Abstract
Citrobacter rodentium is a murine pathogen that is now widely used as an in vivo model for gastrointestinal infections due to its similarities with human enteropathogens, such as the possession of a locus for enterocyte effacement (the LEE island). We studied the lrp gene of C. rodentium and found that it encodes a product highly similar to members of the Lrp (leucine-responsive regulatory protein) family of transcriptional regulators, able to recognize leucine as an effector and to repress the expression of its own structural gene. In enterobacteria, Lrp is a global regulator of gene expression, as it controls a large variety of genes, including those coding for cell appendages and other potential virulence factors. Based on the well-established role of Lrp on the expression of pilus genes in Escherichia coli, we also studied the role of Lrp in controlling the formation of the type I pilus in C. rodentium. Type I pili, produced by the fim system, are virulence factors of uropathogens, involved in mediating bacterial adhesion to bladder epithelial cells. Yeast agglutination assays showed that Lrp is needed for type I pilus formation and real-time PCR experiments indicated that Lrp has a strong leucine-mediated effect on the expression of the fimAICDFGH operon. Mutant studies indicated that this positive action is exerted mainly through a positive control of Lrp on the phase variation mechanism that regulates fimAICDFGH expression. A quantitative analysis of its expression suggested that this operon may also be negatively regulated at the level of transcription.
Members of the Lrp (leucine-responsive regulatory protein) family of transcriptional regulators are small DNA-binding proteins, widely distributed among eubacteria and archaea and active as dimers, tetramers, octamers, or hexadecamers in such diverse organisms as Escherichia coli, Agrobacterium tumefaciens, Pseudomonas putida, Sulfolobus solfataricus, and Pyrococcus furiosus (1, 11, 13, 16, 27). Several Lrp homologues have been studied in detail and have been shown to be involved in the regulation of amino acid metabolism (1).
In enteric bacteria, Lrp appears to play more general role as a global regulator of metabolism (2, 6, 10, 17). The 3,000 dimers of Lrp estimated in E. coli cells were shown by two-dimensional electrophoresis comparison of wild-type and lrp mutant cells to affect the expression of at least 30 genes (5). More recently, a microarray analysis has expanded this view, suggesting that at least 10% of all E. coli genes are under Lrp control (24). In addition to genes involved in amino acid metabolism and transport, most of the genes expressed upon entry into the stationary growth phase belong to this expanded group (24). For some of these genes the interaction with leucine is responsible for the modulation of Lrp action, with cases in which leucine determines or potentiates and others in which it inhibits or reduces the Lrp effect. For a third class of genes, which includes the Lrp structural gene, lrp, leucine has no effect on Lrp action (25).
In pathogenic enterobacteria, Lrp controls some virulence-associated genes. Examples are genes required for conjugal transfer of the virulence plasmid of Salmonella enterica (3), the plasmid-encoded spv gene of S. enterica serovar Typhimurium (15), virulence-associated genes of Proteus mirabilis (8), and various fimbrial genes of E. coli, including the fim, sfa, daa, pap, and fan operons (14, 19).
The fim system is composed of a seven-cistron operon (fimAICDFGH), encoding the structural and regulatory components of the type I pilus, and by two independent genes, fimB and fimE, each encoding a specific recombinase needed to control by phase variation the expression of the fimAICDFGH operon (19). Depending on the orientation of FimS, a 314-bp element containing the promoter of the fimAICDFGH operon, pili are formed (phase ON) or not formed (phase OFF), with the FimB recombinase catalyzing both the ON to OFF and OFF to ON switches and FimE only catalyzing the ON to OFF switch. To allow phase variation, the two recombinases, bound to the two ends of FimS, have to physically interact with each other, which is only possible if the DNA separating them is bent (19). In addition to the fimAICDFGH promoter, the 314-bp element also contains multiple binding sites for the integration host factor (IHF) and for Lrp, two regulatory proteins known to promote DNA bending (19). Mutations in each one of three Lrp binding sites present within FimS, whose effect is to prevent Lrp binding, reduce both FimB- and FimE-dependent switching (7).
We studied the lrp gene and the regulatory role of its product on the expression of fim genes in Citrobacter rodentium, a mouse pathogen that belongs to a family of human and animal pathogens, such as the clinically significant enteropathogenic and enterohemorrhagic E. coli. C. rodentium causes transmissible colonic hyperplasia in mice by means of attaching and effacing lesions through which it colonizes the host gastrointestinal tract (12). Since enteropathogenic and enterohemorrhagic E. coli and other human enteropathogens are not able to colonize mice, C. rodentium has been extensively used as a model of human gastrointestinal pathogens in in vivo experiments and has proven useful in revealing phenotypes for proteins, including fimbrial proteins, not revealed by in vitro infection models (26).
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
C. rodentium ATCC 51459 was the parent of all recombinant strains described below. E. coli DH5α (supE44 ΔlacU169 [φ80ΔlacZM15] hsdR17 recA1) (20) was used for all cloning experiments, while the E. coli strains CV975 (ilvIH::lacZ) and CV1008 (ilvIH::lacZ lrp-35::Tn10) (18) were used for the complementation experiments. Bacteria were grown at 37°C in rich or minimal medium supplemented with thiamine (5 μg/ml), glucose (0.4%), and, when indicated, leucine (100 μg/ml). Antibiotics were 100 μg/ml ampicillin and 35 μg/ml chloramphenicol. For β-galactosidase or β-glucuronidase activity assays, overnight cultures were diluted to an optical density at 600 nm of 0.1 in the appropriate medium, grown in shaking conditions at 37°C up to the log phase (optical density at 600 nm of approximately 0.8), collected by centrifugation, and stored at −80°C until the time of assay.
DNA manipulations.
Plasmid and chromosomal DNA preparations, restriction digestions, ligation, bacterial transformation, and agarose gel electrophoresis were performed as previously described (20). DNA sequencing reactions were performed by using the T7 sequencing kit (USB corporation) with [α-35S]thio-dATP (>1,000 Ci mmol−1; MP Biomedicals). Southern blot experiments were performed according to standard procedures (20) and using as probes DNA fragments labeled by means of Ready-ToGo DNA labeling beads kit (without dCTP) (Amersham) and with [γ-32P]dCTP (MP Biomedicals).
Plasmid pAC12 was obtained by cloning a 1,228-bp amplification product resulting from a PCR performed with C. rodentium chromosomal DNA as the template and synthetic oligonucleotides A and C as the primers (Table 1) into the commercial vector pGEMT-Easy (Promega). Plasmid pAC12 was used to transform competent cells of the E. coli strain CV1008. Ampicillin-resistant clones were screened on LB agar containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and one clone, AC13, checked by nucleotide sequencing reaction, was selected for further study.
TABLE 1.
Synthetic oligonucleotides
| Gene and Name | Sequence (5′-3′)a | Positions of annealingb |
|---|---|---|
| lrp | ||
| A | GGGCCGGTCAGGTC | −732/−719 |
| B | CGACACACGGACCTACG | +427/+443 |
| C | TTAGCGCGTCTTAATAACC | +477/+495 |
| D | AGCTGGGATCCGAGG | +773/+787 |
| E | ACCAGAAGTGACGCATCC | +210/+227 |
| F | GTAGGGAATTTACCGGC | −505/−489 |
| G | GGTGAGAAAATGACGATTTG | −373/−354 |
| H | GTGTTATCTGTGTGTCGC | −252/−234 |
| I | GTCAGGCAGGAATAGGG | −41/−25 |
| L | ATGGTAGATAGCAAGAAGCG | +1/+20 |
| N | GCATGCCTTCTTGCTATCTACCAT | +1/+18 |
| O | TGGATTGTAGGGAATTTACCGGC | −512/−489 |
| P | CCGTTGCCTGACGGC | −148/−133 |
| Q | GTGAGTAAACGTCGTTATCTTACC | +1/+24 |
| X | GAAAATGACGATTTGACGCTGTTGGCAATGAATAACTGGgtgtaggctggagctgcttc | +368/−329 |
| Y | TTGCTCTGTTTGACTTCTTCCATAACGACGTAGGTCCGcatatgaatatcctcctta | +432/+469 |
| Z | CCGGTAAAGAAGTACAGGCTATGATGCTGGCCGCCCGGCATGGCgtgtaggctggagctgcttc | +23/+66 |
| W | CGAATATTACGGTGGCCGAGATAATCCTGGATAAGCCGCGTcatatgaatatcctcctta | +457/+497 |
| c1 | TTATACGCAAGGCGACAAGG (4) | |
| fim | ||
| fimA-rt1 | CGCTGACGCCACCTTCA | +516/+532 |
| fimA-rt2 | GCCCTGAGTCACTCCCTGTCT | +556/+576 |
| fimB-rt1 | CGGGCTACCGCTGGAGAT | +396/+413 |
| fimB-rt2 | GTTTGCCAAAGCGAAACCA | +441/+459 |
| fimE-rt1 | CGCCGCCTGAAAAATGG | +187/+203 |
| fimE-rt2 | AACCGCTTCCCGCTCATC | +232/+249 |
| oNs | AATAAAGAGGAAATATAAATCTGAACAAGTCA | −419/−400 |
| oNa | AACCCCTCACAGGAAGCCAT | −480/−449 |
| sig70s | TCCAGCGTAGAGTCCGAAATC | +259/+279 |
| sig70a | TGCCCATTTCGCGCATAT | +308/+325 |
| A5 | GCTGACGCCACCTTCAAAGT | +517/+536 |
| Cs | CAGGAAATGACGGTG | −12/+3 |
| C4 | GGTGATTGCCCGTGTTTTTTT | +25/+45 |
| Ds | GCCGCCGGGAACCTATCGCG | +219/+238 |
| D1 | CATTGAGGTAGATATCCACGCG | +245/+267 |
| D6 | GGTGCAGCTCAGCGTCACCC | +1583/+1602 |
| Da | GGCTGCCGCTCAGATAGA | +1626/+1634 |
| G5 | TTGTCCTGAAGTTCGAGTGG | −1/+21 |
| G4 | GATGAAATGGTTTCAATCCGG | +318/+338 |
| H3 | CGACGTTCACTTGCGGTGTA | +151/+170 |
Capital and lowercase letters indicate bases of lrp or fim DNA, respectively, and of an unpaired tail carrying a restriction site (underlined).
Position of annealing refers to the lrp or fim sequence, with the first base of the translational initiation codon as +1.
An lrp::gusA translational fusion was constructed by amplifying a 734-bp DNA fragment containing the lrp promoter and 18 nucleotides of the coding region using C. rodentium chromosomal DNA as the template primed with oligonucleotides A and N (Table 1). The PCR product was initially cloned into a pGEMT-Easy vector (Promega) and, upon digestion with SphI, transferred into pGusA, a pGEMT-Easy derivative containing the coding region of the E. coli gusA gene, in frame with the reporter gene. The obtained plasmid, pAC47, was checked by nucleotide sequencing reaction and used to transform wild-type and lrp null mutant C. rodentium (see below), yielding strains AC49 and AC52, respectively.
An entire copy of the lrp gene, released by NotI digestion from plasmid pAC12, was cloned into pAC47 previously linearized by SalI digestion. The noncohesive ends of the DNA fragments were filled by treatment with Klenow fragment (Biolabs), according to the manufacturer's instructions. The resulting plasmid, pAC61, was then used to transform the wild-type C. rodentium, generating strain AC62.
Construction of lrp and fimE null mutations.
Null C. rodentium mutations in the fimE and lrp genes were constructed by using the Datsenko and Wanner (4) method. In brief, the chloramphenicol resistance cassette (cat) of plasmid pKD3 was PCR amplified by using oligonucleotides X and Y for lrp or Z and W for fimE (Table 1). These oligonucleotides were designed with 18 and 17 bases, respectively, complementary to cat sequences at their 3′ ends, next to 40 bases complementary to regions adjacent to the lrp or fimE gene (−368 to −329 for X, +432 to +469 for Y, +23 to +66 for Z, and + 457 to +497 for W, considering the first translated nucleotide as +1). The 1,091-bp (lrp) and 1,099-bp (fimE) amplification products were used to transform C. rodentium strain EM1, generated by transformation of the wild-type strain ATCC 51459 with the low-copy-number plasmid pKD46, expressing the lambda red recombinase and carrying an ampicillin resistance gene, by electroporation. Chloramphenicol-resistant clones were then cured of the pKD46 plasmid by repeated growth cycles at 37°C in the absence of ampicillin. The presence of the cat cassette within the lrp or fimE gene was verified by PCR with oligonucleotides c1 and O (lrp) or Q (fimE) (Table 1) and Southern blot. One positive clone for each transformation, EM2 (lrp) and EM3 (fimE), was selected for further studies.
β-Galactosidase and β-glucuronidase assays.
β-Galactosidase assays were performed as previously described (18). For each strain analyzed, units of activity (nanomoles of o-nitrophenyl galactoside hydrolyzed per minute) were calculated from A420/Ve t, where Ve is the volume of permeabilized cells in ml and t is the time in minutes. Specific activity equals units per A590. β-Glucuronidase assays were performed as previously described (28). For each sample a graph of A405 (y axis) versus time in minutes (x axis) was designed; the slope S of the graph in A405 units per minute was estimated and units of activity (nanomoles of p-nitrophenyl glucuronide hydrolyzed per minute) were calculated from S/Ve × 0.02, where Ve is the volume of permeabilized cells in ml and 0.02 represents the A405 given relative to 1 nmol of product produced. Specific activity equals units per A590. Values reported here were the average of at least three independent experiments. Statistical significance was determined by Student's t test and the significance level was set at P < 0.05.
Isolation of total RNA and RT-PCR.
Total RNA was extracted using the QIAGEN mini kit (QIAGEN, Milan, Italy) using the manufacturer's instructions. Total RNAs were dissolved in 50 μl of RNase-free water containing recombinant porcine RNasin (1U/μg of total RNA; Amersham Pharmacia Biotech) and stored at −80°C until used for the reverse transcription (RT)-PCR analysis. The final concentration and quality of the RNA samples were estimated using either a spectrophotometer or by agarose gel electrophoresis with ethidium bromide staining.
Prior to RT-PCR total RNAs were treated with RNase-free DNase (1 U/μg of total RNA; Turbo DNA-free, Ambion) for 30 min at 37°C and the reaction was stopped with DNase inactivation reagent; 2 μΜ of oligonucleotide E was used to prime 4 μg of total RNAs in a final volume of 15.5 μl. The template and primer were incubated at 70°C for 10 min and then added to a reverse transcriptase reaction mixture containing 1× reaction buffer, 3 mM MgCl2, 0.5 mM each of the deoxynucleoside triphosphates, 1 U/μl recombinant porcine RNasin (1 U/μg of total RNA; Amersham Pharmacia Biotech), and 1 μl Improm II reverse transcriptase (Promega). The mixture was incubated at 25°C for 5 min, at 42°C for 60 min, and at 70° for 10 min to inactivate the enzyme.
After reverse transcription the cDNA obtained was primed with oligonucleotide pairs as indicated in Fig. 1A and 3A. For each oligonucleotide pair, a positive (chromosomal DNA as template) and two negative (no RT and no cDNA) control reactions were performed. PCR conditions were 5 min at 94°C, followed by 30 cycles of 95°C for 30 s, 52°C for 40 s, and 72°C for 1 min, concluding with extension at 72°C for 5 min. The products were separated on 1.0% agarose gels, stained with ethidium bromide, and visualized by UV transillumination.
FIG. 1.
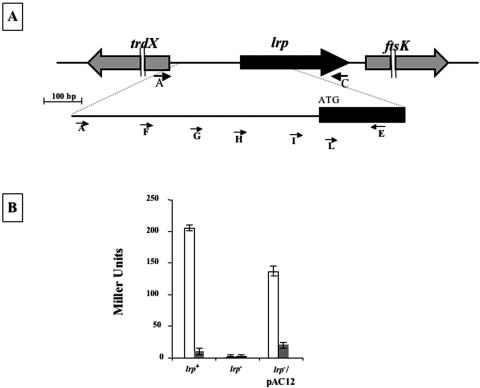
(A) Schematic representation of the trdX-lrp-ftsK region on the C. rodentium chromosome. Short arrows indicate the position of annealing of synthetic oligonucleotides. (B) β-Galactosidase assay performed with the E. coli strains CV975 (ilvIH::lacZ), CV1008 (ilvIH::lacZ lrp::Tn10), and AC13 (CV1008 carrying plasmid pAC12), indicated as lrp+, lrp−, and lrp−/pAC12, respectively. Cells were grown in minimal medium (white bars) and leucine-supplemented minimal medium (gray bars).
FIG. 3.
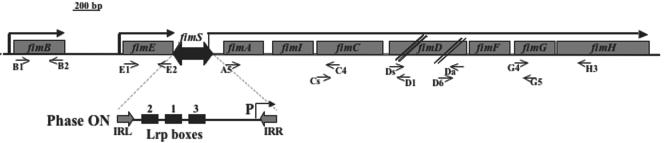
Schematic representation of the fim region on the C. rodentium chromosome. Arrows indicate the transcription orientation; short arrows indicate the position of annealing of synthetic oligonucleotides. Enlarged is the fimS element in the ON orientation. Three Lrp boxes and two inverted repeats (IRL and IRR) are also indicated. The three Lrp boxes are indicated in the order 2-1-3 in homology to the E. coli model (19).
Real-time PCR.
Real-time PCRs were performed by using chromosomal DNA or bulk cDNA samples synthesized from total RNA extracted from C. rodentium cells grown in different growth phases and media, by using SuperScript III reverse transcriptase (Invitrogen) and random hexamers as primers. Templates were always diluted 200-fold. All primers, including those for the normalizing gene, rpoD (Table 1), were designed with ABI PRISM Primer Express software (PE Applied Biosystems). Real-time PCR was performed with each specific primer pair and with cDNA or genomic DNA as the template by using Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen). Reactions were performed with an ABI PRISM 7500 sequence detection system (PE Applied Biosystems) and during the reactions, the fluorescence signal due to SYBER Green intercalation was monitored to quantify the double-stranded DNA product formed in each PCR cycle. The ΔΔCt method was used to calculate the relative amount of specific RNA present in a sample, from which the fold induction of transcription of the gene was estimated by comparison to values relative to the wild-type strain grown in minimal medium at early stationary phase.
Data were expressed as the mean ± standard error of the mean. Statistical significance was determined by Student's unpaired t test and the significance levels are reported in the text.
Primer pair efficiency was tested by looking at how ΔCt (the difference between the two Ct values of two PCRs for the same template amount) varies with template dilution, as suggested by the manufacturer's instruction guide (PE Applied Biosystems). According to the instruction guide, the efficiency of amplicons was considered 100% when plots of log template amount versus Ct originated parallel lines with slope values between −3.1 and −3.7.
Yeast agglutination experiment.
The capacity of bacteria to express an α-d-mannose binding phenotype was assayed by their ability to agglutinate Saccharomyces cerevisiae cells on glass slides, as previously reported (23); 10 μl of liquid bacterial cultures at an optical density of 595 nm of 3 and 10 μl of 3% (wt/vol) yeast cells, dissolved in phosphate-buffered saline, were gently mixed and the resulting suspension was incubated at room temperature for 2 min.
RESULTS
lrp gene of C. rodentium.
In order to study the lrp gene of C. rodentium we compared the nucleotide sequence of the Escherichia coli K-12 chromosomal region containing the lrp coding part and 500 bp upstream and downstream of it (accession number EG10547), with sequences present in the data bank of the Sanger Institute, where the C. rodentium genome sequence has recently been completed but not annotated yet (http://www.sanger.ac.uk/Projects/C_rodentium/). This search identified a C. rodentium genome region highly homologous to the lrp region of E. coli, in which lrp is located between the ftsK and trdX genes, with the latter divergently oriented with respect to the other two (Fig. 1A). A computer-assisted search indicated that the putative product of the C. rodentium lrp gene was homologous to Lrp of various enterobacteria. In particular, it was identical to Lrp of Salmonella enterica serovar Typhimurium LT2 (accession number CB025049) and very similar to Lrp of E. coli K-12 (CB035580), Shigella flexneri (CB015933), Proteus mirabilis (GL086888), and Yersinia enterocolitica (GL082961) with 99.4%, 99.4%, 97.6%, and 93.9% amino acid identity, respectively.
A 1,228-bp PCR product, obtained using synthetic oligonucleotides A and C (Table 1 and Fig. 1A) to prime for the amplification of the lrp gene from the C. rodentium chromosome, was cloned in a pGEM-T-easy (Promega) vector. The recombinant plasmid obtained, pAC12, was used to transform the E. coli strain CV1008 (ilvIH::lacZ lrp::Tn10) (18). The resulting recombinant strain, AC13, contained the lrp-controlled operon ilvIH (18) translationally fused to the lacZ reporter gene on the chromosome and the C. rodentium lrp gene on a plasmid as the only entire copy of this gene, since a Tn10 chromosomal insertion disrupted lrp of E. coli (18). The lrp gene of C. rodentium was able to complement the lrp null mutation of strain CV1008, restoring ilvIH expression to levels similar to those due to endogenous lrp (strain CV975) (Fig. 1B). In addition, like the E. coli protein (5), C. rodentium Lrp responded to the presence of leucine with a strong reduction of the transcriptional activation of the ilvIH operon (Fig. 1B).
To characterize the lrp promoter region we performed a series of RT-PCR experiments. Oligonucleotide E (Table 1, Fig. 1A) was used to prime total RNA with reverse transcriptase. The cDNA obtained was then primed with oligonucleotide pairs as indicated in Fig. 1A. Primer-pairs L-E, I-E, and H-E but not G-E, F-E, and A-E originated an amplification product of the expected size (data not shown), indicating that a DNA region up to 250 bp upstream of the translational start site is transcribed and that the transcriptional start point is in the region between oligonucleotides G and H. A similar situation has been observed for E. coli, where the lrp transcriptional start site has been mapped 267 bp upstream of the translational start site (25).
Lrp negatively controls the expression of its own structural gene.
To study the expression of the lrp gene of C. rodentium we constructed an lrp null mutant by replacing the lrp gene with a chloramphenicol resistance cassette (cat) on the C. rodentium chromosome. The low-copy-number plasmid pKD46, encoding the λ Red recombinase (4), was used to transform wild-type C. rodentium. The resulting strain, EM1, was then transformed with a 1,091-bp PCR product, containing the cat cassette flanked by 40 bp of homologous to DNA adjacent to lrp. Chloramphenicol-resistant clones were the result of a double-crossover event. Several clones were checked by PCR and Southern blot, and one clone, EM2, was selected for further analysis.
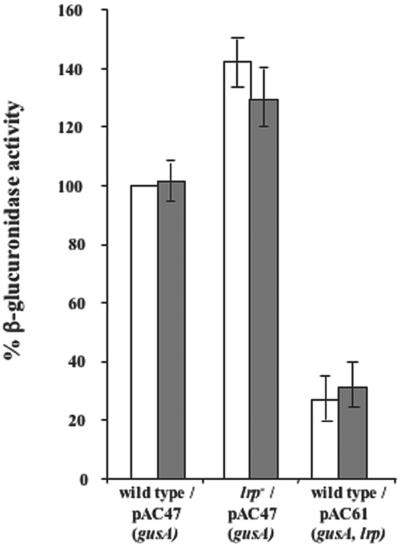
An lrp::gusA translational fusion was then obtained as follows: a 734-bp DNA fragment containing the lrp promoter region and six N-terminal codons of the lrp open reading frame was amplified from C. rodentium chromosomal DNA by using oligonucleotides E and N as primers (Table 1). The PCR product was then fused in frame to the gusA gene of E. coli carried by plasmid pGusA, yielding plasmid pAC47. Plasmid pAC47 was then introduced into the wild-type C. rodentium strain ATCC 51459 and into its isogenic lrp null mutant EM2, yielding strains AC49 and AC52, respectively. AC49 showed a β-glucuronidase activity significantly higher than that observed with ATCC 51459 cells transformed with the vector plasmid pGusA (data not shown), excluding a potential interference of endogenous enzymes in our assays. As shown in Fig. 2, lrp-directed β-glucuronidase activity was slightly higher (P < 0.05) in the lrp null mutant (AC52, white bar) than in the wild type (AC49, white bar), suggesting that Lrp autogenously repressed the expression of its own structural gene. Addition of 100 μg/ml of exogenous leucine to the growth medium did not significantly (P > 0.05) affect β-glucuronidase activity in either strain (Fig. 2, AC49 and AC52, gray bars), suggesting that the Lrp control on lrp was leucine independent.
FIG. 2.
β-Glucuronidase assay performed on C. rodentium strains AC49 (ATCC 51459, wild type carrying plasmid pAC47), AC52 (EM2, lrp null carrying plasmid pAC47), AC62 (ATCC 51459, wild type carrying plasmid pAC61), indicated as wild type/pAC47(gusA), lrp−/pAC47 (gusA), and wild type/pAC61 (gusA lrp), respectively. Cells were grown in minimal medium (white bars) and leucine-supplemented minimal medium (gray bars). The activity value obtained for strain AC49 grown in the absence of leucine was considered 100% activity. All values are the average of at least three independent experiments.
To check whether the modest regulatory effect of Lrp on its structural gene was limited by a titration of Lrp caused by the presence of Lrp target sites on the multicopy plasmid pAC47, we cloned the same DNA fragment used for the complementation experiment of Fig. 1B that contains the entire lrp gene, in plasmid pAC47 also carrying the lrp::gusA fusion, yielding plasmid pAC61. The introduction of pAC61 into a wild-type strain of C. rodentium yielded strain AC62, which showed an lrp-directed β-glucuronidase activity threefold (P < 0.05) lower than that observed with strain AC49 carrying pAC47 (Fig. 2). Since pAC47 and pAC61 showed a similar copy number in C. rodentium (not shown), the result in Fig. 2 (AC62) confirmed the negative role of Lrp in the expression of its own structural gene, indicating that the presence of pAC47 caused an Lrp titration effect in wild-type cells of C. rodentium and, as a consequence, that at least one Lrp binding site is present in pAC47.
Lrp positively controls the expression of fim genes.
In order to verify whether the C. rodentium chromosome contains genes encoding the type 1 pilus, we compared the nucleotide sequence of the fim chromosomal region of Escherichia coli K-12 (accession number U14003) with sequences present in the data bank of the Sanger Institute. This search identified a C. rodentium genome region highly homologous to the fimAICDFGH operon and to the two adjacent fimB and fimE genes of E. coli. In addition, a 314-bp region bounded by left and right inverted repeats (IRL and IRR) and identified as fimS in Fig. 3, located between the fimAICDFGH and fimE genes and containing the fimAICDFGH promoter, was also found. By homology with the same region of E. coli, we assumed that in C. rodentium fimS is involved in a phase variation mechanism controlling fimAICDFGHI expression.
To verify whether the C. rodentium fim genes are transcriptionally active, we performed a series of RT-PCR experiments priming the total RNA of C. rodentium with the synthetic oligonucleotides listed in Table 1 and graphically indicated in Fig. 3. These experiments, summarized in Table 2, indicated that fimB and fimE are transcribed but not cotranscribed (see Table 2, oligonucleotide pairs B1-B2, E1-E2, and B1-E2) and that fimAICDFGH is not cotranscribed with fimE (see Table 2, oligonucleotide pair E1-C4). Indication that fimAICDFGH is transcribed and forms a single transcriptional unit came from DNA sequence data revealing the presence of the open reading frames indicated in Fig. 3 and from RT-PCR data (see Table 2, oligonucleotide pairs A5-C4, Cs-D1, Ds-Da, D6-G5, and G4-H3). These results allowed us to conclude that the fim genes are transcribed and organized into three units of 6,596 bp (fimAICDFGH), 606 bp (fimB), and 597 bp (fimE).
TABLE 2.
RT-PCR experimentsa
| Oligonucleotide pairb | Specific amplificationc |
|---|---|
| B1-B2 | + |
| E1-E2 | + |
| B1-E2 | − |
| E1-C4 | − |
| A5-C4 | + |
| Cs-D1 | + |
| Ds-Da | + |
| D6-G5 | + |
| G4-H3 | + |
C. rodentium total RNA was used to produce cDNA as described in Materials and Methods. cDNA was then amplified with the oligonucleotide pairs here indicated.
For each oligonucleotide pair, a positive (chromosomal DNA as template) and two negative (RT and cDNA not added) control reactions were performed.
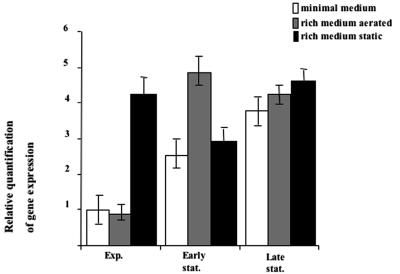
To characterize the expression of the fimAICDFGH transcriptional unit, encoding the major pilin subunit, the tip constituents, and related regulatory proteins, we performed a real-time PCR analysis (see Materials and Methods) using cDNA synthesized from total RNA extracted from C. rodentium cells at different growth phases in minimal or rich medium and in static or aerated conditions. In agreement with observations obtained with other enterobacteria (21), our real-time PCR analysis showed that in cells grown in minimal or rich medium the expression of the fimAICDFGH operon is induced upon entry into the stationary growth phase, with an induction rate (relative expression in early stationary phase/relative expression in exponential phase) of 2.6 (P < 0.05) in minimal medium and 5.4 (P < 0.05) in rich medium (Fig. 4). In addition, fimAICDFGH-specific RNA levels of exponential cells growing in rich medium were significantly higher (P < 0.05) in static conditions than in shaken conditions (Fig. 4). These levels were temporarily reduced in early stationary phase (P < 0.05) and increased again in late stationary phase (P < 0.05) (Fig. 4). The temporary decrease of fimAICDFGH expression in early stationary growth phase, although statistically significant, is difficult to interpret, and additional experiments will be needed to clarify this point.
FIG. 4.
Real-time PCR experiment performed to monitor fimAICDFGH expression in various growth conditions. Cells of the wild-type ATCC 51459 strain were grown in minimal medium (white bars), rich medium in aerated conditions (gray bars), and rich medium in static conditions (black bars) and collected during the exponential growth phase, at entry into stationary growth phase, or after 3 h of stationary growth phase. Total RNA was extracted, and cDNA was synthesized and used in the reactions with an ABI PRISM 7500 sequence detection system (PE Applied Biosystems). The fluorescence signal due to SYBR Green intercalation was monitored to quantify the double-stranded DNA product formed in each PCR cycle. The ΔΔCt method was used to calculate the relative amount of specific RNA present in each sample, and the transcriptional induction was estimated by comparison to values relative to the wild-type strain grown in minimal medium at exponential phase.
Real-time PCR experiments were performed to analyze the effects of Lrp on the expression of the three fim transcriptional units. cDNA was synthesized from total RNA extracted from wild-type and lrp null mutant cells of C. rodentium grown in minimal medium and collected at the onset of stationary phase. As reported in Table 3, fimAICDFGH expression was significantly (P < 0.005) higher in wild-type cells than in the lrp mutant, while fimB- and fimE-specific RNA levels did not differ significantly in the two strains. When wild-type cells were grown in leucine-supplemented minimal medium, fimAICDFGH and fimE expression showed a 2.2-fold (P < 0.005) and a 1.6-fold (P < 0.005) increase, respectively, with respect to transcription levels observed in the absence of leucine, while fimB expression was the same in the two growth conditions (P < 0.005) (Table 3).
TABLE 3.
Real-time PCR analysis of fim gene expression in wild-type and lrp null mutant strainsa
| Gene(s) and medium | Mean expression ± SD
|
|
|---|---|---|
| ATCC 51459 (wild type) | EM2 (lrp) | |
| fimAICDFGH | ||
| Min | 1.00 ± 0.17 | 0.060 ± 0.014 |
| Min + leu | 2.20 ± 0.20 | N.D. |
| fimB | ||
| Min | 1.00 ± 0.20 | 0.90 ± 0.10 |
| Min + leu | 1.20 ± 0.10 | N.D. |
| fimE | ||
| Min | 1.00 ± 0.15 | 1.10 ± 0.26 |
| Min + leu | 1.60 ± 0.22 | N.D. |
Cells were grown in minimal medium (Min) or minimal medium plus leucine (Min + leu) and collected at the onset of stationary growth phase. Data are presented as arithmetic mean ± standard deviation. Data indicate relative levels of transcription compared to the wild-type value in minimal growth conditions for each individual transcriptional unit. N.D., not determined.
These results suggest that Lrp is a positive regulator of fimAICDFGH expression and that its ligand, leucine, potentiates this positive effect. Lrp does not influence fimB expression, while the Lrp-leucine complex, but not Lrp alone, positively affects fimE expression. This last observation could also be explained by an increased stability of the fimE mRNA due to an effect of leucine on the phase variation mechanism, as previously proposed for E. coli (22).
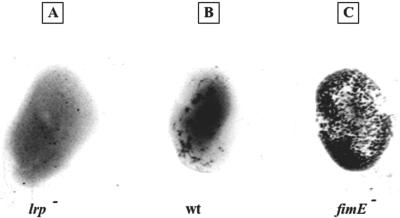
To confirm the transcriptional results in Table 3 and verify the role of Lrp in the formation of the type I pilus, we also performed a yeast agglutination assay (23). This assay is indicative of the presence of type I pili on bacterial cells since their tip is formed by FimH, an adhesin that mediates attachment to mannose-containing receptors, abundantly present on the surface of yeast cells. As shown in Fig. 5, lrp mutant cells of C. rodentium (Fig. 5A) have a reduced ability to agglutinate Saccharomyces cerevisiae cells compared with wild-type cells (Fig. 5B), as evidenced by the smaller number of bacteria-yeast aggregates. This result is in agreement with the transcriptional data summarized in Table 3 and indicate that Lrp has a positive role in the formation of the type I pilus in C. rodentium.
FIG. 5.
Yeast agglutination experiments. We mixed 10 μl of 3% (wt/vol) Saccharomyces cerevisiae yeast cells on a glass slide with the same volume of three bacterial cultures of Citrobacter rodentium EM2 (A), wild-type (wt) (B), and EM3 (C) strains at an optical density of 595 nm.
Lrp regulates fimS orientation.
In E. coli Lrp controls the fim phase variation mechanism through binding to three sites in the fimS region (19). It is possible that in C. rodentium the Lrp role in fimAICDFGH is also played through the control of the phase variation switch. To check this hypothesis, we compared by real-time PCR fimAICDFGH expression and fimS orientation. Real-time PCR experiments were performed on cDNA synthesized from total RNA and chromosomal DNA extracted from wild-type and lrp mutant cells of C. rodentium harvested at entry into the stationary growth phase. In the case of fimS, the synthetic oligonucleotides (ONs and ONa, Table 1) utilized allowed us to monitor only the phase ON orientation, since a computer-assisted search (Primer Express-Software Applied Biosystem) did not identify oligonucleotides to reliably monitor the OFF orientation.
As reported in Table 4, the number of FimS molecules in phase ON was 37-fold (P < 0.05) higher in wild-type cells than in lrp null mutant cells, indicating that Lrp controls the phase variation by favoring the ON orientation. In the same conditions, the number of fimAICDFGH mRNA molecules was 16-fold (P < 0.005) higher in the wild type than in the lrp mutant (Table 4). The different increase in the number of potentially active promoters (FimS in phase ON) and of synthesized mRNA molecules observed in the presence of Lrp suggests that fimAICDFGH expression is not constitutive but somehow negatively regulated.
TABLE 4.
Real-time PCR analysis of fimS orientation and expression of fimAICDFGHa
| Strain | Mean relative expression ± SD
|
|
|---|---|---|
| fimS-ON | fimAICDFGH | |
| ATCC 51459 (wild type) | 1.00 ± 0.25 | 1.0 ± 0.17 |
| EM2 (lrp) | 0.027 ± 0.016 | 0.06 ± 0.014 |
| EM3 (fimE) | 86.3 ± 14.3 | 50.3 ± 6.6 |
Cells were grown in minimal medium and collected at the onset of stationary growth phase. Data are presented as arithmetic means ± standard deviations.
To test this hypothesis, we constructed a fimE null mutant by the same strategy described for the construction of the lrp null mutant. Strain EM1 (see above) was transformed with a 1.099-bp PCR product, containing the cat cassette flanked by 40 bp homologous to DNA adjacent to fimE. Chloramphenicol-resistant clones were the result of a double crossover. Several clones were checked by PCR and Southern blot and one clone, EM3, was selected for further analysis.
Yeast agglutination assays performed with strain EM3 showed that in cells not expressing fimE the agglutination activity is stronger and, as a consequence, the number of type I pili on the cell surface is much higher than on wild-type cells (Fig. 5), suggesting that in C. rodentium, as in E. coli and other enterobacteria, fimE encodes the recombinase that specifically catalyzes the ON-to-OFF switch.
A real-time PCR analysis of fimAICDFGH expression was performed on chromosomal DNA and on cDNA synthesized from total RNA extracted from cells of strain EM3 grown in minimal medium. As reported in Table 4, in EM3 the number of FimS molecules in phase ON was 86-fold (P < 0.05) higher than in wild-type cells, while the number of fimAICDFGH mRNA molecules was 50-fold (P < 0.05) higher than in the wild type. Consistent with data derived from the lrp mutant, the results obtained with strain EM3 (fimE) indicate a different increase in the number of DNA molecules in phase ON and of mRNA molecules synthesized, suggesting that fimAICDFGH expression is not constitutive and is instead regulated by a dual control at the levels of both phase variation and transcription.
Although statistically significant, the difference between the number of fimS DNA and fimAICDFGH mRNA molecules shown in Table 4 could be affected by a different efficiency of the oligonucleotides used to prime the real time PCR amplification. To check the efficiency of our primer pairs, wild-type cells were used to extract chromosomal DNA and total RNA and cDNA were synthesized from the latter. DNA and cDNA were then serially diluted in a range including the dilutions applied in the experiments of Table 4 and used as templates in real-time PCR experiments. Reactions were primed with oligonucleotide pairs ONs and ONa (amplifying fimS in the ON orientation; Table 1), fimA-rt1 and fimA-rt2 (amplifying a coding region of fimA; Table 1), and sig70s and sig70a (amplifying a coding region of the normalizing gene rpoD; Table 1). Parallel straight lines with similar slopes (−3.70 for ONs-ONa; −3.64 for fimA-rt1-fimA-rt2; and −3.35 and −3.55 for sig70s-sig70a amplifying rpoD chromosomal DNA and cDNA, respectively) were obtained with the template dilutions used for all oligonucleotide pairs (data not shown). The slope values were in the range considered optimal (Materials and Methods) and indicated that ΔCt did not vary in different PCR conditions and therefore all oligonucleotide pairs tested were working with the same efficiency.
DISCUSSION
In enterobacteria, members of the Lrp family of transcriptional factors are global regulators controlling genes involved in amino acid metabolism and transport and in the synthesis of various type of cell appendages. In this work we show that the mouse pathogen C. rodentium encodes a structural and functional homologue of Lrp able to complement the Lrp role in the leucine-mediated control of the ilvIH operon in E. coli. The construction of a C. rodentium lrp null mutant allowed us to show that Lrp negatively controls the expression of its own structural gene and to analyze the effect of Lrp on the formation of the type I pilus.
Type I pili, encoded by the fim gene system, are the most common virulence factors of uropathogenic bacteria and allow bacterial adhesion to oligosaccharides containing mannose (21). The adhesive properties of the type I pilus are determined by FimH, a lectin-like protein associated with the fimbrial tip and encoded by the last gene of the fimAICDFGH operon. The type I pilus has recently been identified as a virulence factor of the invasive pathogen Citrobacter freundii (9). However, although they belong to the same genus, C. freundii and C. rodentium have fim genes with different chromosomal organization and low DNA homology. The fim genes of C. freundii are instead similar to those of Salmonella enterica serovar Typhimurium (9) and, as shown here, the C. rodentium ones are instead similar to those of E. coli.
Yeast agglutination assays indicated that lrp null mutant cells of C. rodentium are strongly impaired in the ability to agglutinate yeast cells, showing a positive effect of Lrp on pilus formation. A real-time PCR approach allowed us to observe that the expression of the fimAICDFGH operon, but not that of fimB and fimE, was strongly enhanced by the presence of Lrp, suggesting that the positive role of Lrp on pilus formation is exerted through the control of fimAICDFGH expression. When the Lrp ligand, leucine, is present the positive Lrp effect on fimAICDFGH is enhanced further.
The positive Lrp control of fimAICDFGH expression might either influence the phase variation mechanism by favoring the ON orientation or enhance transcription of the operon. We observed that Lrp strongly favors the ON orientation of the fimS switch and that this effect is not mediated by an action of Lrp on the expression of the fimB or fimE gene, encoding the specific FimS recombinases. Therefore, we propose that the Lrp action on the phase variation mechanism is most probably due to the DNA-bending activity of Lrp (25), which allows the physical interaction of the recombinases bound to the two ends of FimS, as previously proposed for E. coli (19). However, fimAICDFGH transcription does not appear to be constitutive but rather negatively controlled. Whether this transcriptional level of control is also dependent on Lrp remains to be clarified.
Our real time PCR experiments, performed in growth conditions in which fimAICDFGH is highly expressed (Fig. 4), suggested that to an increased number of FimS molecules in the ON orientation did not correspond to an equal increase in the number of fimAICDFGH-specific mRNA molecules. The statistically significant difference between the increase in DNA molecules ready to be transcribed and of fimAICDFGH-specific mRNA molecules induced us to conclude that not all molecules in the ON orientation were transcribed, as expected from an unregulated, constitutive promoter and, as a consequence, that the fimAICDFGH promoter is somehow negatively regulated.
Consistent with this conclusion is the analysis of a mutant that does not produce the recombinase that specifically catalyzes the ON-to-OFF switch. In this mutant the number of FimS molecules in the ON orientation is 86-fold higher than in the isogenic wild type, while the number of fimAICDFGH-specific molecules is only 50-fold higher in the mutant. Taken together, these statistically significant differences suggest that, in addition to the Lrp-mediated control of the fim switch, fimAICDFGH expression is also negatively controlled by a transcriptional mechanism.
Since C. rodentium causes a transmissible colonic hyperplasia in mice similar to that induced in humans by enteropathogenic and enterohemorrhagic E. coli strains (12), it has recently been selected as an in vivo model system to study human infections (26). The characterization in this model organism of Lrp, a transcriptional regulator that affects a large variety of genes including those coding for potential virulence factors, opens a new field of investigation in C. rodentium pathogenesis. The construction of an lrp null mutant will allow the identification of Lrp-controlled virulence factors and a better understanding of the role in the infection process of Lrp and the virulence factors that it controls.
Acknowledgments
We thank S. Lucchini for critical reading of the manuscript, M. R. Oggioni for suggestions on the real-time PCR analysis, and L. Di Iorio for technical assistance.
This work was supported by European Union grant QLK5-CT-2001-01729 to E.R.
REFERENCES
- 1.Brinkman, A. B., T. J. Ettema, W. M. de Vos, and J. van der Oost. 2003. The Lrp family of transcriptional regulators. Mol. Microbiol. 48:287-294. [DOI] [PubMed] [Google Scholar]
- 2.Calvo, J. M., and R. G. Matthews. 1994. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 58:466-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camacho, E. M., and J. Casadesus. 2002. Conjugal transfer of the virulence plasmid of Salmonella enterica is regulated by the leucine-responsive regulatory protein and DNA adenine methylation. Mol. Microbiol. 44:1589-1598. [DOI] [PubMed] [Google Scholar]
- 4.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ernsting, B. R., M. R. Atkinson, A. J. Ninfa, and R. G. Matthews. 1992. Characterization of the regulon controlled by the leucine-responsive regulatory protein in Escherichia coli. J. Bacteriol. 174:1109-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedberg, D., M. Midkiff, and J. M. Calvo. 2001. Global versus local regulatory roles for Lrp-related proteins: Haemophilus influenzae as a case study. J. Bacteriol. 183:4004-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gally, D. L., T. J. Rucker, and I. C. Blomfield. 1994. The leucine-responsive regulatory protein binds to the fim switch to control phase variation of type 1 fimbrial expression in Escherichia coli K-12. J. Bacteriol. 176:5665-5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hay, N. A., D. J. Tipper, D. Gygi, and C. Hughes. 1997. A nonswarming mutant of Proteus mirabilis lacks the Lrp global transcriptional regulator. J. Bacteriol. 179:4741-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hess, P., A. Altenhofer, S. A. Khan, N. Daryab, K. S. Kim, J. Hacker, and T. A. Oelschlaeger. 2004. A Salmonella fim homologue in Citrobacter freundii mediates invasion in vitro and crossing of the blood-barrier in the rat pup model. Infect. Immun. 72:5298-5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hung, S. P., P. Baldi, and G. W. Hatfield. 2002. Global gene expression profiling in Escherichia coli K12. The effects of leucine-responsive regulatory protein. J. Biol. Chem. 277:40309-40323. [DOI] [PubMed] [Google Scholar]
- 11.Jafri, S., S. Evoy, K. Cho, H. G. Craighead, and S. C. Winans. 1999. An Lrp-type transcriptional regulator from Agrobacterium tumefaciens condenses more than 100 nucleotides of DNA into globular nucleoprotein complexes. J. Mol. Biol. 288:811-824. [DOI] [PubMed] [Google Scholar]
- 12.Luperchio, S. A., and D. B. Schauer. 2001. Molecular pathogenesis of Citrobacter rodentium and transmissible murine colonic hyperplasia. Microbes Infect. 3:333-340. [DOI] [PubMed] [Google Scholar]
- 13.Madhusudhan, K. T., N. Huang, and J. R. Sokatch. 1995. Characterization of BkdR DNA binding in the expression of the bkd operon of Pseudomonas putida. J. Bacteriol. 177:636-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahan, M. J., D. M. Heithoff, R. L. Sinsheimer, and D. A. Low. 2000. Assessment of bacterial pathogenesis by analysis of gene expression in the host. Annu. Rev. Genet. 34:139-164. [DOI] [PubMed] [Google Scholar]
- 15.Marshall, D. G., B. J. Sheehan, and C. J. Dorman. 1999. A role for the leucine-responsive regulatory protein and integration host factor in the regulation of the Salmonella plasmid virulence (spv) locus in Salmonella typhimurium. Mol. Microbiol. 34:134-145. [DOI] [PubMed] [Google Scholar]
- 16.Napoli, A., J. van der Oost, C. W. Sensen, R. L. Charlebois, M. Rossi, and M. Ciaramella. 1999. An Lrp-like protein of the hyperthermophilic archaeon Sulfolobus solfataricus which binds to its own promoter. J. Bacteriol. 181:1474-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newman, E. B., and R. Lin. 1995. Leucine-responsive regulatory protein: a global regulator of gene expression in E. coli. Annu. Rev. Microbiol. 49:747-775. [DOI] [PubMed] [Google Scholar]
- 18.Platko, J. V., D. A. Willins, and J. M. Calvo. 1990. The ilvIH operon of Escherichia coli is positively regulated. J. Bacteriol. 172:4563-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roesch, P. L., and I. C. Blomfield. 1998. Leucine alters the interaction of the leucine-responsive regulatory protein (Lrp) with the fim switch to stimulate site-specific recombination in Escherichia coli. Mol. Microbiol. 27:751-761. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N. Y.
- 21.Schwan, W. R., H. S. Seifert, and J. L. Duncan. 1992. Growth conditions mediate differential transcription of fim genes involved in phase variation of type I pili. J. Bacteriol. 174:2367-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sohanpal, B. K., H. D. Kulasekara, A. Bonnen, and I. C. Blomfield. 2001. Orientational control of fimE expression in Escherichia coli Mol. Microbiol. 42:483-494. [DOI] [PubMed] [Google Scholar]
- 23.Stentebjerg-Olsen, B., T. Chakraborty, and P. Klemm. 2000. FimE-catalyzed off-to-on inversion of the type I fimbrial phase variation and insertion sequence recruitment in an Escherichia coli K-12 fimB strain. FEMS Microbiol. Lett. 182:319-325. [DOI] [PubMed] [Google Scholar]
- 24.Tani, T. H., A. Khodursky, R. M. Blumenthal, P. O. Brown, and R. G. Matthews. 2002. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, Q., J. Wu, D. Friedberg, J. Plakto, J. Calvo. 1994. Regulation of the Escherichia coli lrp gene. J. Bacteriol. 176:1831-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiles, S., S. Clare, J. Harker, A. Huett, D. Young, G. Dougan, and G. Frankel. 2004. Organ specificity, colonization and clearance dynamics in vivo following oral challenges with the murine pathogen Citrobacter rodentium. Cell Microbiol. 6:963-972. [DOI] [PubMed] [Google Scholar]
- 27.Willins, D. A., C. W. Ryan, J. V. Platko, and J. M. Calvo. 1991. Characterization of Lrp, and Escherichia coli regulatory protein that mediates a global response to leucine. J. Biol. Chem. 266:10768-10774. [PubMed] [Google Scholar]
- 28.Wilson, K. J., R. A. Jefferson, and S.G. Hughes. 1992. The Escherichia coli gus operon: induction and expression of the gus operon in E. coli and the occurrence and use of GUS in other bacteria, p. 7-23. In S. R. Gallagher (ed.), GUS protocols: using the GUS gene as a reporter of gene expression. Academic Press, Inc., San Diego, Calif.