Abstract
The catalytic core of Escherichia coli DNA polymerase III holoenzyme contains three subunits: α, ε, and θ. The α subunit contains the polymerase, and the ε subunit contains the exonucleolytic proofreading function. The small (8-kDa) θ subunit binds only to ε. Its function is not well understood, although it was shown to exert a small stabilizing effect on the ε proofreading function. In order to help elucidate its function, we undertook a determination of its solution structure. In aqueous solution, θ yielded poor-quality nuclear magnetic resonance spectra, presumably due to conformational exchange and/or protein aggregation. Based on our recently determined structure of the θ homolog from bacteriophage P1, named HOT, we constructed a homology model of θ. This model suggested that the unfavorable behavior of θ might arise from exposed hydrophobic residues, particularly toward the end of α-helix 3. In gel filtration studies, θ elutes later than expected, indicating that aggregation is potentially responsible for these problems. To address this issue, we recorded 1H-15N heteronuclear single quantum correlation (HSQC) spectra in water-alcohol mixed solvents and observed substantially improved dispersion and uniformity of peak intensities, facilitating a structural determination under these conditions. The structure of θ in 60/40 (vol/vol) water-methanol is similar to that of HOT but differs significantly from a previously reported θ structure. The new θ structure is expected to provide additional insight into its physiological role and its effect on the ε proofreading subunit.
The replication of the Escherichia coli chromosome is performed by the DNA polymerase III holoenzyme (HE). This HE is a high-efficiency synthesis system capable of copying the bacterial chromosome at a speed of 500 to 1,000 nucleotides per second. Synthesis occurs coordinately and simultaneously for both the leading and lagging DNA strands. In addition, its synthesis is highly accurate (45). The precise functioning of the HE within the context of the replication fork, including how the enzyme achieves processive synthesis in the leading strand but discontinuous synthesis in the lagging strand, is under active investigation by genetic, biochemical, and structural approaches (for a review, see reference 39). The HE contains two polymerase assemblies, one for each strand. Each core contains three subunits, α, ε, and θ. The α subunit contains the polymerase activity and is encoded by the dnaE gene. The ε subunit provides the proofreading 3′→5′ exonuclease activity and is encoded by the dnaQ gene. Relatively little is known about the 76-amino-acid θ subunit, which is encoded by the holE gene, although in complex the three subunits are bound together in the linear order α-ε-θ (27, 46). Genetically engineered bacteria that lack θ (ΔholE) are viable (46, 48). It was therefore surprising to find that E. coli bacteriophage P1 encodes a homolog of θ (HOT) but few other replicative proteins (34). Although there is apparently some evolutionary advantage to expressing a homolog of θ, the nature of this advantage is not clear.
The θ and HOT protein sequences are 53% identical and share significant homology. Considering only the structured region of HOT, the identity approaches 70% (10). Some structural information on these proteins is already available. A nuclear magnetic resonance (NMR) structure for θ was reported (28), although it was clear from this study, as well as from one of our previous studies (31), that the behavior of θ in solution is far from ideal under the experimental conditions used. We recently determined the solution structure of HOT (protein database [PDB] code 1SE7) (10) and found that the folds were sufficiently different that a DALI search (21) failed to identify a structural relationship. Based on the level of sequence identity, it is surprising to find such a significant structural difference between the two proteins. In order to obtain further insight into the basis for the structural difference, we measured residual dipolar couplings (RDC) for θ and analyzed the results in terms of two structural models: (i) the previously reported structure of θ (PDB code 1DU2) and (ii) a homology model of θ based on the structure of HOT (PDB code 1SE7). The RDC data for θ fit the homology model well (r = 0.78) but not the 1DU2 model (r = 0.02) (10). Based on this result, as well as the generally limited quality of NMR data for θ, we surmised that the reported structure of θ was probably in error (10).
After several initial attempts using high-field NMR data to resolve some of the congested regions of the θ 1H-15N HSQC spectrum were unsuccessful, we sought a variation in solution conditions that would result in improved spectra. As discussed below, we were motivated by the observation that one of the more problematic regions of θ corresponds to a group of residues located toward the end of helix α3 that are not well conserved between HOT and θ. A θ homology model based on the HOT structure predicts complete solvent exposure of several hydrophobic residues—particularly Ile57 and Leu60—in this region of the protein. In order to more effectively solvate these residues, we explored the use of several alcohols as cosolvents for θ. With these modifications, we were able to obtain significant improvements in terms of the peak dispersion and uniformity of peak intensity. Under these solution conditions, we obtained virtually complete assignments for θ, enabling a structural determination. The structure of θ reported here is similar to the fold proposed from the HOT structure, despite the significant difference between the solution conditions used for the two structural determinations. The new θ structure, along with the known structure of ε, may provide further insight into the structural determinants of the ε-θ interaction and its role in the operation of the polymerase III HE and associated proofreading activity.
MATERIALS AND METHODS
Preparation of θ sample.
The θ subunit was overexpressed and purified as previously described (10), except that the protein was labeled with 15N and 13C by growing E. coli BLR(DE3) with 13C glucose as the sole carbon source and 15N ammonium chloride as the sole nitrogen source.
Gel filtration.
Gel filtration experiments were performed with a Superdex 75 column (10 by 300 mm; Amersham) and AKTA fast protein liquid chromatography. The column was equilibrated with 50 mM sodium phosphate buffer (pH 7)-150 mM NaCl and eluted at 0.5 ml/min. The sample volume was 0.1 ml. Chymotrypsinogen A, RNase A, and aprotinin A were used as standards for molecular weight calibration.
NMR spectroscopy.
The NMR experiments were performed with Varian 500 and 800-MHz UNITY/INOVA spectrometers with a cryogenically cooled 5-mm probe at 500 MHz and a room temperature 5-mm Varian 1H{13C,15N} triple-resonance probe with actively shielded triple-axis gradients and variable-temperature capability at 800 MHz. The initial solution buffer was 10 mM NaPi (pH 6.5)-10 μM EDTA-5 mM NaN3 in either 90% H2O-10% D2O or 100% D2O. The 90% H2O-10% D2O buffer was diluted with d3-methanol to 40% methanol (vol/vol), and similarly, the 100% D2O buffer was diluted with d4-methanol to 40% methanol (vol/vol) for the final structural NMR studies. We will generally refer to these buffers at 60% water-40% methanol. Chemical shifts were referenced to an internal 2,2-dimethyl-2-silapentane-5-sulfonate (DSS) standard. The only experiment that was acquired at 800 MHz was the 13C-separated nuclear Overhauser effect spectroscopy (NOESY) in 2H-labeled solvents (43). All NMR experiments were carried out at 25°C. The NMR data were processed with NMRPipe (8), and the spectra were analyzed with NMRView (23) and the module NvAssign (29).
The sequential assignments were determined from a combined analysis of the HNCA (22, 38), HNCACB (40, 50), and CBCA(CO)NH (17, 40) experiments. The HNCA, HNCACB, CBCA(CO)NH, and HNCO experiments were acquired with the aid of Varian's BioPack experiments. The PACES program facilitated the sequential assignment of the backbone resonances (5). More than 80% of the side chain proton and carbon chemical shifts were assigned from a combined analysis of H(CCO)NH total correlated spectroscopy (TOCSY) and (H)C(CO)NH TOCSY (16, 35) experiments, which were acquired with the pulse sequences described by Gardner et al. (14). The side chain chemical shifts of residues preceding proline residues were assigned from a combined analysis of three-dimensional (3D) HCCH TOCSY (1, 25) and 3D 15N-edited NOESY (37, 53) experiments. All NOESY experiments were acquired with a mixing time of 100 ms. The HCCH TOCSY and 3D 15N-edited NOESY experiments were acquired with Varian's hcch_tocsy and gnoesyNhsqc BioPack sequences, respectively. Aromatic chemical shifts from Phe and Tyr were assigned from a combined analysis of (HB) CB(CGCD)HD, (HB)CB(CGCDCE)HE (51), and 1H-13C HSQC experiments. The remaining tryptophan and histidine aromatic resonances were assigned from a 1H-13C HSQC spectrum and NOESY data. The side chain amide chemical shifts of asparagine and glutamine residues were assigned from the 3D 15N-edited NOESY spectrum and low-intensity cross peaks in the HNCACB spectrum.
NOE cross peaks were assigned from 3D 15N- and 13C-edited NOESY-HSQC spectra acquired in 60% water and 40% CD3OH methanol (vol/vol) at 500 MHz. The 3D 15N-separated NOESY experiment is described above. The 3D 13C-edited NOESY-HSQC experiment was acquired with a mixing time of 100 ms, with the CN-NOESY-HSQC experiment (43), obtained from Lewis Kay. In this experiment, the 13C carrier frequency was set to 67.0 ppm to allow observation of NOEs to the aromatic and aliphatic protons. Additionally, the 13C-edited NOESY was acquired at 800 MHz in 60%-40% (vol/vol) D2O-CD3OD to better resolve aliphatic NOEs.
Structure calculations.
The program CNS was used to calculate initial structures (2). Initial structures, used to validate assignments and check for consistency, were calculated with the standard force fields. In all, 332 NOE cross peaks were assigned from the sample in 60:40 water-methanol, and 241 NOEs were assigned from the sample in deuterated water-methanol. There were 74 NOEs that were similarly assigned in both solutions. In all, there were 526 nonredundant NOEs assigned. The distances were calibrated for each experiment separately. In each, the median intensity was assumed to correspond to 2.7 Å and all other intensities scaled following a 1/r6 relationship (23). The bounds were all set to a lower limit of 1.8 Å, and the upper limit was the target distance plus one-eighth times the target distance squared times Å−1. This is essentially the same as the common procedure of sorting the NOEs by intensity and classifying them as strong, medium, and weak but strives for slightly more precision. The intensity of cross peaks either to or from methyl groups was divided by 2 (13). For redundant NOEs that exhibited significantly different intensities, the weaker-intensity NOE was used to calibrate the distance restraint so that the less restrictive restraint was chosen.
The following additional information was included in the structure calculations. The TALOS program (6) was used to predict 104 phi and psi dihedral restraints for 52 residues, which were determined as acceptable according to the published criteria. The carbonyl assignments from the HNCO experiment were included in this analysis (22, 26). Based on the predicted secondary structure from TALOS, 66 restraints were added, corresponding to 33 hydrogen bonds. The distances for the hydrogen bonds were taken from reference 15. As a control, the structure calculations were repeated with and without the hydrogen bond restraints and no significant differences in the fold were found, indicating that the manually determined sequential NOEs are sufficient to define the helices (see the supplemental material). RDC values measured in H2O in a previous study (10) were included where indicated. Thirty restraints were included for the structured region of the protein based on our current assignments.
For a final calculation, the ARIA program (v 1.2) was used primarily to utilize the force fields and structure calculations in explicit solvent (32, 41). It was not used to make assignments or calibrate distances. This protocol was found to improve the percentage of residues in allowed Ramachandran space and the number of side chains with common rotamers as assessed by MOLPROBITY (7). No attempt was made to designate the chirality of long side chains, but the ARIA protocols swap various rotamers to search for the lowest-energy conformer. The program calculated 100 structures and further refined the 10 best individually in an explicit water solvent. Although the data for structure determination were acquired in a mixed water-methanol solvent, the models presented here clearly benefited from the final refinement in H2O in terms of rotamer and main chain conformation based on analysis with the MOLPRO BITY program (7, 36). Structures were analyzed and visualized with PyMol (www.pymol.org) and MOLMOL (30). The 10 best structures can be accessed at the PDB, code 2AE9.
RESULTS
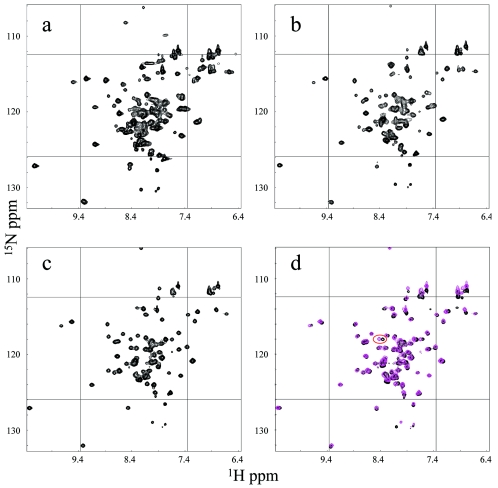
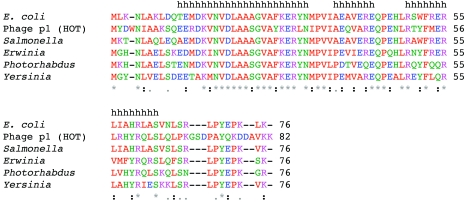
The 1H-15N HSQC spectrum of θ obtained in aqueous solution is characterized by several regions of poor resolution and broad resonances (Fig. 1a). This broadening presumably arises from conformational exchange and/or protein aggregation. One of the most problematic regions of the NMR spectra of θ corresponds to the sequence L56IAHRL61 (Fig. 2), as the resonances corresponding to these residues could not be assigned in the previous NMR study of θ (28). Further, this is also the region of lowest homology in the structured domain between θ and the phage-encoded θ homolog HOT (Fig. 2). The corresponding sequence in HOT is L57RHYRQ62. In the solution structure of HOT, the side chains of residues R58 and Q62 extend into solution, so that in the θ homology model based on this structure, the side chains of I57 and L61 are similarly extended into solution (10). We were motivated to utilize more-hydrophobic solution conditions in order to reduce the aggregation and/or conformational exchange problems resulting from these possibly solvated hydrophobic side chains.
FIG. 1.
1H-15N HSQC spectra of θ under different solution conditions. Panels: a, 100% water; b, 80% water-20% methanol; c, 60% water-40% methanol; d, overlaid spectra obtained in 60% water-40% methanol (magenta) and 65% water-35% ethanol (black). In panel d, the amide resonances of L20 are circled.
FIG. 2.
ClustalW alignment of proteins similar to θ. E. coli, Salmonella, Erwinia, Photorhabdus, and Yersinia θ proteins and the phage P1 HOT protein were aligned by ClustalW (49). The letters are colored coded as follows: blue, D and E; green, N, Q, G, H, Y, S, and T; magenta, K and R; red, M, A, I, L, V, W, F, and P. The black lowercase letters h at the top indicate helical residues in θ. An asterisk indicates identity, a colon indicates strong similarity, and a period indicates some similarity.
As discussed previously, circular dichroism (CD) spectra of θ obtained in 25% ethanol show an increased α-helical content (see, in particular, Fig. 1) (31). Analysis of the secondary-structure content with the SELCON program (24, 47) gave a calculated α-helical percentage that increased from 51.9% in the reference buffer at pH 5.5 (31) to 57.9% in 25% ethanol. This result is consistent with the above hypothesis that stabilization of helix 3, which contains the problematic residues, might be achieved by the use of a more-hydrophobic solvent system. In contrast, increasing the ionic strength by the addition of various salts or organic ions tended to reduce the α-helical content calculated on the basis of the CD data (data not shown). However, despite these relatively small quantitative changes, the CD spectra for θ obtained in 40% ethanol or methanol were very similar to those obtained in water, suggesting no major structural change in the protein.
Alcohol titration.
To assess the effect of alcohols on the NMR spectra of the protein, both d3-methanol and d5-ethanol were titrated into a solution containing [15N]-θ and the resulting 1H-15N HSQC spectra were evaluated. Figure 1a to c show the results of the methanol titration. Given the poor quality of the initial spectra, they are displayed individually instead of as overlays. At an 80% water-20% methanol ratio, the θ spectrum shows improved uniformity of peak intensities and peak dispersion (Fig. 1b). There was a significant further improvement in resonance uniformity upon increasing the methanol content to 40% (Fig. 1c). The spectra from buffers containing 65% water-35% ethanol and 60% water-40% methanol are shown overlaid in Fig. 1d. There are very few differences, the most significant being the peak that was eventually assigned to L20 (see below). In our previous structural homology model of θ (10), the side chain of L20 is solvent exposed. We interpret this result to indicate that L20 likely interacts with the alcohol cosolvent. Other noticeable differences are in the region of C-terminal residues (Fig. 1d, bottom center) and side chain NH2 resonances (Fig. 1d, top right), which are generally solvent exposed and very likely to show chemical shift changes due to changes in solvent. Given the improved spectral quality, we attempted to proceed with assignments and structure determination in 60% water-40% methanol. The choice of methanol over ethanol was simply based on methanol representing a smaller perturbation to the system compared to ethanol, which is more hydrophobic.
Gel filtration.
In order to better understand the physical basis for the effect of alcohols on the NMR spectra of θ, we examined the elution profile of θ on a size exclusion column. First, we examined the elution profile of θ injected at different concentrations. A sample of 100 μM θ eluted at 27.7 min, while a sample of 10 μM θ eluted at 29.6 min. These elution times correspond to Mrs of 16,700 and 10,800, respectively, compared to standard proteins (chymotrypsinogen A, RNase A, and aprotinin A) chromatographed under the same conditions. The sequence of θ predicts a molecular mass of 8.9 kDa. The elongated shape of the structure may account for the discrepancy in elution times and the predicted molecular weight at the lower concentration, but aggregation also may be a factor. At the higher concentration, the elution time is close to the molecular weight predicted for a θ dimer. The buffer was diluted with methanol to a level of 40%, and the experiment was repeated. Under these conditions, a sample of 100 μM θ eluted at 31.9 min while a sample of 10 μM θ eluted at 32.0 min. Thus, in the methanol-water buffer there does not appear to be a significant change in elution time due to the concentration of the protein. In addition, the elution time corresponds more closely to the molecular weight expected for a θ monomer; however, the reliability of the comparison is limited since the interaction properties of the column may be affected by the high alcohol concentration. Determining a standard molecular weight curve in 60% water-40% methanol was problematic, as these conditions denature most proteins. In conclusion, these results suggest that alcohol disrupts either a dimer or an aggregated state of θ that is concentration dependent in 100% water.
NMR assignments.
The protein spectrum was nearly completely assigned in the 60% water-40% methanol buffer, as described in Materials and Methods; there were no missing assignments from residues 10 to 76. Residues 1 to 9 appear to be exchange broadened under these conditions. In contrast, the resonances from residues 63 to 67 at the C terminus show generally more intense cross peaks, indicating that they are likely very dynamic on a fast time scale. The chemical shift assignments have been submitted to the BMRB BioMagResBank database and assigned accession number 6571.
The TALOS program was used to assess the probable secondary structure based on the similarity between the primary sequence and chemical shifts of θ and a database of known structure and chemical shifts. The program predicts three contiguous segments with torsion angles characteristic of alpha helices. The corresponding residues are 11 to 30, 37 to 43, and 49 to 65 (Fig. 2). Interestingly, G24 is included in helix α1, although glycine residues are uncommon in helices (4). This behavior may be influenced by the adjacent alanine residues, which have a strong propensity to form helices; the sequence containing this residue is A21-A22-A23-G24-V25-A26. TALOS predicts residues 64 and 65 to be helical, but the intensities of the resonances of these residues in the 1H-15N HSQC spectrum are noticeably stronger than the intensities for other residues in the helix, indicting that these residues are highly dynamic. Therefore, the boundary of the helix is probably 63 and not 65. Hydrogen bond restraints were added to the structure calculations based on the assigned secondary structure above.
NOESY analysis.
θ structural models were calculated by an iterative procedure. NOESY cross peaks were analyzed first for unambiguous assignment, and any ambiguities were first resolved by searching for symmetry-related cross peaks in the 15N and/or 13C separated NOESY. The previous homology model developed for θ (10) was occasionally used to resolve ambiguities. The NOEs that were assigned can be characterized as shown in Table 1. In total, there were 526 nonredundant NOEs from all of the data that were used in the calculation.
TABLE 1.
Structural statistics
| Parameter | Value |
|---|---|
| NOEs | |
| Amide-amide | 42 |
| Amide-aliphatic | 215 |
| Aliphatic-aliphatic | 269 |
| H bonds | 66 |
| Total | 592 |
| RDCa | |
| RMS error (Hz) | 0.84 |
| R factor | 0.08 |
| Pearson's r | 0.99 |
| Ensemble RMSD (Å)b | |
| 2° Structure (bb)e | 0.90 |
| 2° Structure (heavy) | 1.74 |
| Backbone (11-63) | 0.97 |
| Heavy atoms (11-63) | 1.95 |
| Avg violations per structurec | |
| NOEs and/or H bonds | 2.3 |
| Dihedrals | 0.4 |
| RMSD (experimental)c | |
| NOE (Å) | 0.75 |
| H bonds (Å) | 0.30 |
| Dihedral angles (°) | 0.86 |
| RMSD (covalent geometry)c | |
| Bonds (Å) | 0.005 |
| Angles (°) | 0.708 |
| Impropers (°) | 2.00 |
| Ramachandran spaced (%) | |
| Favored | 77 |
| Allowed | 99 |
Structural models.
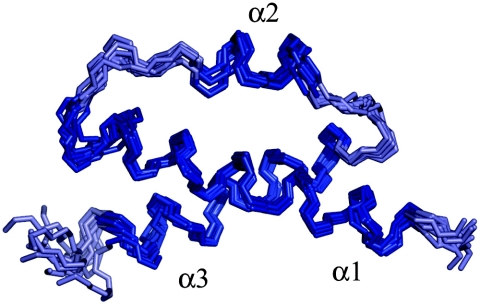
The structures of θ were calculated with the ARIA program, although the program was not used to make any assignments or calibrate distances. The ARIA protocols are able to deal with the ambiguity that results from the lack of stereochemical assignments. Additionally, ARIA provides a high-quality refinement protocol in explicit solvent. One hundred structures were calculated, and each of the 10 structures with the lowest energies were further refined. Figure 3 shows the ensemble of these 10 structures. The average pairwise alignment of residues 11 to 63 yields a 0.97-Å root mean square deviation (RMSD). The termini are largely unstructured in the models, in agreement with the lack of long-range NOEs and the strong intensity of cross peaks in the C terminus. Residues 1 to 9 were not assigned. The secondary structural elements align slightly better (0.90-Å RMSD). If helix 2 is excluded, the alignment is further improved to a 0.74-Å RMSD, indicating that helix 2 is the least well restrained. Presumably, this is due to a combination of its small size and a paucity of long-range NOEs.
FIG. 3.
Ensemble of calculated structures. The 10 lowest-energy structures of θ, based on the experimental data, are shown aligned over residues 11 to 63. The residues displayed are 9 to 68. The helices are dark blue, and the loops are light blue. The coordinates of this ensemble are available from the PDB, code 2AE9.
Table 1 shows structural statistics related to the calculated structures. The statistics indicate that while the RMSD may be slightly high, the structures are very good in quality. Indeed, looking at the lowest-energy structure, 77% of the residues are in the favored region of Ramachandran space and 99% are in the allowed region. The side chain rotamers were analyzed with the MOLPROBITY program. Only five side chains are in conformations that are uncommon in high-resolution crystal structure data. Each of these is solvent exposed, so the intensity of the NOE results in a calibrated distance that is probably shorter than it should be. The quality of the structures is also a testament to the high quality of the refinement protocols (32, 42).
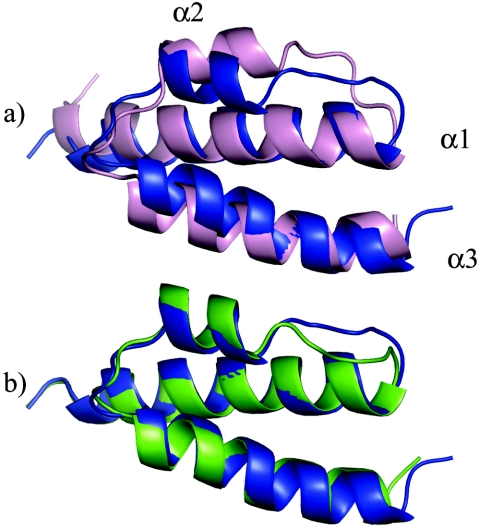
The θ structures calculated from the experimental data are very similar to the previously reported HOT structure (Fig. 4) and to the HOT-based θ homology model (see the supplemental material). For simplicity, the comparisons made here are all to the lowest-energy structure that was calculated. Figure 4 also shows that the topology and orientation of the helices are almost identical in all cases. Helices 1 and 3, which have a parallel orientation, are long and coil around each other slightly. Helix 2 is short and runs in the opposite direction of helices 1 and 3, creating the up-down-up topology. Residues 11 to 63 of θ fit the model of HOT with a 2.4-Å RMSD. Most of the differences can be accounted for by the loops and the slight difference in orientation of helix 2. When only the elements of secondary structure are aligned, the RMSD decreases to 1.8 Å. The sequence of helix 2 in θ is AEAVERE (residues 37 to 43), while in HOT it is AEQVARE (residues 38 to 44). The switched position of the neighboring alanine when comparing V40 (θ) to V41 (HOT) is responsible for the slight pivot of the helix. Also noticeable is the slight misalignment at the beginning of helix 3. This appears to be due to the substitution of Y52 in HOT for W51 in θ and the associated difference in size of the tryptophan and tyrosine side chains.
FIG. 4.
Structure alignments of θ. (a) θ protein (blue ribbon) aligned over residues 11 to 63 with HOT (pink, residues 12 to 64). (b) θ calculated with RDC restraints (blue) and without RDC restraints (green).
The addition of dipolar coupling restraints (10) only marginally affects the calculated structures or the orientation of helix 2, as shown in Fig. 4b. This indicates that helix 2 is positioned largely by NOE restraints, albeit few of them. The lowest-energy structure (residues 11 to 63) calculated without RDC restraints aligns with the corresponding lowest-energy structure by using the RDC restraints with an RMSD of 1.1 Å. Note that this is near the total ensemble RMSD of 0.97 Å. The alignment of the structures calculated with and without RDC improves slightly to a 0.97-Å RMSD when considering only elements of secondary structure. This is an important point, because the RDC restraints were measured in 100% H2O (10), with the assignments from Keniry et al. (28), while the θ structure, as described, was obtained in 60% water-40% methanol. Attempts to measure RDCs in buffers containing 40% methanol generally failed, as these conditions either precipitate f1 phage or are not conducive to bicelle formation. The structures calculated without the RDCs still fit the RDC data reasonably well; the correlation coefficient of the calculated versus experimental RDC is 0.59. Finally and importantly, this also indicates that the global fold of θ is generally not strongly affected by the presence of methanol, since the RDCs were measured in 100% H2O. As noted above, this conclusion is also consistent with the similarity of the CD spectra obtained under the two solution conditions.
DISCUSSION
The structural analysis of the θ subunit of E. coli DNA polymerase III has presented an extremely challenging NMR problem. Despite the protein's small size, our lab and others have had substantial difficulty assigning the entire protein (28, 31). Even with improved NMR hardware, we were unable to confidently assign large sections of the main chain. These limitations have also been noted by Keniry et al., who were unable to assign resonances for residues 22, 23, 27, 28, and 55 to 61 (28). Homology modeling of θ based on the recently determined structure of the θ homolog HOT suggested that one of the reasons for the exchange broadening of some of the resonances of θ, particularly in the segment from I57 to L61, resulted from solvent exposure of the hydrophobic I57 and L61 side chains. The corresponding residues in HOT are hydrophilic. In order to make the folded conformation of θ more energetically favorable, we explored the use of alcohol cosolvents and found a significant improvement in the quality of the 1H-15N HSQC spectra of θ. The spectra obtained in the mixed-solvent system were, in fact, so significantly improved that the resonance assignments became facile. The structure of θ that we derived is very different from the previously reported structure of θ (28) but is very similar to our recently reported structure of HOT, the θ homolog in bacteriophage P1 (10).
Although the structural differences between the θ fold determined here and that reported previously (28) could, in principle, arise from different solution conditions, several lines of evidence indicate that this is most likely not the case. First, the RDC constants for the assigned residues obtained in water are consistent with the present structure determined in the mixed solvent but not with the previously reported structure. Second, we do not see the dramatic changes in the 1H-15N HSQC spectra that would be expected if θ were to adopt two very different solution-dependent conformations. Third, the CD spectra also show only a small increase in α-helical content upon the addition of ethanol. The stability of the structure with variations in solvent composition is also consistent with the high melting temperature of 56°C derived from CD measurements (10).
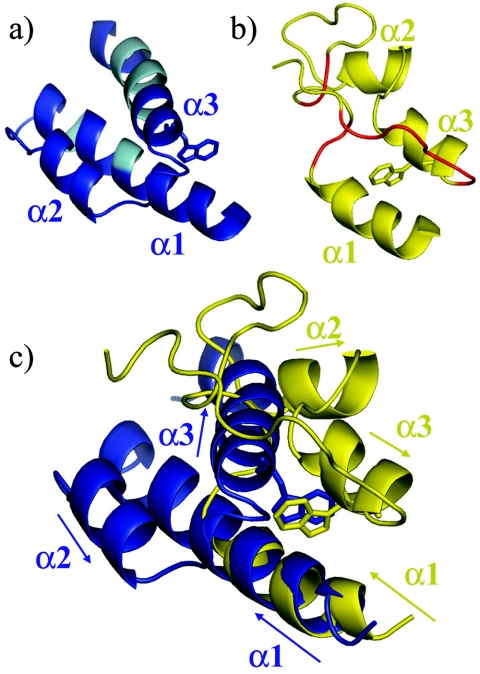
Despite the structural difference, our results are consistent with most of the previous assignments and, remarkably, with many of the previously assigned NOE interactions as well. The reported assignments and secondary structure (28) for the residues that were assigned are generally consistent with the present results. The previous study correctly identified W51 as a crucial residue for the core of the protein. Figure 5a and b compare the present and previously reported structures of θ. Figure 5c shows a superposition of the structures in Fig. 5a and b, in which residues 11 to 19 in the two structures have been aligned. Note that the W51 side chain occupies similar positions in both models and is able to interact with V18 on α1, as indicated by Keniry et al. (28). The remaining structural differences can be understood based on the absence of the critical assignments for A22, A23, and L55 to L61. Interestingly, one can imagine detaching helix 3 in the 1DU2 structure and pivoting about the W51 side chain. This would place helix 3 in approximately the correct position.
FIG. 5.
Comparisons with the previously published θ structure (1DU2). Ribbon diagrams are displayed for residues 11 to 65 of the structure determined in this work (a, blue) and the previously reported structure 1DU2 (28) (b, yellow). Residues that were unassigned in the previous study are cyan in panel a and red in panel b. The structures in panels a and b are in approximately the same orientation as in panel c. In making the overlay in panel c, only residues 11 to 19 of the two structures were aligned. The arrows indicate the direction of the primary sequence. Note that W51, shown as a stick figure, is in a similar position, although helix 3 of 1DU2 runs in the opposite direction relative to helix 2. The insufficient assignment data in the 1DU2 structure led to an antiparallel orientation of α1 and α3, compared with the roughly parallel alignment in the determined structure.
The previous paper identified two other long-range interactions which positioned helix 2 relative to helix 3: Tyr31 to Glu43 and Tyr31 to Gln44 (28). These NOEs are inconsistent with the HOT-based θ homology model (10), as well as with the structure of θ reported here. Based on our more complete resonance assignment of the 1H, 13C, and 15N shifts, we can now suggest that these NOEs were actually aromatic-aromatic NOEs. The amide proton shifts of Glu43 and Gln44 are 7.2 and 7.5 ppm, respectively, in 40% methanol, which is in the region of overlap between aromatic and aliphatic 1H-amide chemical shifts. We assigned NOEs from Tyr31 to both Phe27 and Tyr71, which have aromatic proton shifts assigned at 7.2, 7.0, and 6.8 ppm. Unfortunately, Phe27 was one of the residues that were previously unassigned, so this was likely the error that oriented helix 3 incorrectly and mistakenly positioned helix 2 far from helix 1.
Potential interactions of θ with ε.
Knowledge of the solution structure of θ, in conjunction with the reported crystal structure of ε (20), will permit us to investigate the ε-θ interaction, focusing on the role of θ in stabilizing ε and in facilitating the ε proofreading activity. Clearly, ε by itself is a rather unstable protein, undergoing significant precipitation at concentrations required for NMR studies (11, 19), while the ε-θ complex can be readily studied (9, 31, 44). NMR studies of labeled ε186 in the presence of unlabeled θ have demonstrated that the interface involves a number of hydrophobic residues (9). The recent observation that the ε-θ complex can be broken up by simple alcohols is fully consistent with the ε-θ interaction being maintained by hydrophobic forces (18). Although the ε-θ complex is significantly more stable than ε, there are no detailed structural data for the ε-θ complex. Analysis of the hydrophobic surfaces of θ may provide insight into its possible interaction with ε.
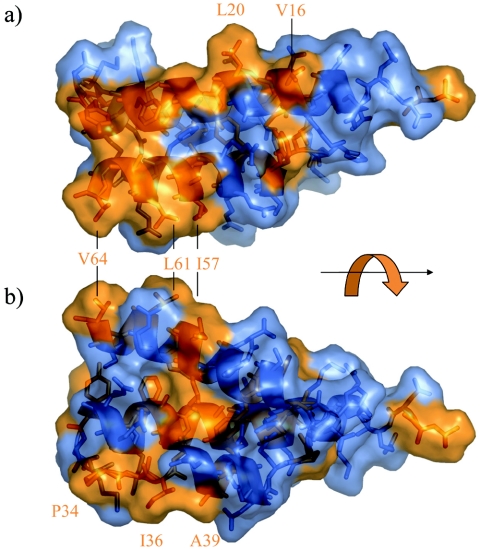
Figure 6 shows the hydrophobic surface of θ. In the semitransparent surface rendering, Gly, Ala, Val, Ile, Leu, Phe, Tyr, Trp, and Pro are colored orange and all other residues are colored blue. Several clusters of hydrophobic residues are apparent. For example P34, I36, and A39 protrude into solution in Fig. 6b. The NOE spectra show no long-range interactions for I36, consistent with this observation. Similarly, L20 and V16, displayed in Fig. 6a, show no long-range interactions. We note also that the amide shift of L20 differs significantly between the ethanol and methanol solutions (Fig. 1d). Including V16, there are five hydrophobic residues (V16, L20, P34, I36, and A39) that are both solvent exposed and identical in θ and HOT. These residues are primary candidates for interaction with ε. Keniry et al. (28) reported chemical shift changes in θ upon binding with ε for residues 21 to 27 (AAAGVAF), also suggesting that this part of helix 1 is involved with the ε interaction. Overall, these data suggest that the “open” side of helix 1, the loop between helix 1 and helix 2, and possibly the first part of helix 2 form a hydrophobic surface that may be the primary determinant for interaction with ε.
FIG. 6.
Transparent-surface renderings of θ showing exposed hydrophobic residues. Residues Gly, Ala, Val, Ile, Leu, Phe, Tyr, Trp, and Pro are orange, and all other types are blue. Panels a and b are rotated approximately 180 ° about the axis indicated by the black arrow. Disordered residues 1 to 7 and 66 to 76 are not shown.
Exposed hydrophobic residues I57 and L61, which in our model initially motivated the use of an alcohol cosolvent, are perhaps not involved in the interaction with ε since they correspond to R58 and Q62 in HOT. Thus, if these residues constitute part of the θ-ε interface, the corresponding HOT-ε interface would be unstable. Hence, although alcohols have recently been reported to promote dissociation of θ-ε (18), the interactions most probably do not involve these nonconserved residues.
Two resonances appear in the glycine region (δ15N, <110 ppm) of the 1H-15N HSQC spectrum (Fig. 1a), although there is only one glycine residue (Gly24) in θ. The intensity ratio of the two glycine peaks apparently is solvent dependent, so that the more intense resonance in water is no longer detected in 60:40 water-methanol (Fig. 1d). The dependence on solvent and the position of Gly24 on the external face of the helix suggest that it may be involved in the interaction with ε. The position of Gly24 near the center of α-helix 1 is unexpected since this residue generally functions as a helix breaker (4). However, Smith et al. have shown that glycine residues play a major role in mediating helix-helix interactions in membrane proteins (12, 33). They note that glycine has a high occurrence in membrane-spanning helices, where it facilitates helix association by acting as a molecular notch (12). Since our previous chemical shift mapping studies have indicated that helices 1 and 2 of ε interact with θ, one attractive possibility is that helices 1 and/or 2 on ε bind to helix 1 of θ with Gly24, helping to accommodate the interaction.
Genetic experiments have shown that lack of θ, while not affecting the viability of E. coli, causes a modest increase in the spontaneous mutation rate, indicating that θ plays a role in replicative fidelity, presumably by affecting the in vivo stability of the proofreading ε subunit (48). A more dramatic effect of θ is seen in strains carrying a structurally impaired ε subunit (dnaQ mutator mutants). For example, in the temperature-sensitive dnaQ49 (V96G) mutant the absence of θ leads to a more-than-1,000-fold increase in the mutation frequency at 28°C, signifying, in essence, a complete loss of proofreading capability (48). In contrast, little or no effect of θ is seen with other dnaQ mutator mutants that are characterized by a largely catalytic ε defect (48). Thus, the effects of θ may be twofold: first, gross stabilization of the overall ε fold and second, a more localized effect on ε, even when well structured, affecting the workings of the exonucleolytic site. Indeed, NMR analysis of the θ interaction surface on ε has revealed a broad area of interaction, affecting structurally and catalytically important helix 7 on ε, as well as other areas presumably important for maintaining the proper functioning of the catalytic site (9).
Interestingly, further experiments with E. coli strains in which the holE gene (encoding θ) has been replaced with the P1 hot gene (encoding HOT) have shown that HOT can readily replace θ in its function of stabilizing the unstable dnaQ49 mutant (3). In fact, HOT appears significantly more effective than θ in stabilizing this dnaQ mutant, at least paralleling the observed greater stability of HOT compared to θ in the NMR and CD experiments (10). Interestingly, HOT is not capable of substituting for θ in all dnaQ mutants (3). Thus, the precise interactions of ε with θ and HOT likely differ in certain aspects, and these differences may reflect the small deviations between the θ and HOT structures described above. It is likely that a careful analysis of the ε-θ and ε-HOT interactions at the structural level, combined with genetic analyses described above, will provide important insight into the mechanism(s) by which θ may affect the efficiency of the editing process.
Supplementary Material
Acknowledgments
We are grateful to Devon Allen for discussions related to the CD data.
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bax, A., G. M. Clore, and A. M. Gronenborn. 1990. H-1-H-1 correlation via isotropic mixing of C-13 magnetization, a new 3-dimensional approach for assigning H-1 and C-13 spectra of C-13-enriched proteins. J. Magn. Reson. 88:425-431. [Google Scholar]
- 2.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 3.Chikova, A. K., and R. M. Schaaper. 2005. Bacteriophage P1 hot gene product can substitute for the Escherichia coli DNA polymerase III θ subunit. J. Bacteriol. 187:5528-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou, P. Y., and G. D. Fasman. 1974. Prediction of protein conformation. Biochemistry 13:222-245. [DOI] [PubMed] [Google Scholar]
- 5.Coggins, B. E., and P. Zhou. 2003. PACES: Protein sequential assignment by computer-assisted exhaustive search. J. Biomol. NMR 26:93-111. [DOI] [PubMed] [Google Scholar]
- 6.Cornilescu, G., F. Delaglio, and A. Bax. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13:289-302. [DOI] [PubMed] [Google Scholar]
- 7.Davis, I. W., L. W. Murray, J. S. Richardson, and D. C. Richardson. 2004. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32:W615-W619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delaglio, F., S. Grzesiek, G. W. Vuister, G. Zhu, J. Pfeifer, and A. Bax. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277-293. [DOI] [PubMed] [Google Scholar]
- 9.DeRose, E. F., T. Darden, S. Harvey, S. Gabel, F. W. Perrino, R. M. Schaaper, and R. E. London. 2003. Elucidation of the epsilon-theta subunit interface of Escherichia coli DNA polymerase III by NMR spectroscopy. Biochemistry 42:3635-3644. [DOI] [PubMed] [Google Scholar]
- 10.DeRose, E. F., T. W. Kirby, G. A. Mueller, A. K. Chikova, R. M. Schaaper, and R. E. London. 2004. Phage like it HOT: solution structure of the bacteriophage P1-encoded HOT protein, a homolog of the theta subunit of E. coli DNA polymerase III. Structure 12:2221-2231. [DOI] [PubMed] [Google Scholar]
- 11.DeRose, E. F., D. Li, T. Darden, S. Harvey, F. W. Perrino, R. M. Schaaper, and R. E. London. 2002. Model for the catalytic domain of the proofreading epsilon subunit of Escherichia coli DNA polymerase III based on NMR structural data. Biochemistry 41:94-110. [DOI] [PubMed] [Google Scholar]
- 12.Eilers, M., A. B. Patel, W. Liu, and S. O. Smith. 2002. Comparison of helix interactions in membrane and soluble alpha-bundle proteins. Biophys. J. 82:2720-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fletcher, C. M., D. N. M. Jones, R. Diamond, and D. Neuhaus. 1996. Treatment of NOE constraints involving equivalent of nonstereoassigned protons in calculations of biomacromolecular structures. J. Biomol. NMR 8:292-310. [DOI] [PubMed] [Google Scholar]
- 14.Gardner, K. H., R. Konrat, M. K. Rosen, and L. E. Kay. 1996. An (H)C (CO)NH-TOCSY pulse scheme for sequential assignment of protonated methyl groups in otherwise deuterated N-15,C-13-labeled proteins. J. Biomol. NMR 8:351-356. [DOI] [PubMed] [Google Scholar]
- 15.Grishaev, A., and A. Bax. 2004. An empirical backbone-backbone hydrogen-bonding potential in proteins and its applications to NMR structure refinement and validation. J. Am. Chem. Soc. 126:7281-7292. [DOI] [PubMed] [Google Scholar]
- 16.Grzesiek, S., J. Anglister, and A. Bax. 1993. Correlation of backbone amide and aliphatic side-chain resonances in 13C/15N-enriched proteins by isotropic mixing of 13C magnetization. J. Magn. Reson. Ser. B. 101:114-119. [Google Scholar]
- 17.Grzesiek, S., and A. Bax. 1992. Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J. Am. Chem. Soc. 114:6291-6293. [Google Scholar]
- 18.Gupta, R., S. M. Hamdan, N. E. Dixon, M. M. Sheil, and J. L. Beck. 2004. Application of electrospray ionization mass spectrometry to study the hydrophobic interaction between the epsilon and theta subunits of DNA polymerase III. Protein Sci. 13:2878-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamdan, S., S. E. Brown, P. R. Thompson, J. Y. Yang, P. D. Carr, D. L. Ollis, G. Otting, and N. E. Dixon. 2000. Preliminary X-ray crystallographic and NMR studies on the exonuclease domain of the epsilon subunit of Escherichia coli DNA polymerase III. J. Struct. Biol. 131:164-169. [DOI] [PubMed] [Google Scholar]
- 20.Hamdan, S., P. D. Carr, S. E. Brown, D. L. Ollis, and N. E. Dixon. 2002. Structural basis for proofreading during replication of the Escherichia coli chromosome. Structure (London) 10:535-546. [DOI] [PubMed] [Google Scholar]
- 21.Holm, L., and C. Sander. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233:123-138. [DOI] [PubMed] [Google Scholar]
- 22.Ikura, M., L. E. Kay, and A. Bax. 1990. A novel approach for sequential assignment of 1H, 13C, and 15N spectra of proteins: heteronuclear triple-resonance three-dimensional NMR spectroscopy. application to calmodulin. Biochemistry 29:4659-4667. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, B. A., and R. A. Blevins. 1994. NMRVIEW: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4:603-614. [DOI] [PubMed] [Google Scholar]
- 24.Kabsch, W., and C. Sander. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577-2637. [DOI] [PubMed] [Google Scholar]
- 25.Kay, L. E., G. Y. Xu, A. U. Singer, D. R. Muhandiram, and J. D. Formankay. 1993. A gradient-enhanced HCCH TOCSY experiment for recording side-chain H-1 and C-13 correlations in H2O samples of proteins. J. Magn. Reson. Ser. B 101:333-337. [Google Scholar]
- 26.Kay, L. E., G. Y. Xu, and T. Yamazaki. 1994. Enhanced-sensitivity triple-resonance spectroscopy with minimal H2O saturation. J. Magn. Reson. Ser. A 109:129-133. [Google Scholar]
- 27.Kelman, Z., and M. O'Donnell. 1995. Structural and functional similarities of prokaryotic and eukaryotic DNA polymerase sliding clamps. Nucleic Acids Res. 23:3613-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keniry, M. A., H. A. Berthon, J. Y. Yang, C. S. Miles, and N. E. Dixon. 2000. NMR solution structure of the theta subunit of DNA polymerase III from Escherichia coli. Protein Sci. 9:721-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirby, N. I., E. F. DeRose, R. E. London, and G. A. Mueller. 2004. NvAssign: protein NMR spectral assignment with NMRView. Bioinformatics 20:1201-1203. [DOI] [PubMed] [Google Scholar]
- 30.Koradi, R., M. Billeter, and K. Wuthrich. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14:51-55. [DOI] [PubMed] [Google Scholar]
- 31.Li, D., D. L. Allen, S. Harvey, F. W. Perrino, R. M. Schaaper, and R. E. London. 1999. A preliminary CD and NMR study of the Escherichia coli DNA polymerase III theta subunit. Proteins 36:111-116. [DOI] [PubMed] [Google Scholar]
- 32.Linge, J. P., and M. Nilges. 1999. Influence of non-bonded parameters on the quality of NMR structures: a new force field for NMR structure calculation. BMJ 13:51-59. [DOI] [PubMed] [Google Scholar]
- 33.Liu, W., E. Crocker, W. Zhang, J. I. Elliott, B. Luy, H. Li, S. Aimoto, and S. O. Smith. 2005. Structural role of glycine in amyloid fibrils formed from transmembrane alpha-helices. Biochemistry 44:3591-3597. [DOI] [PubMed] [Google Scholar]
- 34.Lobocka, M. B., D. J. Rose, G. Plunkett III, M. Rusin, A. Samojedny, H. Lehnherr, M. B. Yarmolinsky, and F. R. Blattner. 2004. Genome of bacteriophage P1. J. Bacteriol. 186:7032-7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Logan, T. M., E. T. Olejniczak, R. X. Xu, and S. W. Fesik. 1993. A general method for assigning NMR spectra of denatured proteins using 3D HC (CO)NH-TOCSY triple resonance experiments. J. Biomol. NMR 3:225-231. [DOI] [PubMed] [Google Scholar]
- 36.Lovell, S. C., I. W. Davis, W. B. Arendall III, P. I. de Bakker, J. M. Word, M. G. Prisant, J. S. Richardson, and D. C. Richardson. 2003. Structure validation by Cα geometry: φ, ψ and Cβ deviation. Proteins 50:437-450. [DOI] [PubMed] [Google Scholar]
- 37.Marion, D., P. C. Driscoll, L. E. Kay, P. T. Wingfield, A. Bax, A. M. Gronenborn, and G. M. Clore. 1989. Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heteronuclear 1H-15N Hartmann-Hahn-multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy: application to interleukin 1β. Biochemistry 28:6150-6156. [DOI] [PubMed] [Google Scholar]
- 38.Matsuo, H., E. Kupce, H. Li, and G. Wagner. 1996. Increased sensitivity in HNCA and HN(CO)CA experiments by selective Cβ decoupling. J. Magn. Reson. Ser. B 113:91-96. [DOI] [PubMed] [Google Scholar]
- 39.McHenry, C. S. 2003. Chromosomal replicases as asymmetric dimers: studies of subunit arrangement and functional consequences. Mol. Microbiol. 49:1157-1165. [DOI] [PubMed] [Google Scholar]
- 40.Muhandiram, D. R., and L. E. Kay. 1994. Gradient-enhanced triple-resonance three-dimensional NMR experiments with improved sensitivity. J. Magn. Reson. Ser. B 103:203-216. [Google Scholar]
- 41.Nilges, M. 1997. Ambiguous distance data in the calculation of NMR structures. Folding Design 2:S53-S57. [DOI] [PubMed] [Google Scholar]
- 42.Nilges, M., and S. O'Donoghue. 1998. Ambiguous NOEs and automated NOE assignment. Prog. NMR Spectrosc. 32:107-139. [Google Scholar]
- 43.Pascal, S. M., R. D. Muhandiram, T. Yamazaki, J. D. Forman-Kay, and L. E. Kay. 1994. Simultaneous acquisition of 15N- and 13C-edited NOE spectra of proteins dissolved in H2O. J. Magn. Reson. Ser. B 103:197-201. [Google Scholar]
- 44.Pintacuda, G., M. A. Keniry, T. Huber, A. Y. Park, N. E. Dixon, and G. Otting. 2004. Fast structure-based assignment of 15N HSQC spectra of selectively 15N-labeled paramagnetic proteins. J. Am. Chem. Soc. 126:2963-2970. [DOI] [PubMed] [Google Scholar]
- 45.Schaaper, R. M. 1993. Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. J. Biol. Chem. 268:23762-23765. [PubMed] [Google Scholar]
- 46.Slater, S. C., M. R. Lifsics, M. O'Donnell, and R. Maurer. 1994. holE, the gene coding for the θ subunit of DNA polymerase III of Escherichia coli: characterization of a holE mutant and comparison with a dnaQ (ε-subunit) mutant. J. Bacteriol. 176:815-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sreerama, N., and R. W. Woody. 1993. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal. Biochem. 209:32-44. [DOI] [PubMed] [Google Scholar]
- 48.Taft-Benz, S. A., and R. M. Schaaper. 2004. The θ subunit of Escherichia coli DNA polymerase III: a role in stabilizing the ε proofreading subunit. J. Bacteriol. 186:2774-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wittekind, M., and L. Mueller. 1993. HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the α- and β-carbon resonances in proteins. J. Magn. Reson. Ser. B. 101:201-205. [Google Scholar]
- 51.Yamazaki, T., J. D. Forman-Kay, and L. E. Kay. 1993. Two-dimensional NMR experiments for correlating C-13-β and H-1-δ/ε chemical shifts of aromatic residues in C-13-labeled proteins via scalar couplings. J. Am. Chem. Soc. 115:11054-11055. [Google Scholar]
- 52.Yang, D., R. A. Venters, G. A. Mueller, W. Y. Choy, and L. E. Kay. 1999. TROSY-based HNCO pulse sequences for the measurement of 1HN-15N, 15N-13CO, 13CO-13CA, and 1HN-13CA dipolar couplings in 15N, 13C, 2H-labeled proteins. J. Biomol. NMR 14:333-343. [Google Scholar]
- 53.Zhang, O., L. E. Kay, J. P. Olivier, and J. D. Forman-Kay. 1994. Backbone 1H and 15N resonance assignments of the N-terminal SH3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed field gradient NMR techniques. J. Biomol. NMR 4:845-858. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.