing proteins (30). While this number of targets exceeds those for any known prokaryotic small RNA, we were interested in determining whether there were additional targets. Because RyhB leads to rapid degradation of the mRNAs of its targets, and thus to decreased levels of these messages, we could use microarray analysis to identify RyhB mRNA targets.
For the arrays, RyhB was expressed from a plasmid under the control of the inducible araBAD promoter, in cells lacking the chromosomal copy of ryhB. This had a number of advantages. The effects of RyhB expression could be assayed after a short time (15 min in most cases), and the effects of RyhB expression could be examined in the presence of functional Fur repressor, without perturbing cellular iron levels. As shown in Fig. 1, the level of RyhB achieved upon arabinose induction of cells was somewhat higher than the level of RyhB produced by wild-type cells in response to iron limitation.
FIG. 1.
Northern blot analysis of RyhB levels in different bacterial backgrounds. The wild-type cells grown in the presence of the iron chelator 2,2′-dipyridyl (dip) at 250 μM express the small RNA RyhB from the endogenous gene. In comparison, the cells (ΔryhB::cat strain) carrying the pBAD-ryhB plasmid express the small RNA only from the arabinose-inducible pBAD promoter in the presence of 0.1% arabinose (ara). The difference in RyhB levels after 15 min of induction is about 10 times more in pBAD-ryhB plus ara than in wild type plus dip.
Cells carrying pBAD-ryhB plasmid were grown to mid-log phase in LB and treated with 0.1% arabinose for 15 min. Two sets of negative-control conditions were used for comparison. In one, the vector control, pNM12 was induced with arabinose for 15 min; in the second, cells containing the pBAD-ryhB plasmid were grown without arabinose induction. Total RNA was extracted from all cultures and processed for microarray analysis (see Materials and Methods). The gene expression profiles were compared in two ways: (i) induced pBAD-ryhB-containing cells were compared to induced vector-containing cells, and (ii) uninduced pBAD-ryhB-containing cells were compared to induced pBAD-ryhB-containing cells. The values for these two types of comparisons showed consistent results, with the exception of the expected differences for arabinose-inducible genes. These comparisons were performed for three distinct experiments: (i) fur+ cells grown in LB; (ii) Δfur::kan cells grown in LB; and (iii) fur+ cells grown in LB supplemented with 50 μM FeSO4. The results of these experiments are shown in Tables 1, 2, and 3. Tables 1 and 2 describe the major down-regulated genes, and Table 3 shows the few moderately up-regulated genes. In each table, results are shown for fur+ cells grown in LB, fur mutant cells grown in LB, and fur+ cells grown in LB with added FeSO4. The array signals for all genes with arabinose (either with the vector or with pBAD-ryhB) are available at the following URL: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE3105.
Direct and indirect regulation of genes: iron sparing by RyhB.
We first describe two examples of RyhB-regulated genes that we have categorized as direct (Table 1) or indirect (Table 2) RyhB targets. sodB is known to be a direct target of RyhB (30, 47, 15). It was the most stringently down-regulated transcript seen in the arrays. It was down-regulated 19-fold in fur+ cells expressing RyhB, 11-fold in fur mutant cells, and 21-fold in fur+ cells grown in the presence of additional FeSO4. Given our previous demonstration that the mRNA for sodB totally disappears shortly after RyhB expression (29), these results were expected and confirm that we can use the microarrays to monitor this process. Other genes in Table 1 showed a similar pattern, that is, a down-regulation upon RyhB expression that was independent of fur or added FeSO4. The genes in this table are likely direct targets of RyhB.
The fhuF gene (Table 2) shows a pattern of expression that is consistent with an indirect effect of RyhB. fhuF encodes a protein involved in acquiring iron from ferrioxamine and is known to be repressed by Fur (45). However, fhuF and other Fur-regulated genes (fhuACDB operon, exbBD, fec operon; see Table 2) were significantly (14-fold) down-regulated upon RyhB expression. We reasoned that RyhB overproduction, by down-regulating the synthesis of many iron-binding proteins, may increase the intracellular iron concentration, thereby leading to more complete Fur repression of these genes. The two sets of growth conditions shown in the tables for fur mutant cells grown in LB and fur+ cells grown in LB with added FeSO4 were developed to test this hypothesis.
The first strategy was to overproduce RyhB from pBAD-ryhB, in a fur ryhB background. Under these conditions, transcripts affected indirectly by RyhB effects on Fur-mediated repression would not be changed in response to RyhB expression. The results for the fur mutant in Table 2 show that this prediction is supported for the Fur-repressed genes described above and others in a total of 10 operons. In the fur mutant, the vector/pBAD-ryhB signal ratio was close to 1 for all of these genes (Table 2). As expected, the basal signal for these genes (in the absence of RyhB expression) was higher in the fur mutant than in fur+ cells (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE3105). Genes that showed this pattern (decreased expression in fur+ cells after RyhB expression but little or no effect of RyhB in fur cells) were placed in Table 2; those that were still decreased in the fur mutant upon RyhB expression are found in Table 1. The significant increase in Fur-dependent repression after RyhB expression supports the role of RyhB in elevating intracellular iron concentrations for incorporation into essential enzymes by decreasing its use by other proteins.
The second strategy used to test the role of Fur in RyhB-dependent down-regulation was to add excess iron to the media, which we presumed would keep intracellular Fur-iron levels sufficient to maintain full repression of Fur-regulated genes (see results for LB plus FeSO4 in Tables 1, 2, and 3). In general, excess iron decreased the effect of RyhB for genes in Table 2 but not as completely as that seen in the fur mutant. In addition, because the expression of some of the genes in Table 2 was very low in the presence of excess iron (as expected), this was not as sensitive a test. It is worth noting that some of the genes in Table 2 still show some regulation by RyhB in the presence of excess FeSO4. This suggests that the iron-sparing effect of RyhB is more effective at raising intracellular iron concentrations than the addition of extracellular iron and/or that the amount of iron necessary to repress these genes is particularly high.
Not all Fur-repressed genes are present in Table 2, in part because the level of expression was not sufficiently high to be detected under our growth conditions or because the degree of Fur repression is not as dramatic; nonetheless, the iron-sparing effect can be seen for some of these genes as well. For instance, the tonB gene, encoding a protein necessary for ExbBD function, is known to be weakly repressed by Fur (31). Consistent with this, we observed a 3.7-fold increase in the signal from tonB in a fur mutant compared to fur+, in the absence of RyhB expression (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE3105). When RyhB was overproduced in fur+ cells, we observed a modest iron-sparing effect (1.8-fold reduction) in the tonB signal (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE3105). However, no effect was observed on tonB signal under conditions of RyhB production in a fur mutant (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE3105). Thus, tonB shows a pattern of regulation by RyhB similar to that seen with exbBD, the genes encoding other components of iron entry, which are included in Table 2.
Time course of RyhB effect on sodB (RyhB-direct) and exbBD (Fur-regulated) mRNAs.
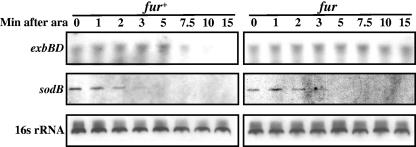
A prediction made on the basis of the iron-sparing model for regulation of the genes in Table 2 is that the RyhB-dependent repression of Fur-regulated genes is a response to decreased synthesis of iron-containing proteins. Therefore, the kinetics of RyhB repression of the indirect targets should be slower than that for the direct targets. Our previous work showed significant degradation of direct targets such as sodB mRNA in less than 3 min after RyhB expression (29). In those experiments RyhB was expressed from its own promoter after the addition of an iron chelator (2,2′-dipyridyl; see Fig. 1 for comparison with pBAD-ryhB expression). To determine the kinetics of loss of message of a direct RyhB target (sodB) and Fur-regulated (exbBD) genes, a time course of mRNA levels was examined with fur+ and fur strains after arabinose induction of RyhB from pBAD-ryhB. The results (Fig. 2) showed that while sodB mRNA was almost totally gone by 3 min after RyhB expression, the exbBD mRNA level dropped only after 7.5 min of RyhB expression in the fur+ background. As in the array (at the 15-min time point), the exbBD mRNA levels remained unchanged in a fur mutant at all times. These results are consistent with RyhB directly targeting sodB mRNA for rapid degradation by RNase E (29, 33) while only indirectly repressing exbBD by increasing available iron and therefore increasing Fur repression. This iron-sparing effect of RyhB expression, while necessarily slower than regulation of direct targets, is still very rapid, suggesting that even a few minutes of shutoff of synthesis of iron-binding proteins (from 3 min after RyhB expression to 7.5 min) is sufficient to increase available iron in the cell, demonstrating the efficacy of using a regulator like RyhB (see Fig. 3 for a model).
FIG. 2.
Northern blot analysis of time course effect of RyhB overproduction on exbBD and sodB mRNAs. fur+ and fur cells carrying the pBAD-ryhB plasmid were grown to an optical density at 600 nm of 0.5 and then treated with 0.1% arabinose (ara) to express RyhB. Total RNA was extracted at the indicated time by the hot phenol technique and probed for sodB or exbBD mRNAs with biotinylated oligonucleotides EM33 and EM120, respectively (for probe descriptions, see Materials and Methods). The EM12 probe hybridizes with the loading control 16S rRNA.
FIG. 3.
Model for indirect RyhB effect on Fur-regulated genes. RyhB causes rapid degradation of the target mRNAs it pairs with. In this model, the rapid block of synthesis of many iron-utilizing proteins increases intracellular levels of Fe (iron sparing), improving repression by Fur. After 15 min of RyhB expression, Fur-regulated mRNAs show a pattern of disappearance similar to those seen with RyhB-regulated genes. The loss of a RyhB effect in fur mutants supports the iron-sparing model.
Regulation of multiple operons by RyhB.
The observation of a significant level of iron sparing (increased Fur-dependent repression) upon RyhB expression suggests that during exponential growth in LB, there are abundant iron-binding proteins that are down-regulated by RyhB. The array results support this proposal.
Genes of intermediary metabolism.
As mentioned above, regulation of the previously identified target, sodB, is easily seen on arrays. Two other previously identified targets, fumA and sdhCDAB, were also significantly down-regulated (two- to eightfold under the three sets of growth conditions; Table 1) (30). In addition, fumarate reductase, the enzyme catalyzing the reaction opposite to that catalyzed by succinate dehydrogenase, is encoded by the frdABCD operon; like the sdh operon, the frd mRNA was also down-regulated by RyhB (Table 1). While frdABCD is most highly expressed under anaerobic conditions (24), it could be detected on the arrays. fumB and fumC, alternative enzymes to fumA, were too poorly expressed under these growth conditions to be detected on the arrays.
Another target previously described for RyhB is acnA, the stationary-phase aconitase (7, 30). In the arrays, the signal for acnA was modestly (1.3× to 3.8× under the various growth conditions) reduced by RyhB expression. However, the message for acnB, the major aconitase during exponential growth, was also found to be significantly reduced after RyhB expression (1.4× to 4.4× under the three sets of growth conditions). Thus, for these steps in intermediary metabolism, alternative enzymes under somewhat different transcriptional regulation conditions are coregulated by RyhB.
The effects on intermediary metabolism were even more widespread than those seen with these TCA cycle enzymes. A number of genes encoding other Fe-S-containing metabolic enzymes were also found to be down-regulated after RyhB expression. The large nuo operon, encoding the first enzyme of the respiratory chain NADH dehydrogenase, showed a two- to eightfold decrease in signal across the operon under all three sets of growth conditions. The formate dehydrogenase operon (fdo) is also down-regulated three- to sixfold under all growth conditions. pflA, encoding a pyruvate formate lyase-activating enzyme, is also moderately down-regulated. ydbK is a gene of unknown function but is predicted on the basis of homology to be an Fe-S oxidoreductase; we suggest that its down-regulation by RyhB supports this assignment. Less stringently regulated is the hyb operon, encoding an anaerobic hydrogenase also containing Fe-S clusters and its maturation components (32, 39). Similarly weakly regulated is cueO, part of the copper homeostasis system but with some in vitro activity with iron as well (16).
In addition to the genes discussed above and some others discussed in the next section, two other genes of unknown function were identified as RyhB regulated. YdhD, a conserved protein of unknown function, and yggG, another gene of unknown function annotated as a possible metalloprotease (ExPASy [http://au.expasy.org]), would both be predicted to be involved in iron homeostasis on the basis of their sensitivity to RyhB expression.
Genes of Fe-S metabolism and iron-protein maturation.
Also present in Table 1 are the genes of the isc cluster, involved in assembly of Fe-S clusters (14, 41). There are two possible explanations for this observation. One is that this operon is also a direct RyhB target (implying base pairing of RyhB with the isc mRNA). If this were the case, it would imply that the isc genes are not required under iron-limiting conditions that would lead to RyhB expression and therefore that the isc gene products are not required for assembly of Fe-S clusters in essential enzymes. An alternative explanation lies in isc regulation by iscR. IscR is an iron-binding repressor (42), and it is plausible that changes in intracellular iron availability upon RyhB expression could lead to increased IscR repression activity. However, this model is not supported by the unchanged regulation of these genes in the presence of excess iron (Table 1). Therefore, we favor a direct action of RyhB on the isc operon, although it is still possible that the higher level of intracellular iron resulting when RyhB is active is more effective in potentiating IscR-dependent repression than is the addition of external FeSO4.
E. coli contains another set of genes involved in Fe-S cluster assembly, the suf genes (46, 35). The suf operon has recently been shown to be under Fur repression (31), and, consistent with that finding, we found indirect RyhB-dependent down-regulation of suf genes in the fur+ background (Table 2). We note that RyhB has a modest effect on suf operon, even in a fur mutant (Table 2), suggesting possible additional (possibly also indirect) regulatory effects of the small RNA. Nevertheless, our results with respect to RyhB regulation would suggest that suf, rather than isc, should be the primary source of Fe-S clusters under iron-limiting conditions, consistent with the findings and conclusions of Outten et al. (35). These authors examined the physiology of cells mutant with respect to either the suf or isc genes and concluded that while isc genes play a major role in Fe-S center assembly during normal growth, the suf-encoded proteins are required during iron starvation. Thus, our data are consistent with a model where RyhB helps to shut down both synthesis of nonessential Fe-S proteins and Fe-S assembly for these proteins; the existence of a second Fe-S cluster assembly system allows insertion of Fe-S clusters in essential proteins under Fe-limiting conditions.
The pepB and sseB transcripts, located immediately downstream of the iscRSUA-hscBA-fdx-iscX operon, were also down-regulated upon RyhB expression, suggesting that these genes might be part of the same operon. To confirm this, the RNA was reverse transcribed and the resulting DNA amplified by PCR. The results show a PCR product spanning fdx-iscX-pepB-sseB, which indicates that at least some transcripts extend through these genes (data not shown).
A number of other genes associated with Fe-S clusters and/or metabolism are also found in Table 1. mrp, a gene implicated in formation and repair of Fe-S clusters in Salmonella spp. (43), is regulated by RyhB. MsrB, a methionine sulfoxide reductase (17), is in an operon with the unknown gene yeaC; both are modestly regulated by RyhB, suggesting a possible connection between RyhB targets and sulfur metabolism. However, because not much is known about the transcriptional regulation of these genes, indirect effects of RyhB on these genes cannot yet be ruled out.
Genes positively regulated by Fur via RyhB.
Positive regulation by Fur and iron has been described for acnA, fumA, ftnA, bfr, and sodB (22, ). The positive regulation of these genes as well as sdhCDAB is mediated by RyhB (30). Because RyhB is normally negatively regulated by Fur and iron, negative regulation by RyhB of this large number of targets would also be expected to be detected as positive regulation by Fur and iron in global examinations of Fur-regulated genes. In such a previous search for iron-regulated genes, McHugh and collaborators found that a large number of genes involved in energy metabolism and iron storage were decreased in expression in the presence of the iron chelator 2,2′-dipyridyl or in a fur mutant (31), conditions which should lead to high-level RyhB expression. In common with the genes listed in Table 1, they found this type of Fur-dependent positive regulation with the frd operon, the nuo operon, and sodB. Our results are entirely consistent with their findings and extend them to demonstrate that the positive regulation by Fur and iron can be mimicked by ectopic expression of RyhB alone. Of the other genes positively regulated by Fur and iron identified in their study, the cydA operon shows a RyhB-sensitive pattern of regulation (see Table 1).
Comparison between the results of the study by McHugh et al. and those of ours revealed some genes that were positively regulated by Fur and iron in their experiments but apparently not affected by RyhB expression in our experiments, suggesting that other mechanisms for positive regulation by Fur and iron must exist. The cyoA operon and the narG operon, found to be positively regulated in the study by McHugh et al. (31), also showed strong positive regulation by Fur in our experiments but no effect of RyhB (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE3105). Expression of narK was more complicated; RyhB increased rather than decreased its expression (data not shown). Thus, the overall down-regulation of energy metabolism genes is due in large part, but not entirely, to RyhB.
Fur-regulated genes and iron sparing.
With one exception, the genes listed in Table 2 had previously been either shown to be Fur regulated or suggested to be, on the basis of previous arrays (see table for references). The yojI gene, predicted to be an ABC-type transporter, was not previously characterized as a Fur-regulated gene (40). However the microarray signal of yojI increased 2-fold in the fur background compared to the results seen with fur+ and 5.5-fold in the fur background compared to the results seen with cells with added FeSO4 (data not shown). The numbers in Table 2 also suggest yojI is down-regulated by RyhB only in the presence of Fur, 14-fold in fur+ and 0.9-fold in fur cells. The Fur-binding consensus contains three or four imperfect adjacent hexamers 5′-GATAAT-3′ (10, 27), although an alternative consensus sequence (5′-TGATAATNATTATCA-3′) has been proposed recently (4). We looked at potential Fur-binding consensus sequences in the upstream region of yojI, and the sequence 5′-GAATAAAAATAAGAATTATTATT-3′ showed the closest resemblance to a Fur-binding sequence. Thus, we believe that yojI is most likely a member of the Fur regulon.
Operons that are regulated by Fur at the transcriptional level could, in principle, also be regulated by RyhB. bfr, encoding bacterioferritin, may be such a case. Bfr is the second gene in an operon with bfd; bfd has been proposed to regulate iron release from bacterioferritin (37) and is known to be regulated by Fur (31). The signal for bfd is increased 12-fold in the fur host carrying the vector relative to fur+; the bfr signal is increased 4.7-fold (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE3105), consistent with Fur regulation of these genes. However, bfr had previously been reported to be positively regulated by Fur and iron (20, 31), and we previously demonstrated regulation by RyhB (30). The bfr mRNA signal in the arrays was sensitive to RyhB in fur+ cells, although the signal level was relatively low and the severalfold change was not enough to qualify it for a directly regulated gene by the criteria used for Table 1. Thus, it remains to be determined whether bfr is in fact a direct target for RyhB in E. coli. In Pseudomonas aeruginosa, a bacterioferritin homolog but not the major bacterioferritin was found to be regulated by small RNAs that are functional homologs of RyhB (48).
Genes positively regulated by RyhB expression.
Positive regulation of transcripts by RyhB was relatively rare; all operons in which expression increased by at least twofold in one of the three experiments are shown in Table 3. It is quite possible that all cases of positive regulation by RyhB are indirect, and we note that for a number of the genes in Table 3, the increase is not consistent under different experimental conditions.
One unexpected entry in Table 3 was ftnA. ftnA, encoding the major E. coli ferritin, was previously reported to be positively regulated by Fur and iron (20) and found to be a target of RyhB (30). Previous experiments demonstrated high-level expression of ftnA in the absence of RyhB in M63 minimal medium but not in its presence (30). However, in our arrays, ftnA mRNA was fourfold more abundant when RyhB was overproduced (Table 3). We have examined this discrepancy in more detail.
There are three major differences between previous experiments and those reported here. In the current experiments, cells were grown in LB medium rather than minimal medium, RyhB expression was manipulated independently of Fur or iron, and the effects of RyhB expression were examined after only 15 min rather than in a steady state. Positive regulation of ftnA by Fur was detectable in the arrays done with a fur+ host grown in LB, consistent with previous reports (20). In the absence of RyhB (vector columns, Table 3), ftnA mRNA expression was highest in the presence of added FeSO4 and lowest in fur mutants (showing a 2.5-fold decrease). The surprising result was the significant increase in the ftnA signal in the presence of RyhB but only in fur+ cells (Table 3). None of the other genes listed in Table 3 show a similar pattern of expression.
One explanation for this could be that ftnA expression is controlled by a Fur-repressed negative regulator that is not RyhB. Expression of RyhB could result in better Fur repression of this putative regulator by the iron-sparing mechanism (Fig. 3). In our previous experiments examining ftnA expression, cells were grown in glucose minimal medium, and ftnA transcript levels were compared in ryhB+ and ryhB mutant cells. We know that under these growth conditions, iron levels are low and thus, Fur repression is incomplete; RyhB is expressed constitutively at relatively high levels. We do not currently understand how this situation leads to an apparent negative regulation of ftnA by RyhB, but the long-term expression may modulate the intracellular levels of iron in ways that perturb ftnA regulation. Future experiments will address the mechanism of ftnA regulation.
General conclusions.
RyhB causes rapid degradation of the target mRNAs it pairs with, allowing us to use microarrays to monitor its direct effects on the cell. Ectopic expression of RyhB in the presence and absence of the Fur regulator has enabled us to define the surprisingly extensive network of genes affected by this small RNA. Because expression of RyhB was limited to 15 min, we did not expect to see long-term indirect effects of the small RNA. Nonetheless, we find that even within 15 min, there is enough decrease in the synthesis of Fe-binding proteins to lead to increased Fur repression, most probably by intracellular iron sparing. In this model (Fig. 3), the rapid cessation of synthesis of many iron-binding proteins increases intracellular levels of Fe, improving repression by Fur. Because Fur blocks the expression of genes at the promoter level, Fur-regulated mRNAs with short half-lives could show a pattern of disappearance similar to those of RyhB-regulated genes in these experiments. While the direct target sodB mRNA disappeared within 3 min after RyhB expression, the decrease in exbBD mRNA (due to increased Fur repression) was detectable by 7 min. The loss of the RyhB effect on presumed indirect targets such as the exbBD mRNA in fur mutants (Fig. 2) provided further evidence for the iron-sparing model. Thus, while showing a direct in vivo interaction between a small RNA and its putative target mRNA requires a demonstration of base pairing and an examination of kinetics of action, a knowledge of likely regulatory consequences of small RNA action can provide tools for differentiating direct from indirect effects of regulatory RNAs. In our experiments, the indirect effects of iron sparing could be seen because LB medium contains iron levels that are apparently sufficient only for partial repression of Fur-regulated genes.
The genes regulated by RyhB include some encoding proteins involved in the TCA cycle (acnA, acnB, sdhCDAB, fumA), glycolysis (pflA), oxidative stress (sodB, msrB), iron-sulfur cluster formation (isc, mrp), aerobic respiration (nuo, fdo), and anaerobic respiration (hyb, frd). Thus, the range of action of the small RNA is much wider than previously thought. Some of the proteins are expressed under aerobic (sdh, nuo, fdo) or anaerobic (hyb, pflA, fdo, frd) conditions or both (acnA, acnB, fumA, sodB). Direct interaction of RyhB with many of these messages is predicted from the existence of possible pairing regions (data not shown); these will be examined further in future work. It is very likely that RyhB expression under anaerobic conditions might uncover yet further targets.
It is apparent from the changes in expression of not only direct RyhB targets, but also members of the Fur regulon, that RyhB significantly changes intracellular iron use. How these patterns may change with longer expression of RyhB remains to be investigated. However, this study clearly demonstrates the wide-ranging effects of one small RNA and the complexity of the cellular response to changes in iron availability.
Acknowledgments
We thank N. Majdalani for pNM12 and C. Rosenow for initial microarrays not included in the analysis here. We appreciate the help of O. Aprelikova, and G. V. Chandramouli for suggestions on the microarray technique and analysis and G. Storz, D. FitzGerald, and N. Majdalani for editorial comments.
This work was supported in part by a postdoctoral fellowship of the Canadian Institute for Health Research and startup funds of the Université de Sherbrooke to E.M. E.M. is a scholar from the Fonds de la Recherche en Santé du Québec (FRSQ). C. K. Vanderpool was supported by a postdoctoral fellowship from the American Cancer Society.
REFERENCES
- 1.Aiba, H., S. Adhya, and B. de Crombrugghe. 1981. Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 256:11905-11910. [PubMed] [Google Scholar]
- 2.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 3.Angerer, A., and V. Braun. 1998. Iron regulates transcription of the Escherichia coli ferric citrate transport genes directly and through the transcription initiation proteins. Arch. Microbiol. 169:483-490. [DOI] [PubMed] [Google Scholar]
- 4.Baichoo, N., and J. D. Helmann. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 184:5826-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, V., S. Mahren, and M. Ogierman. 2003. Regulation of the FecI-type ECF sigma factor by transmembrane signalling. Curr. Opin. Microbiol. 6:173-180. [DOI] [PubMed] [Google Scholar]
- 6.Buchanan, S. K., B. S. Smith, L. Venkatramani, D. Xia, L. Esser, M. Palnitkar, R. Chakraborty, D. van der Helm, and J. Deisenhofer. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 6:56-63. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham, L., M. J. Gruer, and J. R. Guest. 1997. Transcriptional regulation of the aconitase genes (acnA and acnB) of Escherichia coli. Microbiology 143(Pt. 12):3795-3805. [DOI] [PubMed] [Google Scholar]
- 8.Eick-Helmerich, K., and V. Braun. 1989. Import of biopolymers into Escherichia coli: nucleotide sequences of the exbB and exbD genes are homologous to those of the tolQ and tolR genes, respectively. J. Bacteriol. 171:5117-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst, J. F., R. L. Bennett, and L. I. Rothfield. 1978. Constitutive expression of the iron-enterochelin and ferrichrome uptake systems in a mutant strain of Salmonella typhimurium. J. Bacteriol. 135:928-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1998. Binding of the fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J. Mol. Biol. 283:537-547. [DOI] [PubMed] [Google Scholar]
- 11.Fee, J. A. 1991. Regulation of sod genes in Escherichia coli: relevance to superoxide dismutase function. Mol. Microbiol. 5:2599-2610. [DOI] [PubMed] [Google Scholar]
- 12.Ferguson, A. D., R. Chakraborty, B. S. Smith, L. Esser, D. van der Helm, and J. Deisenhofer. 2002. Structural basis of gating by the outer membrane transporter FecA. Science 295:1715-1719. [DOI] [PubMed] [Google Scholar]
- 13.Ferguson, A. D., E. Hofmann, J. W. Coulton, K. Diederichs, and W. Welte. 1998. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science 282:2215-2220. [DOI] [PubMed] [Google Scholar]
- 14.Flint, D. H. 1996. Escherichia coli contains a protein that is homologous in function and N-terminal sequence to the protein encoded by the nifS gene of Azotobacter vinelandii and that can participate in the synthesis of the Fe-S cluster of dihydroxy-acid dehydratase. J. Biol. Chem. 271:16068-16074. [PubMed] [Google Scholar]
- 15.Geissmann, T. A., and D. Touati. 2004. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 23:396-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grass, G., and C. Rensing. 2001. Genes involved in copper homeostasis in Escherichia coli. J. Bacteriol. 183:2145-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimaud, R., B. Ezraty, J. K. Mitchell, D. Lafitte, C. Briand, P. J. Derrick, and F. Barras. 2001. Repair of oxidized proteins. Identification of a new methionine sulfoxide reductase. J. Biol. Chem. 276:48915-48920. [DOI] [PubMed] [Google Scholar]
- 18.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hantke, K. 1981. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol. Gen. Genet. 182:288-292. [DOI] [PubMed] [Google Scholar]
- 20.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 21.Hantke, K., and V. Braun. 1997. Control of bacterial iron transport by regulatory proteins, p. 11-44. In S. Silver and W. Walden (ed.), Metal ions in gene regulation. Chapman and Hall, New York, N.Y.
- 22.Hantke, K., and V. Braun. 2000. The art of keeping low and high iron concentrations in balance, p. 275-288. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 23.Higgs, P. I., P. S. Myers, and K. Postle. 1998. Interactions in the TonB-dependent energy transduction complex: ExbB and ExbD form homomultimers. J. Bacteriol. 180:6031-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones, H. M., and R. P. Gunsalus. 1987. Regulation of Escherichia coli fumarate reductase (frdABCD) operon expression by respiratory electron acceptors and the fnr gene product. J. Bacteriol. 169:3340-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kammler, M., C. Schon, and K. Hantke. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 175:6212-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larsen, R. A., M. G. Thomas, G. E. Wood, and K. Postle. 1994. Partial suppression of an Escherichia coli TonB transmembrane domain mutation (delta V17) by a missense mutation in ExbB. Mol. Microbiol. 13:627-640. [DOI] [PubMed] [Google Scholar]
- 27.Lavrrar, J. L., and M. A. McIntosh. 2003. Architecture of a Fur binding site: a comparative analysis. J. Bacteriol. 185:2194-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Litwin, C. M., and S. B. Calderwood. 1993. Role of iron in regulation of virulence genes. Clin. Microbiol. Rev. 6:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Massé, E., F. E. Escorcia, and S. Gottesman. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17:2374-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Massé, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McHugh, J. P., F. Rodriguez-Quinones, H. Abdul-Tehrani, D. A. Svistunenko, R. K. Poole, C. E. Cooper, and S. C. Andrews. 2003. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 278:29478-29486. [DOI] [PubMed] [Google Scholar]
- 32.Menon, N. K., C. Y. Chatelus, M. Dervartanian, J. C. Wendt, K. T. Shanmugam, H. D. Peck, Jr., and A. E. Przybyla. 1994. Cloning, sequencing, and mutational analysis of the hyb operon encoding Escherichia coli hydrogenase 2. J. Bacteriol. 176:4416-4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moll, I., T. Afonyushkin, O. Vytvytska, V. R. Kaberdin, and U. Blasi. 2003. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA 9:1308-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Brien, I. G., and F. Gibson. 1970. The structure of enterochelin and related 2,3-dihydroxy-N-benzoylserine conjugates from Escherichia coli. Biochim. Biophys. Acta 215:393-402. [DOI] [PubMed] [Google Scholar]
- 35.Outten, F. W., O. Djaman, and G. Storz. 2004. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol. Microbiol. 52:861-872. [DOI] [PubMed] [Google Scholar]
- 36.Outten, F. W., C. E. Outten, J. Hale, and T. V. O'Halloran. 2000. Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, cueR. J. Biol. Chem. 275:31024-31029. [DOI] [PubMed] [Google Scholar]
- 37.Quail, M. A., P. Jordan, J. M. Grogan, J. N. Butt, M. Lutz, A. J. Thomson, S. C. Andrews, and J. R. Guest. 1996. Spectroscopic and voltammetric characterisation of the bacterioferritin-associated ferredoxin of Escherichia coli. Biochem. Biophys. Res. Commun. 229:635-642. [DOI] [PubMed] [Google Scholar]
- 38.Rosenow, C., R. M. Saxena, M. Durst, and T. R. Gingeras. 12. November 2001, posting date. Prokaryotic RNA preparation methods useful for high density array analysis: comparison of two approaches. Nucleic Acids Res. 29:E112. [Online.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sargent, F., S. P. Ballantine, P. A. Rugman, T. Palmer, and D. H. Boxer. 1998. Reassignment of the gene encoding the Escherichia coli hydrogenase 2 small subunit—identification of a soluble precursor of the small subunit in a hypB mutant. Eur. J. Biochem. 255:746-754. [DOI] [PubMed] [Google Scholar]
- 40.Saurin, W., M. Hofnung, and E. Dassa. 1999. Getting in or out: early segregation between importers and exporters in the evolution of ATP-binding cassette (ABC) transporters. J. Mol. Evol. 48:22-41. [DOI] [PubMed] [Google Scholar]
- 41.Schwartz, C. J., O. Djaman, J. A. Imlay, and P. J. Kiley. 2000. The cysteine desulfurase, IscS, has a major role in in vivo Fe-S cluster formation in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:9009-9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwartz, C. J., J. L. Giel, T. Patschkowski, C. Luther, F. J. Ruzicka, H. Beinert, and P. J. Kiley. 2001. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc. Natl. Acad. Sci. USA 98:14895-14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skovran, E., and D. M. Downs. 2003. Lack of the ApbC or ApbE protein results in a defect in Fe-S cluster metabolism in Salmonella enterica serovar Typhimurium. J. Bacteriol. 185:98-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stancik, L. M., D. M. Stancik, B. Schmidt, D. M. Barnhart, Y. N. Yoncheva, and J. L. Slonczewski. 2002. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J. Bacteriol. 184:4246-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stojiljkovic, I., A. J. Baumler, and K. Hantke. 1994. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. J. Mol. Biol. 236:531-545. [DOI] [PubMed] [Google Scholar]
- 46.Takahashi, Y., and U. Tokumoto. 2002. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J. Biol. Chem. 277:28380-28383. [DOI] [PubMed] [Google Scholar]
- 47.Vecerek, B., I. Moll, T. Afonyushkin, V. Kaberdin, and U. Blasi. 2003. Interaction of the RNA chaperone Hfq with mRNAs: direct and indirect roles of Hfq in iron metabolism of Escherichia coli. Mol. Microbiol. 50:897-909. [DOI] [PubMed] [Google Scholar]
- 48.Wilderman, P. J., N. A. Sowa, D. J. FitzGerald, P. C. FitzGerald, S. Gottesman, U. A. Ochsner, and M. L. Vasil. 2004. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. USA 101:9792-9797. [DOI] [PMC free article] [PubMed] [Google Scholar]