Abstract
Cryoelectron microscope tomography (cryoEM) and a fluorescence loss in photobleaching (FLIP) assay were used to characterize progression of the terminal stages of Caulobacter crescentus cell division. Tomographic cryoEM images of the cell division site show separate constrictive processes closing first the inner membrane (IM) and then the outer membrane (OM) in a manner distinctly different from that of septum-forming bacteria. FLIP experiments had previously shown cytoplasmic compartmentalization (when cytoplasmic proteins can no longer diffuse between the two nascent progeny cell compartments) occurring 18 min before daughter cell separation in a 135-min cell cycle so the two constrictive processes are separated in both time and space. In the very latest stages of both IM and OM constriction, short membrane tether structures are observed. The smallest observed prefission tethers were 60 nm in diameter for both the inner and outer membranes. Here, we also used FLIP experiments to show that both membrane-bound and periplasmic fluorescent proteins diffuse freely through the FtsZ ring during most of the constriction procession.
The early stages of bacterial cytokinesis, involving septal ring assembly and constriction, are better understood than the terminal stages. In this work we investigate the late stages of cytokinesis in the gram-negative bacterium Caulobacter crescentus. We use fluorescence loss in photobleaching (FLIP) and cryoelectron microscope tomography (cryoEM) to determine (i) the geometry of the inner and outer membranes at various stages of cell division and (ii) whether the FtsZ ring and associated cell division machinery hinder diffusion of membrane-bound or periplasmic molecules through the division plane in predivisional cells.
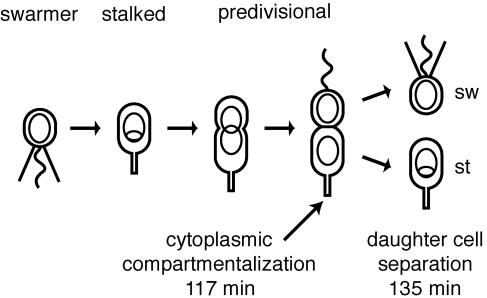
Caulobacter crescentus divides asymmetrically, producing two distinct daughter cells: the swarmer cell and the stalked cell (Fig. 1). These cells differ in their transcription programs, protein composition, and behavior (3, 20, 28, 32). The daughter cells also differ in size; the nascent swarmer-cell compartment is about two-thirds as long as the nascent stalked-cell compartment. After cell division, swarmer cells undergo an initial motile phase, and then they differentiate into stalked cells; they lose their flagellum and pili, and DNA replication is initiated. The compartmentalization of the predivisional cell is an important step in the process of creating daughter cells with different cell fates. Immediately after cytoplasmic compartmentalization, the complement of cytoplasmic proteins in the two compartments can begin to diverge, launching the two daughter cells on different developmental paths. The physical compartmentalization of the Caulobacter cytoplasm well before cell division triggers a phosphotransport-based switch mechanism that initiates the differential regulation of development in the nascent daughter cells (19, 21).
FIG. 1.
Schematic of C. crescentus cell cycle. The motile swarmer (sw) cell has a polar flagellum (wavy line) and several pili (straight lines) and does not replicate its DNA (nonreplicating DNA represented as a ring). During differentiation into a stalked (st) cell, the flagellum is shed, the pili are retracted, a stalk is built at the same pole, and DNA replication initiates (theta structure). The stalked cell becomes a predivisional cell when it begins to constrict. Cell division yields a swarmer cell and a stalked cell.
Many proteins involved in cytokinesis are known in Escherichia coli and Bacillus subtilis (1, 17, 35) and to a lesser extent in Caulobacter crescentus (17, 18, 26, 27, 29). In each of these species, the widely conserved tubulin-like FtsZ protein initiates cell division by polymerizing into a ring at the future division site. After initial formation of the ring, additional protein components of the division apparatus are recruited into the ring at various times during progression to cell division. The details of the function and protein composition of the FtsZ ring in late stages of bacterial cytokinesis are both less understood than earlier stages and more variable across different species (13). In Caulobacter, the FtsZ ring forms at the division plane about 90 min into a 180-min cell cycle; about 30 min later, constriction of the cell is visible by light microscopy. The time from first visible constriction to cell separation is about 60 min (27).
Conceptual models of the FtsZ ring depict the ring located close to the inner side of the inner membrane and attached to the inner membrane in some manner to effect the inward-directed force that constricts the cell at the division plane (for example, see Fig. 2 of reference 1). In E. coli, both FtsA and ZipA function to attach FtsZ to the inner membrane (23). In mutant E. coli strains that form a short partial spiral, rather than a circumferential ring, of FtsZ, sharp indentation in the cell envelope is observed along the spiral arc in scanning electron microscopy (EM) images (2). This observation shows that the constrictive force is a local phenomenon along segments of the FtsZ ring, rather than being the consequence of constriction by a “purse string” structure. So, although details of the physical association of the ring and the inner membrane are unknown, the connection is sufficiently strong and continuous in E. coli cells that the ring can pull the cell envelope sharply inward along the length of the ring. The E. coli FtsZ proteins are located at the leading edge of the septal invagination (4). One can easily imagine that the FtsZ ring might be a physical obstacle to diffusion for membrane-bound proteins, effectively partitioning the inner membrane surface into two compartments after formation of the ring. This possibility motivated our FLIP assay to examine the diffusion of fluorescently tagged membrane-bound proteins through the division plane. We observed that membrane-bound and periplasmic proteins diffused freely past the FtsZ ring throughout most of the cell constriction process. A FLIP assay was previously used to show that the Caulobacter crescentus cytoplasm is compartmentalized about 18 min before cell separation in a 135-min cell cycle (15).
FIG. 2.
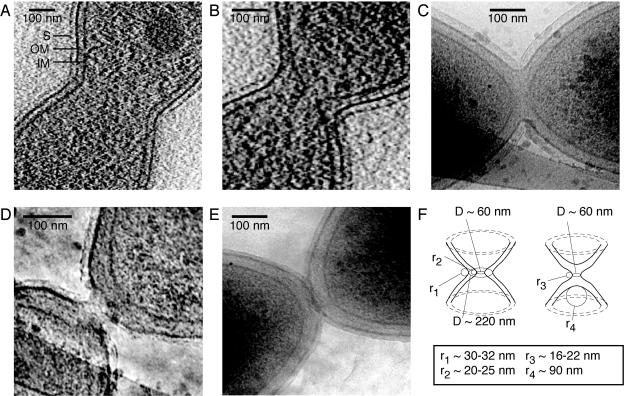
CryoEM images of dividing Caulobacter cells. (A) Midcell slice of a tomogram of a predivisional cell showing rounded invagination forming at the division site. The inner (IM) and outer (OM) membranes are clearly visible, surrounded by the fainter image of the S layer (S). (B) Slice of a tomogram of a late-predivisional cell showing widening of the inner membrane-outer membrane spacing in the region of the constriction. The cytoplasm of the two nascent daughter cells is connected by a small tubular region. (C) Transmission EM image of a cell after fission of the inner membrane and additional constriction of the outer membrane. The continuity of the periplasmic space surrounding the newly separated cytoplasmic compartments is evident. (D) Transmission EM image of cell nearing fission of the outer membrane and cell separation. Restricted periplasmic diffusion may still be possible at this stage. (E) Divided cells. No division “scar” is visible at the poles of these cells. Note that we are assuming here that these two cells in close polar proximity are newly divided sibling cells, but we cannot be absolutely sure that they are not two older cells caught in close proximity at the instant of freezing. (F) Drawing illustrating the minimum observed constrictions and radii of curvature of the inner and outer membranes. The left drawing shows a cell with a highly constricted inner membrane and a less constricted outer membrane, as in panel B. r1 and r2 are the radii of curvature of the outer and inner membranes in this cell, respectively. The right drawing shows a cell in which the inner membrane has already separated and the outer membrane is highly constricted, as in panel D. r3 is the radius of curvature of the outer membrane in this cell. r4 is the radius of curvature of the separated inner membrane in the same cell.
We also used high-resolution (∼6 nm resolution) cryoEM tomography to image Caulobacter cells at successive stages of cell division. It is well known that some gram-negative bacterial species, including C. crescentus, divide by a constrictive process that “pinches” the cell in two, while others, including E. coli, form a septum (24). The high-resolution images provided here, together with the previous FLIP studies (15), show how the late stages of constriction and fission of the Caulobacter inner and outer membrane are distinct events occurring one after the other, distinctively separated in time and space.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Strain LS4026 is strain PV2398 with the plasmid pEJ178. PV2398 is strain CB15N with the pilA gene fused to the N terminus of enhanced green fluorescent protein (EGFP) under the control of the pilA promoter, integrated into the chromosome at the pilA locus (Patrick Viollier, unpublished). Plasmid pEJ178 is pJS14 based and contains the tdimer2 gene (8) under the control of the xylose promoter, with the xylose promoter in the same orientation as the lac promoter.
Strain LS4029 is strain CB15N with plasmids pEJ178 and pEJ204. Plasmid pEJ204 is pMR10 based and contains the CC2909 gene fused to the EGFP gene under the control of the xylose promoter, with the xylose promoter in the opposite orientation from the lac promoter. CC2909 is a histidine kinase of unknown function with two predicted transmembrane domains.
Strain LS4032 is strain LS3008 with the plasmid pEJ216. LS3008 is strain CB15N, with the EGFP gene under the control of the xylose promoter, integrated into the chromosome at the xylose locus. Plasmid pEJ216 is pJS14 based, with the signal sequence of torA (9, 30, 34) from E. coli fused to the N terminus of the tdimer2 gene, under the control of the xylose promoter, with the xylose promoter in the same orientation as the lac promoter.
Strain LS2677, used as a control in the fractionation experiments, is CB15N with the pJS14 plasmid.
Bacterial growth media.
Caulobacter crescentus strains were grown in PYE complex medium (12) or M2G minimal medium at 28°C. M2G was made as previously described (12) but using 8.7 g/liter Na2HPO4, 5.3 g/liter KH2PO4, 0.2% glucose, and 0.5 mM MgSO4 in place of 0.5 mM MgCl2. Media were supplemented with kanamycin (5 μg/ml) or chloramphenicol (1 μg/ml) as necessary. Transcription from the xylose promoter was induced by adding 0.3% xylose to the growth media. Cells were grown overnight in PYE. They were then washed and diluted in M2G and incubated until the A660 reached ∼0.2. Cells were then diluted to an A660 of ∼0.05 in M2GX (M2G plus 0.3% xylose) to induce transcription from the xylose promoter. Strain LS4032 was induced for 3 to 6 h prior to imaging. Strains LS4026 and LS4029 were induced for at least 8 h prior to imaging. During induction, cultures were diluted as necessary to keep the A660 at <0.2.
An aliquot of these cells was harvested and spread on a pad of 1% agarose (Sigma, A-0169) in M2GX medium, mounted on a 25-mm by 75-mm glass slide. The slide was sealed with valap (1:1:1 vaseline:lanolin:paraffin). The cells grew and divided on the slides, and data collection lasted no more than 1 h on each slide.
Cell fractionation.
Separation of C. crescentus cell lysates into soluble and membrane fractions was performed as previously described (9), except cells were grown until the A600 reached 0.3 to 0.4, and incubation with lysozyme lasted for 5 min at room temperature and 5 min on ice.
Periplasmic proteins were isolated using the same spheroplast formation method as previously described (31), except that EDTA and lysozyme were added to final concentrations of 1 mM and 10 μg/ml, respectively.
Fluorescence microscopy and photobleaching.
Fluorescence microscopy was performed on a Nikon Diaphot 200 inverted microscope with a Nikon PlanApo ×100 objective with a numerical aperture of 1.4. Images were recorded with a Princeton Instruments MicroMax charge-coupled device (CCD) camera (NTE/CCD-512-EBFT.GR-1). A Novalux Protera frequency-doubled semiconductor laser with a wavelength of 488 nm was used for illumination. A lens of focal length 30 cm (KPX112 AR.14; Newport) was placed 30 cm from the back focal plane of the microscope objective on a mount that allowed the lens to be moved in and out of the beam path. With the lens in the path, the laser formed a Gaussian spot with a full-width-half-maximum size of 9 μm. Without the lens, the full-width-half-maximum size was 0.4 μm. The compartmentalization assay was performed as follows: a cell, immobilized on an agarose pad, was positioned with one end at the focal point of the laser. The cell was imaged in wide field mode with the lens in the beam path. The lens was removed from the path, and the cell was bleached with the focused beam. The lens was put back into the beam, and the cell was imaged again. The exposure time for each wide field image was 1 s.
To image both the tdimer2 and the EGFP signals simultaneously, we used two Q570LP dichroic mirrors (Chroma Technology Corp.) to separate the two colors and direct them to different regions of the CCD. To reduce cross talk, we used an HQ525/50 M bandpass filter (Chroma) (strains LS4026, LS4029, and LS4032) or a 535DF55 bandpass filter (Omega Optical Inc.) (strain LS4032) in the EGFP beam path and an E580LPM filter (Chroma) in the tdimer2 beam path. The dichroic mirror in the microscope (to separate the excitation and emission light) was a Q495LP (Chroma).
For experiments on strain LS4026, the laser power was 100 μW entering the microscope. The laser was attenuated using a neutral density filter with an optical density (OD) of 1 during the bleaching pulse. The bleaching pulse was 30 s long, followed by a 20-s pause before the second image was taken.
For experiments on strain LS4029, the laser power was 200 μW entering the microscope. The laser was attenuated using a neutral density filter with an OD of 1.5 during the bleaching pulse. The bleaching pulse was 240 s long, followed by a pause of approximately 20 s while the microscope was refocused, before the second image was taken.
For experiments on strain LS4032, the laser power was 200 μW entering the microscope. The laser was attenuated using a neutral density filter with an OD of 1 during the bleaching pulse. The bleaching pulse was 5 s long, followed by a 2-s pause before the second image was taken.
Fluorescence image analysis.
Image analysis was performed as previously described, using the Matlab image processing toolbox (15). For compartmentalization assay images, the average fluorescence intensity in the cell compartment distal to the focused laser spot was computed before and after the bleaching pulse for both the red and the green channels. The percent changes in average intensity (ΔId_red, ΔId_green) were calculated. Images from the control experiment, in which the bleaching laser was focused outside of the cell, were analyzed by computing the average intensity of the whole cell before and after “bleaching” (ΔIred, ΔIgreen). The percent decrease in fluorescence intensity of these control cells after the “bleaching” pulse was calculated. The decrease in fluorescence signal in these control cells is due to nonspecific bleaching by the tails of the laser spot and by the wide field illumination used for imaging.
For strain LS4026, the ΔIred value ranged from 8% to 54%, and the ΔIgreen value ranged from 2% to 39%. LS4026 cells for which the ΔId_red value was <55% were said to have a compartmentalized cytoplasm. If the ΔId_red value was >55%, the cytoplasm was deemed not compartmentalized. Cells for which the ΔId_green value was <40% were said to have a compartmentalized inner membrane. If the ΔId_green value was >40%, the inner membrane was deemed not compartmentalized.
For strain LS4032, the ΔIred value ranged from 13% to 43% and the ΔIgreen value ranged from 4% to 42%. LS4032 cells for which the ΔId_red value was <50% were said to have a compartmentalized periplasm. If the ΔId_red value was >50%, the periplasm was not compartmentalized. Cells for which the ΔId_green value was <50% were said to have a compartmentalized cytoplasm. If the ΔId_green value was >50%, the cytoplasm was not compartmentalized.
Cryoelectron microscopy.
Caulobacter crescentus cells were grown in liquid PYE media at 30°C for six to eight hours until reaching an A610 between 0.4 and 0.7. Cells were synchronized as described previously (14) and resuspended in M2G to a final A610 between 0.4 and 0.5. The synchronized cells were grown for 120 min at 30°C. The cell cycle was approximately 135 min long under these growth conditions. Aliquots of 5 μl were then taken directly from the culture and placed onto glow-discharged lacey carbon grids (Ted Pella 01881). The grids were manually blotted, plunged into liquid ethane, and then stored in liquid nitrogen. Enough water was left so that cell cross-section distortion by water surface tension was minimized.
All images were acquired in a JEOL-3100-FEF electron microscope with a field emission gun electron source operating at 300 kV. The instrument was equipped with an energy filter, a CCD camera (2,048 × 2,048 pixels) (Gatan 795), and a cryotransfer stage. Cells were imaged at 80 K using liquid nitrogen cooling.
Tomographic tilt series were acquired under low-dose conditions, typically over an angular range between +65° and −65°, ±5° with increments of 1.5° or 2°. Between 70 and 90 images were recorded for each series. All data sets were acquired using DigitalMicrograph (Gatan). Two-dimensional projections of Caulobacter cells at different stages of the cell division cycle were acquired using doses between 1,600 electrons per nm2 (e−/nm2) and 5,000 e−/nm2 per image.
All images were recorded using a magnification of 32K (nominal value of 25K with postcolumn magnification of ×1.4), giving a pixel size of 1 nm at the specimen, underfocus of 6.4 μm ± 0.5 um, and energy filter widths of 32 eV to 50 eV. The maximum dose used per complete tilt series was 8,000 e−/nm2, with typical values of approximately 7,000 e−/nm2. A total of 12 tomographic data sets were acquired.
The work flow of the EM imaging was as follows. A preliminary image was taken with a short exposure and examined to determine which cells to image within the radiation exposure limit for a particular sample. About 250 cells were imaged; this comprised 10 to 15% of the several thousand cells in all the samples that were inspected. Cells were selected for imaging based on (i) the need to get examples of different stages, (ii) quality of preservation, and (iii) whether they were obscured by the carbon support structure or debris.
The samples for the cryoEM images were drawn from synchronized cell populations at a time estimated to be near the time of cell division. Only a small percentage of cells had the small residual constrictions (which we call “tethers”) in the inner membrane (IM) or outer membrane (OM) (See Results, below). We imaged every cell exhibiting the small tethers.
The IMOD package (16) was used for image processing and tomographic reconstructions by back projection. The program ImageJ (NIH [http://rsb.info.nih.gov/ij/]) was used for analysis of the two-dimensional image projections. Volume rendering, surface rendering, and image analysis used the packages AVS Express (AVS Advanced Visual Systems [http://www.avs.com/index_wf.html]) and VolView (KitWare).
RESULTS
Electron microscope images of the dividing cell.
Three-dimensional images of whole Caulobacter cells at various points in the late stages of Caulobacter cytokinesis were collected using cryoEM tomography. Figure 2 shows a series of cryoEM images at successive stages of Caulobacter cell division. Additional images are in the supplemental material. Fig. 2A and B are slices from three-dimensional tomographic images. Figures 2C, D, and E are two-dimensional transmission cryoEM images. Initial stages of Caulobacter cytokinesis involve an extended pinching of the cell (Fig. 2A). Away from the constriction, the distance between the inner and outer membranes is constant at approximately 30 nm. Even in the higher-quality source images, we are unable to reliably identify the murein sacculus layer in the region near the division plane, so we were not able to determine how it associates with either the inner or outer membrane in the late stages of division. In later stages of division (Fig. 2B and 3A), constriction of the inner membrane proceeds ahead of outer membrane constriction, and the intermembrane spacing at the constriction site increases. This may involve dissolution of intermembrane structural components to enable separation of the membranes. The constriction of the inner membrane continues until the inner membranes of the two nascent cell compartments are connected only by a small tubular section (Fig. 2B and 3A). The three-dimensional cryoEM tomographic data facilitate viewing this small passage connecting the two cytoplasmic compartments. Figure 3A shows a volume-rendered view of the cell with the inner and outer membranes segmented for clarity. A dynamic three-dimensional view of the cell is available in the supplemental material. Next, the inner membrane parts, and the cytoplasm is separated into two compartments (Fig. 2C), while the outer membrane remains continuous for a while. After the fission of the inner membrane, constriction of the outer membrane continues (Fig. 2D) until there is only a small connection between the two daughter cells. Finally, the outer membrane parts completely, and cytokinesis is complete (Fig. 2E).
FIG. 3.
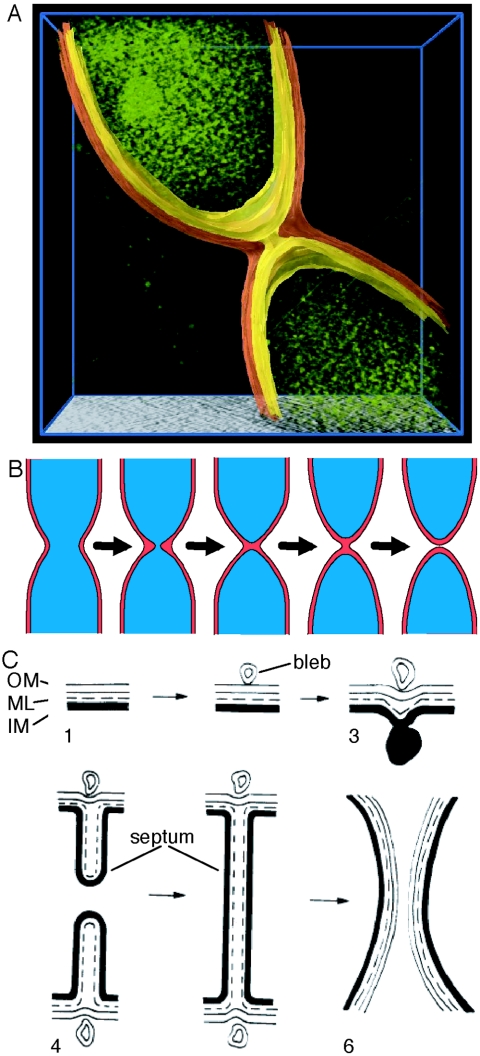
(A) Three-dimensional rendering of a dividing cell just before inner membrane fission, constructed from the tomographic image data of the cell shown in Fig. 2B. The cytoplasm is shown in green, the inner membrane in yellow, and the outer membrane in orange. The 60-nm-diameter channel connecting the cytoplasm of the nascent daughter cells is clearly visible. The spots within the cytoplasm are localized sites of high absorption that may be ribosomes. (B) Schematic in consistent scale showing stages of cell division corresponding to Fig. 2A to E. Red, periplasm; blue, cytoplasm. (C) Schematic of the progression of Escherichia coli cell division (from Fig. 32 of the work of Burdett and Murray, 1974 [6]). OM, outer membrane; ML, murein layer; IM, inner membrane.
From several thousand of these cells examined, we observed only 5 IM tethers with diameters less than ∼100 nm and 15 OM tethers with diameters less than ∼300 nm. Thus, the small constrictions were present in only 20 cases or so out of several thousand cells that were examined. The combination of the small fraction of the tethers observed and the bias of the cell sample toward late stages of cell division implies that the cells spend only a small amount of time in these transitional stages, perhaps only a few seconds, and that the few cases we observed were frozen in place while undergoing fairly rapid changes in membrane topology.
An introduction to the structures of the E. coli bacterial cell envelope and the machinery that produces and divides it is given by Nanninga (22). While the biophysics of the cell envelope and the tensions, stresses, and pressures that determine its shape are beyond the scope of this paper, we note the potential of the quantitative high-resolution whole-cell cross-sections available from cryoEM tomograms to provide data sets for detailed analysis. While surveying this literature, we were struck by the similarity of the shapes of the small tubular terminal connections we observed between the dividing IM and OM and the fluid-membrane tethers described in reference 25. This similarity motivated us to call these connections “tethers.” To constrain future biophysical models of membrane fission, we measured the smallest observed constriction width and radius of curvature of the inner and outer membranes (Fig. 2F). We observed five IM tethers with diameter less than ∼100 nm. The smallest inner membrane connection we observed (Fig. 2B) was 60 nm in diameter, and the radii of curvature of the inner membrane at the constriction were 20 nm and 25 nm (two values in the same cell because the cell was slightly bent). The radius of curvature of the inner membrane at the cell pole after fission (Fig. 2C) was approximately 90 nm. We observed 15 OM tethers with diameters less than ∼300 nm. The smallest outer membrane connection we observed (Fig. 2D) was 60 nm in diameter, and the radii of curvature of the outer membrane at that constriction were 22 nm and 16 nm.
FLIP assay with fluorescently tagged periplasmic and inner membrane proteins.
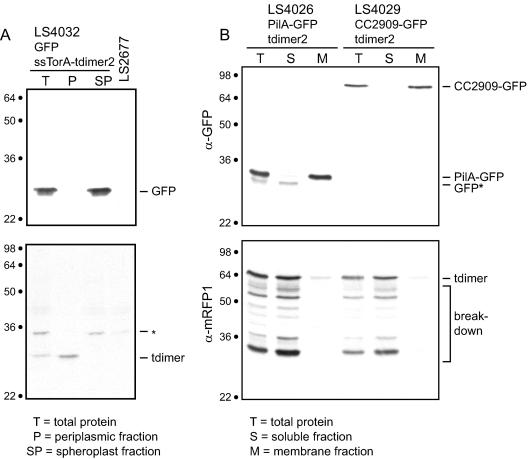
To determine if the FtsZ ring impeded diffusion of membrane-bound proteins and to compare the timing of the compartmentalization of the cytoplasm and the periplasm, we used the FLIP compartmentalization assay on strain LS4032, expressing both cytoplasmic EGFP and periplasmic tdimer2. tdimer2 was fused to the signal sequence of the E. coli TorA protein (ssTorA) to cause it to be exported to the periplasm. Western blots of fractionated cells (Fig. 4A) show that ssTorA-tdimer2 is in the Caulobacter periplasm and that soluble EGFP remains in the spheroplast fraction. We performed immunoblot analysis on spheroplast and periplasmic fractions from LS4032 cells (Fig. 4A). Probing with anti-green fluorescent protein (GFP) antibody revealed that EGFP was in the spheroplast fraction, while probing with anti-tdimer2 antibody showed that tdimer2 was in the periplasmic fraction.
FIG. 4.
(A) ssTorA-tdimer2 is in the periplasmic fraction of Caulobacter cells. LS4032 cells were separated into periplasmic and spheroplast fractions as described in Materials and Methods. The total protein is also shown. These fractions were probed with an anti-GFP antibody (top panel) and with an anti-tdimer2 antibody (bottom panel). The tdimer2 protein is two copies of the dimeric red fluorescent protein dimer2 (8) fused with a polypeptide linker. There is a band in the periplasmic fraction the size of the dimer2 protein, indicating that the ssTorA-tdimer2 protein is exported to the periplasm of Caulobacter cells and then cleaved into two parts. LS2677 is a control strain containing no fluorescent proteins. The asterisk indicates a nonspecific band. (B) PilA-EGFP and CC2909-GFP are in the membrane fraction of Caulobacter cells. Cells containing pilA-EGFP (LS4026) and CC2909-EGFP (LS4029) were separated into membrane and soluble fractions as described in Materials and Methods. The total protein is also shown. These fractions were probed with an anti-GFP antibody (top panel) and with an anti-tdimer2 antibody (bottom panel). In strain LS4026, PilA-GFP is visible in the membrane fraction. A weaker and slightly smaller band (GFP*), presumably a breakdown product of PilA-GFP, is seen in the cytoplasmic fraction. Numbers on the left of each blot show approximate molecular masses in kDa.
For the FLIP compartmentalization assay (15), the red (ssTorA-tdimer2) and green (EGFP) fluorescence signals were imaged in single cells. The cells were then bleached with a laser focused at one end of the cell (Fig. 5, left panels), and a second fluorescence image was taken of the bleached cells (Fig. 5, center panels). Cells that appeared compartmentalized in the second image were imaged again 10 min after bleaching (Fig. 5B, C, and D, right panels). We refer to the end of the cell that received the laser pulse as the proximal end and the other end of the cell as the distal end. In a cell that is not compartmentalized, proteins initially located throughout the cell will diffuse through the focused laser beam during the bleaching pulse and be photobleached. This cell will appear dark in the second fluorescence image, since all the fluorescent proteins in the cell have then been bleached. In a compartmentalized cell, proteins in the distal portion of the cell cannot diffuse into the proximal portion of the cell, and therefore they do not get bleached by the focused laser. In the compartmentalized cell, the distal portion of the cell will remain fluorescent in the second fluorescence image. Some decrease in fluorescence signal will occur in all cells due to nonspecific bleaching by the tails of the laser spot and by the wide field illumination used for imaging.
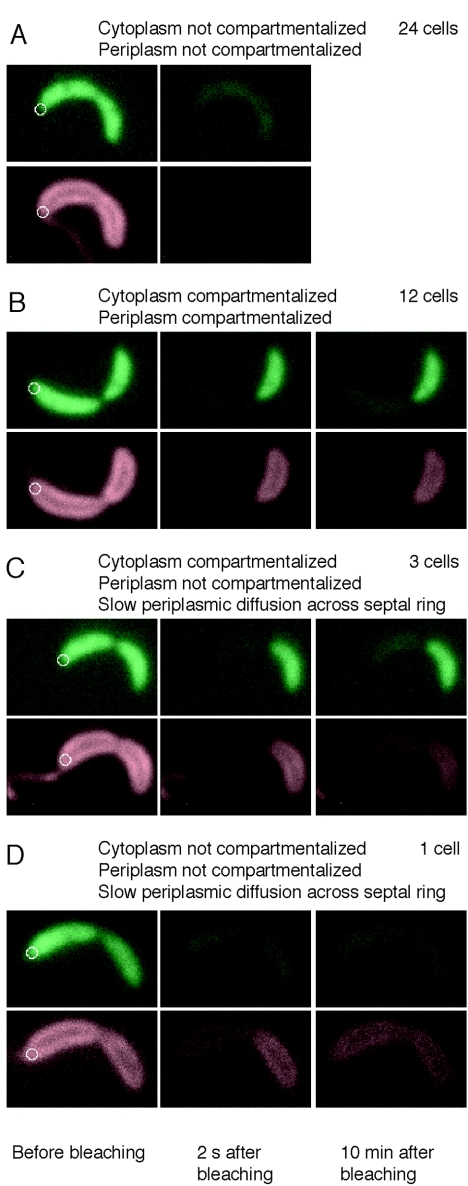
FIG. 5.
Compartmentalization of the periplasm. Four representative FLIP experiments using strain LS4032, containing cytoplasmic EGFP and periplasmic ssTorA-tdimer2. Images are false colored; green represents the fluorescence signal from EGFP, and red represents the florescence signal from ssTorA-tdimer2. (A) A LS4032 cell before (left panels) and 2 s after (second-from-left panels) photobleaching. The cellwide loss of fluorescence in both channels indicates that neither the cytoplasm nor the periplasm is compartmentalized. (B) The same experiment performed on another cell. The right panel shows an image of the cell taken 10 min after photobleaching. In this case, retention of fluorescence in both channels in the portion of the cell distal to the focused laser beam shows that both the membrane and the cytoplasm of this cell are compartmentalized. (C) In this cell, the cytoplasm is compartmentalized but the periplasm is not, based on the picture taken 10 min after bleaching. (D) The cytoplasm of this cell is not compartmentalized. The periplasm appears to be compartmentalized in the image taken 2 s after bleaching. However, the image taken 10 min after bleaching shows that ssTorA-tdimer2 has diffused from the distal portion of the cell into the proximal portion, indicating that the periplasm is not fully compartmentalized. White circles: focused laser used for bleaching. The diameter of the circles is 0.4 μm, the full-width half-maximum size of the laser beam.
By performing this FLIP assay on cells in which the cytoplasm and the periplasm were labeled with differently colored fluorescent proteins, we determined the compartmentalization state of both the periplasm and the cytoplasm in individual cells. LS4032 cells were bleached for 5 s, and the second image was taken 2 s after the end of the bleaching pulse. After bleaching, a cell in which the cytoplasm is not yet compartmentalized will display no EGFP fluorescence signal, and a cell in which the periplasm is not yet compartmentalized will display no tdimer2 signal. A cell in which the cytoplasm is compartmentalized will retain an EGFP signal in the distal compartment, and a cell in which the periplasm is compartmentalized will retain a tdimer2 signal in the distal compartment. We performed this two-color assay on late predivisional cells (identified as cells that appeared pinched in the light microscope). Of the 40 LS4032 cells treated with the FLIP assay, 24 had neither the cytoplasm nor the periplasm compartmentalized (Fig. 5A), while 12 cells had compartmentalized cytoplasm and periplasm (Fig. 5B). In the 24 cells that were not compartmentalized, the labeled periplasmic protein was able to diffuse across the constriction site. Therefore, we conclude that the FtsZ ring, present at the constriction site, did not block the diffusion of periplasmic proteins in these cells.
In three cells, both the cytoplasm and the periplasm appeared compartmentalized 2 s after bleaching, but the red fluorescence signal had spread to the proximal part of the cell in the third image, taken after 10 min, indicating that the periplasm was not fully compartmentalized (Fig. 5C). One cell had red fluorescence, but not green fluorescence, remaining in the distal portion of the cell 2 s after bleaching. However, when this cell was imaged again after 10 min, the red fluorescence signal was again present in the proximal part of the cell (Fig. 5D), indicating that slow periplasmic diffusion was still occurring between the two cell compartments and that the periplasm was not fully compartmentalized.
We interpret results from these four cells as suggestive that at some point in the progression of the division process, diffusion in the periplasm past the constriction site is slowed but not completely stopped. The four examples that we observed are clear evidence that this slowing of diffusion can occur. Since the fraction of cells exhibiting this effect was small, we also believe that the slowing of diffusion is associated with a short time period in the cell cycle. Three of the four cases we observed showed more inhibition of cytoplasmic than periplasmic diffusion between the nascent compartments. The number of examples is too small to conclude with statistical confidence from the FLIP assay alone that the cytoplasm is compartmentalized before the periplasm in the manner that the geometry of the late-stage dividing cells seen in the cryoEM images suggests would occur.
FLIP assay with fluorescently tagged membrane-bound protein and cytoplasmic protein.
To compare the timing of the compartmentalization of the cytoplasm and the inner membrane and to determine if the FtsZ ring blocks diffusion of inner membrane proteins, we performed a FLIP compartmentalization assay similar to the one described above on two strains, LS4026 and LS4029, each containing cytoplasmic tdimer2 (8) and EGFP fused to an inner membrane-bound protein. Strain LS4026 contains cytoplasmic tdimer2 and membrane-bound pilA-EGFP. PilA is a pilus subunit that is predicted to have a single transmembrane domain (33). Strain LS4029 contains cytoplasmic tdimer2 and membrane-bound CC2909-EGFP. CC2909 is a histidine kinase of unknown function with two predicted transmembrane domains. Western blots of fractionated cells (Fig. 4B) show that both PilA-EGFP and CC2909-EGFP were found in the membrane fraction and that tdimer2 remains in the cytoplasm of Caulobacter cells. We performed immunoblot analysis on soluble and membrane fractions from LS4026 and LS4029 cells (Fig. 4A). Probing with anti-GFP antibody revealed that PilA-GFP and CC2909-GFP were in the membrane fraction, while probing with anti-tdimer2 antibody showed that tdimer2 was in the soluble fraction.
All of the 50 LS4026 cells observed with the FLIP assay had either both the cytoplasm and inner membrane compartmentalized or neither the cytoplasm nor the inner membrane compartmentalized (images not shown). This result suggests that the inner membrane and the cytoplasm become compartmentalized at the same time. Twenty-eight of the 30 LS4029 cells had either both the cytoplasm and inner membrane compartmentalized or neither the cytoplasm nor the inner membrane compartmentalized. However, two of the LS4029 cells had the inner membrane compartmentalized but not the cytoplasm (images not shown). This result suggests that the inner membrane could be compartmentalized before the cytoplasm. There may be a barrier related to the constriction apparatus that forms shortly before the compartmentalization of the cytoplasm. Perhaps diffusion of CC2909-EGFP (an 88-kDa protein) past the division site is more inhibited than that of the smaller PilA-EGFP (a 33-kDa protein). It also possible that these results arise from the differences in the physical geometry of the dividing cells combined with the much slower diffusion in the membrane compared to the cytoplasm. The cryoEM images described above show that at one point in the division process, cells have a highly constricted, but not completely closed, inner membrane. In the two LS4029 cells in which the membrane appeared to be compartmentalized but not the cytoplasm, it may be that the membrane was not fully compartmentalized but that diffusion in the membrane past the division site was slowed due to the constriction of the membrane. If the diffusion past the division site was slowed enough, the membrane would appear compartmentalized in our assay. These results, however, are suggestive rather than definitive because of the limited number of cases observed.
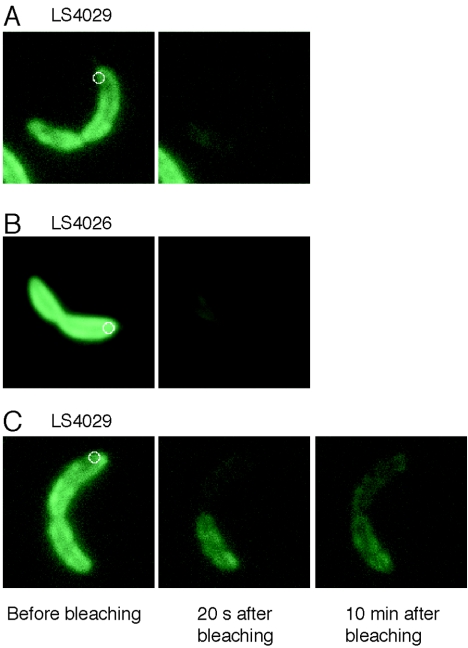
In both strains LS4026 and LS4029, we observed diffusion of inner membrane proteins past the constriction site in about half of the predivisional cells observed (Fig. 6A and B). The FtsZ ring is present at the division site prior to the start of constriction, so we conclude that the FtsZ ring alone does not block diffusion of inner membrane proteins over most of the cell division process. However, six of the LS4029 cells assayed showed green fluorescence, but no red fluorescence, in the distal half of the cell 20 s after bleaching, but when imaged again 10 min after bleaching, the remaining green fluorescence had spread from the distal half of the cell into the proximal half (Fig. 6C). We cannot determine the precise stage of cell constriction with the light microscope, but some small fraction of the cells in the observed sample would be nearing inner membrane separation. Again, we interpret the observation of a small fraction of the cells showing slow diffusion of inner membrane-bound proteins across the constriction site as evidence that diffusion is slowed near the time of cell division, consistent with the cryoEM images that show a highly constricted inner membrane (Fig. 2B and 3A).
FIG. 6.
FLIP experiments on strains LS4026, containing PilA-EGFP (in the inner membrane) and cytoplasmic tdimer2, and LS4029, containing CC2909-EGFP (in the inner membrane) and cytoplasmic tdimer2. Images are false colored; green represents the fluorescence signal from CC2909-EGFP or PilA-EGFP. (A) shows a fluorescence image of a LS4029 cell before (left panels) and 20 seconds after (second-from-left panels) photobleaching. The cellwide loss of fluorescence indicates that CC2909-EGFP can diffuse past the constriction site. (B) The same experiment performed on a LS4026 cell. The cellwide loss of fluorescence indicates that PilA-EGFP can diffuse past the constriction site. (C) The same experiment performed on another LS4029 cell. The membrane appears to be compartmentalized in the image taken 20 s after bleaching. However, the image taken 10 min after bleaching shows that CC2909-EGFP has diffused from the distal portion of the cell into the proximal portion. The diameter of the circles is 0.4 μm, the full-width half-maximum size of the laser beam.
DISCUSSION
Our cryoEM images of the dividing Caulobacter cells show that in early stages of cell division, the inner and outer membranes constrict simultaneously, initially maintaining the 30-nm separation seen in regions distant from the constriction. As cell division progresses, the IM constricts faster, creating a growing distance between the inner and outer membranes near the division plane. Fission of the inner membrane creates a cell containing two inner membrane-bound cytoplasmic compartments surrounded by a single continuous outer membrane.
The constriction in each membrane becomes remarkably small (as small as 60 nm in diameter) before unknown terminal events effect complete closure. These highly constricted membranes are also tightly bent, with a radius of curvature of 20 nm or less. The bent membrane (inner membrane in Fig. 2B) may break spontaneously and then reseal around the cytoplasm of the nascent daughter cell, with a significantly larger radius of curvature (inner membranes in Fig. 2C). In other words, the final fission event may occur simply because the two-surface configuration is a lower energy state for the lipid membrane structure. After inner and outer membrane fission, we did not observed any evidence for a residual “scar” in either membrane (Fig. 2C and E).
Since the constriction of the IM occurs earlier and well separated spatially from the lagging OM constriction, distinct molecular processes must control inner and outer membrane constriction in the later stages of cell division. Early constriction of the inner membrane is effected by the FtsZ ring and associated proteins (13, 35). Separate protein structures controlling late constriction of the outer membrane have not yet been identified.
The FLIP experiments show that both inner membrane and periplasmic proteins are able to freely diffuse past the site of constriction throughout most of the constriction process. The FtsZ ring and associated proteins form a ring-like structure attached to the inner membrane at the constriction site (13, 35). The mode of attachment to the membrane is unknown. It is also unknown whether the FtsZ ring is a continuous ring. It is possible that the FtsZ ring is continuous but only attached to the inner membrane at discrete points. Alternatively, there may be breaks in the ring that allow membrane proteins to diffuse through. FtsZ is known to turn over rapidly, which may cause transient breaks in the ring. The slow diffusion of membrane and periplasmic proteins through the division site observed for a small fraction of the FLIP experiments could be due to the very small dimension of the remaining connection as the compartments near separation in these cells (Fig. 2B, D, and F), or the constriction machinery may congest the connection as the constriction nears separation.
The measured diffusion coefficient for the inner membrane protein PleC-EYFP is D = (12 ± 2) × 10−3 μm2/s (10). This is roughly 200 times smaller than the D value of (2,500 ± 600) × 10−3 μm2/s measured by fluorescence recovery after photobleaching for a cytoplasmic protein of similar mass (GFP fused to a maltose-binding protein domain, 72 kDa) in E. coli (11). Our results reported here for the FLIP experiments are consistent with slower diffusion of proteins in the inner membrane than in the cytoplasm. All the cytoplasmic EGFP in a cell could be bleached with a laser focused at one end of the cell for 5 s (10). The membrane-bound EGFP, however, required localized bleaching for from 30 s (for PilA-EGFP) to 240 s (for CC2909-EGFP) to completely bleach the cell independent of the laser intensity. This indicates that the membrane-bound EGFP fusion proteins took longer to diffuse from the far end of the cell into the laser beam than did the cytoplasmic EGFP.
Electron microscope images show that in E. coli and B. subtilis, the inner membrane and the cell wall invaginate together, forming a septum, and the outer membrane constricts later (5-7). Figure 3B shows the progression of Caulobacter cell division schematically based on the images in Fig. 2 with an approximately consistent scale. For comparison, Fig. 3C shows a schematic of progression of E. coli cell division (adapted with permission from Fig. 32 in the work of Burdett and Murray [6]). (The exterior blebs and interior mesosomes seen in the E. coli images are probably artifacts of the fixation and slicing procedures involved at the time in producing the EM images on which the diagram in Fig. 3C is based. We have not found published cryoEM images of either E. coli or B. subtilis cytokinesis.) The differences between the septum-forming division process in E. coli and the constrictive division process in Caulobacter are quite clear. In E. coli, inner membrane constriction also precedes outer membrane constriction, but the constriction of the inner membrane involves physical association of the two inner membranes of the nascent daughter cells to form the septal disk. Later, after closure of the septum, the E. coli outer membrane constricts between the two inner membrane surfaces and separates the cell. In other EM images of E. coli, it appears that the inner and outer membranes invaginate together (4). The geometric differences in the cell constriction and separation process between Caulobacter, E. coli, and B. subtilis suggest there are differences at the molecular level between the Caulobacter terminal cell constriction and closure mechanisms compared to the septum-forming bacteria. The spatial separation of the Caulobacter inner and outer membrane constrictive rings will facilitate labeling and identification of the distinct proteins involved in IM and OM constriction.
Supplementary Material
Acknowledgments
E.M.J. and H.H.M. were supported by Office of Naval Research grant N00014-02-1-053. The Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy provided support under contract no. DE-AC03-76SF00098 (L.R.C. and K.D.), grant DE-FG03-01ER63219-A001 (H.H.M.), and grant DE-FG02-04ER63777 (W.E.M.). The NIH provided support under grant NIH 1 P20 HG003638-01 (H.H.M.). J.C.C. was supported by National Institutes of Health grant F32 G067472.
VolView was used under a personal free license from KitWare. The movies and isosurfaces shown in the supplemental material were done in collaboration with the Scientific Visualization Group at LBNL. We thank Patrick Viollier (Case Western University) for providing the PilA-GFP strain.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Addinall, S. G., and B. Holland. 2002. The tubulin ancestor, FtsZ, draughtsman, designer and driving force for bacterial cytokinesis. J. Mol. Biol. 318:219-236. [DOI] [PubMed] [Google Scholar]
- 2.Addinall, S. G., and J. Lutkenhaus. 1996. FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol. Microbiol. 22:231-237. [DOI] [PubMed] [Google Scholar]
- 3.Ausmees, N., and C. Jacobs-Wagner. 2003. Spatial and temporal control of differentiation and cell cycle progression in Caulobacter crescentus. Annu. Rev. Microbiol. 57:225-247. [DOI] [PubMed] [Google Scholar]
- 4.Bi, E. F., and J. Lutkenhaus. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161-164, 1991. [DOI] [PubMed] [Google Scholar]
- 5.Burdett, I. D. 1979. Electron microscope study of the rod-to-coccus shape change in a temperature-sensitive rod− mutant of Bacillus subtilis. J. Bacteriol. 137:1395-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burdett, I. D., and R. G. Murray. 1974. Electron microscope study of septum formation in Escherichia coli strains B and B-r during synchronous growth. J. Bacteriol. 119:1039-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burdett, I. D., and R. G. Murray. 1974. Septum formation in Escherichia coli: characterization of septal structure and the effects of antibiotics on cell division. J. Bacteriol. 119:303-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, J. C., P. H. Viollier, and L. Shapiro. 2005. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol. Microbiol. 55:1085-1103. [DOI] [PubMed] [Google Scholar]
- 10.Deich, J., E. M. Judd, H. H. McAdams, and W. E. Moerner. 2004. Visualization of the movement of single histidine kinase molecules in live Caulobacter cells. Proc. Natl. Acad. Sci. USA 101:15921-15926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elowitz, M. B., M. G. Surette, P. E. Wolf, J. B. Stock, and S. Leibler. 1999. Protein mobility in the cytoplasm of Escherichia coli. J. Bacteriol. 181:197-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ely, B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204:372-384. [DOI] [PubMed] [Google Scholar]
- 13.Errington, J., R. A. Daniel, and D. J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evinger, M., and N. Agabian. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Judd, E. M., K. R. Ryan, W. E. Moerner, L. Shapiro, and H. H. McAdams. 2003. Fluorescence bleaching reveals asymmetric compartment formation prior to cell division in Caulobacter. Proc. Natl. Acad. Sci. USA 100:8235-8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kremer, J. R., D. N. Mastronarde, and J. R. McIntosh. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71-76. [DOI] [PubMed] [Google Scholar]
- 17.Margolin, W. 2003. Bacterial division: the fellowship of the ring. Curr. Biol. 13:R16-R18. [DOI] [PubMed] [Google Scholar]
- 18.Martin, M. E., M. J. Trimble, and Y. V. Brun. 2004. Cell cycle-dependent abundance, stability and localization of FtsA and FtsQ in Caulobacter crescentus. Mol. Microbiol. 54:60-74. [DOI] [PubMed] [Google Scholar]
- 19.Matroule, J. Y., H. Lam, D. T. Burnette, and C. Jacobs-Wagner. 2004. Cytokinesis monitoring during development; rapid pole-to-pole shuttling of a signaling protein by localized kinase and phosphatase in Caulobacter. Cell 118:579-590. [DOI] [PubMed] [Google Scholar]
- 20.McAdams, H. H., and L. Shapiro. 2003. A bacterial cell-cycle regulatory network operating in time and space. Science 301:1874-1877. [DOI] [PubMed] [Google Scholar]
- 21.McGrath, P. T., P. Viollier, and H. H. McAdams. 2004. Setting the pace: mechanisms tying Caulobacter cell-cycle progression to macroscopic cellular events. Curr. Opin. Microbiol. 7:192-197. [DOI] [PubMed] [Google Scholar]
- 22.Nanninga, N. 1998. Morphogenesis of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:110-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pichoff, S., and J. Lutkenhaus. 2005. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol. Microbiol. 55:1722-1734. [DOI] [PubMed] [Google Scholar]
- 24.Poindexter, J. S., and J. G. Hagenzieker. 1981. Constriction and septation during cell-division in Caulobacters. Can. J. Microbiol. 27:704-719. [DOI] [PubMed] [Google Scholar]
- 25.Powers, T. R., G. Huber, and R. E. Goldstein. 2002. Fluid-membrane tethers: minimal surfaces and elastic boundary layers. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 65:041901. [DOI] [PubMed] [Google Scholar]
- 26.Quardokus, E. M., and Y. V. Brun. 2002. DNA replication initiation is required for mid-cell positioning of FtsZ rings in Caulobacter crescentus. Mol. Microbiol. 45:605-616. [DOI] [PubMed] [Google Scholar]
- 27.Quardokus, E. M., N. Din, and Y. V. Brun. 2001. Cell cycle and positional constraints on FtsZ localization and the initiation of cell division in Caulobacter crescentus. Mol. Microbiol. 39:949-959. [DOI] [PubMed] [Google Scholar]
- 28.Ryan, K. R., and L. Shapiro. 2003. Temporal and spatial regulation in prokaryotic cell cycle progression and development. Annu. Rev. Biochem. 72:367-394. [DOI] [PubMed] [Google Scholar]
- 29.Sackett, M. J., A. J. Kelly, and Y. V. Brun. 1998. Ordered expression of ftsQA and ftsZ during the Caulobacter crescentus cell cycle. Mol. Microbiol. 28:421-434. [DOI] [PubMed] [Google Scholar]
- 30.Santini, C. L., A. Bernadac, M. Zhang, A. Chanal, B. Ize, C. Blanco, and L. F. Wu. 2001. Translocation of jellyfish green fluorescent protein via the Tat system of Escherichia coli and change of its periplasmic localization in response to osmotic up-shock. J. Biol. Chem. 276:8159-8164. [DOI] [PubMed] [Google Scholar]
- 31.Schierle, C. F., M. Berkmen, D. Huber, C. Kumamoto, D. Boyd, and J. Beckwith. 2003. The DsbA signal sequence directs efficient, cotranslational export of passenger proteins to the Escherichia coli periplasm via the signal recognition particle pathway. J. Bacteriol. 185:5706-5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shapiro, L., H. H. McAdams, and R. Losick. 2002. Generating and exploiting polarity in bacteria. Science 298:1942-1946. [DOI] [PubMed] [Google Scholar]
- 33.Skerker, J. M., and L. Shapiro. 2000. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 19:3223-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas, J. D., R. A. Daniel, J. Errington, and C. Robinson. 2001. Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway in Escherichia coli. Mol. Microbiol. 39:47-53. [DOI] [PubMed] [Google Scholar]
- 35.Weiss, D. S. 2004. Bacterial cell division and the septal ring. Mol. Microbiol. 54:588-597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.