Abstract
The planar and anchoring residues of the family IIIa cellulose binding domain (CBD) from the cellulosomal scaffolding protein of Clostridium cellulovorans were investigated by site-directed mutagenesis and cellulose binding studies. By fusion with maltose binding protein, the family IIIa recombinant wild-type and mutant CBDs from C. cellulovorans were expressed as soluble forms. Cellulose binding tests of the mutant CBDs indicated that the planar strip residues played a major role in cellulose binding and that the anchoring residues played only a minor role.
Clostridium cellulovorans produces a cellulase enzyme complex (cellulosome) containing a variety of cellulolytic subunits attached to a nonenzymatic scaffolding component termed CbpA (2). Binding to cellulose is considered to be the initial process of cellulose degradation by the cellulosome (9, 14). CbpA contains a family IIIa cellulose binding domain (CBD), and the binding of cellulosome to cellulose is considered to be mediated mainly by the CbpA-CBD (5).
Previously, we performed site-directed mutagenesis analyses to determine the structure-function relationship in the CbpA-CBD (4, 5). In these studies, the recombinant wild-type and the engineered CbpA-CBDs were expressed as insoluble forms; therefore, we were required to denature and renature the recombinant products. These processes were time-consuming, and after renaturation from the insoluble state, we could not be certain that the protein was in a completely native configuration. Therefore, a soluble expression system for the CbpA-CBD was preferable for site-directed mutagenesis analysis. In this study, we expressed CbpA-CBD as a soluble protein by fusion with the maltose binding protein (MBP), which is known to help express passenger proteins in soluble forms (7, 12, 19). With the MBP fusion system, we did site-directed mutagenesis studies on the amino acid residues in the planar strip (D57, H58, Y68, R116, W122) and the anchoring residues (N17, Q114), which are considered to have essential roles in cellulose binding, to test the role of these residues in cellulose binding.
Soluble expression of CbpA-CBD fused with MBP by Escherichia coli.
The CbpA-CBD gene was amplified by PCR with the primers CbpA-CBD N (from amino acid number 7 of the CbpA-CBD sequence) (Fig. 1), which had a PstI site at its 5′ end, and primer CbpA-CBD C (from amino acid number 163) (Fig. 1), which had an EcoRI site at its 5′ end. The genomic DNA of C. cellulovorans was prepared as described previously (15) and used as a template for PCR. The amplified gene was inserted into the PstI and EcoRI sites of the vector pMAL p2x (New England Biolabs) to generate pMAL-CbpA-CBD. The CbpA-CBD expressed by this vector was fused with the N-terminal MBP. Then, E. coli TOP10 (Invitrogen) was transformed with the pMAL-CbpA-CBD. Recombinant strains were cultivated in Luria-Bertani medium supplemented with ampicillin (50 μg/ml). After induction by 400 μM IPTG (isopropyl-β-d-thiogalactopyranoside), the CbpA-CBD fused with the MBP (MBP-CbpA-CBD) was expressed as a dominant protein in the soluble fraction of E. coli cells (data not shown). This result indicated that the MBP-CbpA-CBD was expressed successfully as a soluble form.
FIG. 1.
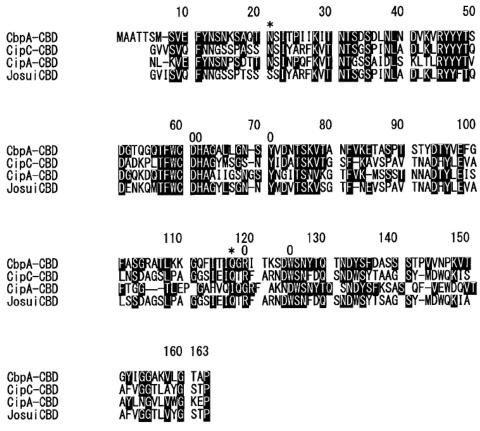
Amino acid sequence alignment of family IIIa CBDs. Conserved amino acids are indicated by white letters in black boxes. Asterisks indicate the amino acids in the planar strip. Circles indicate the anchoring residues. Abbreviations and accession numbers: CbpA-CBD, C. cellulovorans CbpA-CBD (15), P38058; CipC-CBD, C. cellulolyticum CipC-CBD (10), PC6006; CipA-CBD, C. thermocellum CipA-CBD (18), S36859; JosuiCBD, C. josui CipA-CBD (6), T30433.
The expressed MBP-CbpA-CBD was purified by maltose affinity chromatography. The MBP-CbpA-CBD in the E. coli cell extract was bound onto a maltose affinity column and was then eluted by elution buffer including 10 mM maltose. The sodium dodecyl sulfate-polyacrylamide gel electrophoresis pattern of the eluted fraction showed a homogenous band at around 60 kDa, which was in good agreement with the theoretical molecular mass of the MBP-CbpA-CBD (43 kDa for the MBP plus 18 kDa for the CbpA-CBD equals 61 kDa; data not shown). Therefore, the MBP-CbpA-CBD was considered to be purified almost to homogeneity by one-step maltose affinity chromatography. With the E. coli cells from 1 liter of culture broth, 2.4 mg of the purified MBP-CbpA-CBD was obtained.
Cellulose binding affinity of MBP-CbpA-CBD.
To determine the effect of the MBP fusion on the cellulose binding affinity of the CbpA-CBD, we determined the cellulose binding affinity (dissociation constant [Kd]) of the MBP-CbpA-CBDs by performing cellulose binding isotherm tests established previously (5). The Kds of the MBP and bovine serum albumin (BSA) were also determined as negative controls. The purified MBP-CbpA-CBDs (5 μg to 50 μg) were mixed with 1 ml of 1% Avicel type PH-101 (FMC Corporation) in PCM buffer (pH 7, 50 mM KH2PO4 and 10 mM sodium citrate). Since the cellulose binding affinities of the MBP and BSA were much lower than that of the MBP-CbpA-CBP, the Kds of these proteins were determined by use of 10% Avicel. To eliminate specific interactions between the MBP and cellulose, 10 mM of maltose was added to the reaction mixtures for the cellulose binding assay. After incubation at 37°C for 2 h with shaking, the Avicel was removed, and the free protein concentration in the supernatant ([FP]) was measured by the method of Bradford (1) with a protein assay kit from Bio-Rad. The concentration of the protein bound to the Avicel ([PC]) was calculated by subtracting the [FP] from the total protein concentration. Adsorption parameters were obtained by use of the equation described previously, [PC] = [FP][PC]max/Kd + [FP], where Kd and [PC]max are the equilibrium dissociation constant and the maximum amount of protein bound to Avicel, respectively.
The Kds of the purified MBP-CbpA-CBD, MBP, and BSA are shown in Table 1. The Kds were compared to that of the recombinant CbpA-CBD determined previously (5). The results showed that the Kd of the MBP-CbpA-CBD was at a similar level to that of the recombinant CbpA-CBD; on the other hand, the Kd of the MBP was much larger than those of the MBP-CbpA-CBD and the recombinant CbpA-CBD and at a similar level to that of BSA. These results indicated that the MBP did not bind specifically to cellulose, and therefore, the fusion of MBP did not have a large effect on the cellulose binding affinity of the CbpA-CBD.
TABLE 1.
Dissociation constants of MBP-CbpA-CBD and its mutants
| Dissociation constant protein | Mean ± SD Kd ± SD (mM; n = 3)c | Position of replaced residues |
|---|---|---|
| MBP-CbpA-CBD | 0.383 ± 0.048 | Wild type |
| MBP-CbpA-CBD N21A | 1.47 ± 0.18*** | Anchoring residue |
| MBP-CbpA-CBD D60Aa | 16.2 ± 7.3** | Planar strip |
| MBP-CbpA-CBD H61Aa | 14.4 ± 4.4** | Planar strip |
| MBP-CbpA-CBD Y71Aa | 19.5 ± 5.4** | Planar strip |
| MBP-CbpA-CBD Q117A | 0.765 ± 0.033 | Anchoring residue |
| MBP-CbpA-CBD R119Aa | 10.7 ± 4.2*** | Planar strip |
| MBP-CbpA-CBD W125Aa | 12.8 ± 2.8*** | Planar strip |
| MBP | 108 ± 16* | |
| BSA | 129 ± 16* | |
| Recombinant wild-type CbpA-CBDb | 0.6 |
The KDs were estimated by using the [PC]max of MBP-CBD.
The KD was determined previously (5).
*, P < 0.05; **, P < 0.01; ***, P < 0.001 (against the KD of the MBP-CbpA-CBD).
The maximum amount of the MBP-CbpA-CBD bound onto cellulose was 0.12 μmol/g cellulose and 17 times less than that of the recombinant CbpA-CBD (2.1 μmol/g Avicel). This value seemed very reasonable based on the following estimation. The overall dimension of the CipA-CBD is shown to be 48 by 28 by 23 Å (13). Therefore, the cellulose area occupied by the CbpA-CBD is roughly estimated to be at most a circle with a diameter of 48 Å. The overall dimensions of the MBP are 30 by 40 by 65 Å (16). As described above, the MBP was considered not to bind specifically on cellulose. Therefore, the MBP in the MBP-CbpA-CBD may move in a circle where the CbpA-CBD is at the center of the circle. The diameter of this circle should be at most 178 Å (48 Å for the CbpA-CBD plus 2 × 65 Å for the MBP). The area of a circle with a diameter of 65 Å is 13.8 times less than that of a circle with a diameter of 178 Å. Therefore, the amount of MBP-CbpA-CBD which could bind to a certain area of cellulose surface was roughly estimated to be 13.8 times less than that for the recombinant CbpA-CBD and in good agreement with the experimental results described above.
Cellulose binding analysis of CBD mutants.
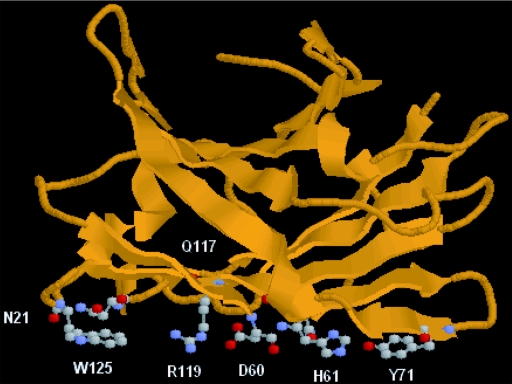
So far, four family IIIa CBDs (17), the CbpA-CBD from C. cellulovorans (14), the CipA-CBD from C. thermocellum (3), the CipA-CBD from C. josui (6), and the CipC-CBD from C. cellulolyticum (10), have been isolated (13). Among them, the crystal structures of the CipA-CBD from C. thermocellum (18) and the CipC-CBD (13) have been determined. Both structures have a planar strip of hydrophobic amino acid residues, which is a linear arrangement of amino acids on the interacting surface of the CBD (18), and two adjacent anchoring residues. Tormo et al. proposed that family IIIa CBDs would bind to crystalline cellulose via the planar strip and the anchoring residues (18), although there is no direct evidence for the function of the anchoring residues. They proposed that the planar strip could bind to six consecutive glucose residues in a cellulose chain (18). The Y71, H61, and W125 were predicted to stack, respectively, the first, the second, and the sixth glucose residues by an aromatic stacking interaction with glucose rings. The fourth glucose residue was predicted to interact with D60 and R119 by salt bridges. They also suggested the contribution of two uncharged polar residues (N21 and Q117, anchoring residues) to cellulose binding (18). These anchoring residues were parallel to the planar strip and were predicted to interact with the cellulose chain bound to the planar strip. Our model structure of the CbpA-CBD (Fig. 2) suggests that CbpA-CBD may also have the planar strip and the anchoring residues. The six residues in the planar strip and one of two anchoring residues (Q117) were conserved in all four family IIIa CBDs (Fig. 1). Another anchoring residue (N21) was also conserved in three family IIIa CBDs out of four CBDs, including the CbpA-CBD. Based on this information, each one of the 6 amino acids in the planar strip and the 2 anchoring residues was replaced by alanine, and the Kds of these engineered CBDs were determined.
FIG. 2.
Model structure of CbpA-CBD. The structure was modeled with the program Swiss-Model version 3.5 at the Expasy server (11). The structure of CipA-CBD from C. thermocellum (18) was used as the modeling template. The structure model was visualized with the cartoon mode of the Protein Explorer software (8). The similarity of amino acid sequences between the CbpA-CBD and the CipA-CBD was 49.7%. The amino acid residues in the planar strip and the anchoring residues are visualized with the ball-and-stick mode.
By fusion with the MBP, all the engineered CbpA-CBDs were expressed as soluble proteins by E. coli. All the engineered MBP-CbpA-CBDs were readily purified almost to homogeneity by one-step chromatography with a maltose affinity column (data not shown). From the E. coli cells of 1-liter culture broth, 2.0 to 3.8 mg of the purified proteins was obtained.
The Kds of the engineered MBP-CbpA-CBDs were determined as described above. Since the cellulose binding affinities of the D57A, H58A, Y68A, R116A, and W122A proteins were much lower than that of the MBP-CbpA-CBP, the Kds of these proteins were determined by use of 10% Avicel. The Kds of the engineered MBP-CbpA-CBDs are shown in Table 1. The alanine replacements of the residues of the planar strip (D60, H61, Y71, R119, or W125) caused a dramatic decrease in cellulose binding affinity. These results suggested that all planar strip residues were essential to bind to cellulose. In contrast to the substitutions of the residues in the planar strip, alanine replacements at the anchoring residues (N21 or Q117) did not have large effects on cellulose affinity, and their Kds were only 1.5 to 2.5 times less than that of the wild type. The direct contributions of anchoring residues to cellulose binding were considered to be limited. However, it is possible that these anchoring residues have supportive roles in binding cellulose to the planar strip. Although the Kds of the engineered MBP-CbpA-CBDs, the D60A, H61A, Y71A, R119A, and W125A CBDs, decreased dramatically, their Kds were still 5 to 10 times higher than those of the MBP and BSA. These results suggested that the engineered MBP-CBDs, in which one of the amino acid residues in the planar strip was replaced, retained specific cellulose binding abilities.
In this study, we developed a soluble expression system of the CbpA-CBD by fusion with the MBP. By this system, the engineered CbpA-CBDs as well as the recombinant wild-type CbpA-CBD were expressed as soluble forms. Also, the expressed proteins were readily purified to homogeneity by one-step maltose affinity chromatography. Therefore, this system should help in ready preparations of purified recombinant wild-type CbpA-CBD and its mutants not only for future protein engineering studies but also for crystal structure analyses.
Acknowledgments
We thank Andrew J. Fisher and Wilfred P. dela Cruz for helpful discussion.
The research was supported in part by the U.S. Department of Energy grant DDF03-92ER20069.
REFERENCES
- 1.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 2.Doi, R. H., A. Kosugi, K. Murashima, Y. Tamaru, and S. O. Han. 2003. Cellulosomes from mesophilic bacteria. J. Bacteriol. 185:5907-5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerngross, U. T., M. P. Romaniec, T. Kobayashi, N. S. Huskisson, and A. L. Demain. 1993. Sequencing of a Clostridium thermocellum gene (cipA) encoding the cellulosomal SL-protein reveals an unusual degree of internal homology. Mol. Microbiol. 8:325-334. [DOI] [PubMed] [Google Scholar]
- 4.Goldstein, M. A., and R. H. Doi. 1994. Mutation analysis of the cellulose-binding domain of the Clostridium cellulovorans cellulose-binding protein A. J. Bacteriol. 176:7328-7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldstein, M. A., M. Takagi, S. Hashida, O. Shoseyov, R. H. Doi, and I. H. Segel. 1993. Characterization of the cellulose-binding domain of the Clostridium cellulovorans cellulose-binding protein A. J. Bacteriol. 175:5762-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kakiuchi, M., A. Isui, K. Suzuki, T. Fujino, E. Fujino, T. Kimura, S. Karita, K. Sakka, and K. Ohmiya. 1998. Cloning and DNA sequencing of the genes encoding Clostridium josui scaffolding protein CipA and cellulase CelD and identification of their gene products as major components of the cellulosome. J. Bacteriol. 180:4303-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapust, R. B., and D. S. Waugh. 1999. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 8:1668-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martz, E. 2002. Protein Explorer: easy yet powerful macromolecular visualization. Trends Biochem. Sci. 27:107-109. [DOI] [PubMed] [Google Scholar]
- 9.Murashima, K., A. Kosugi, and R. H. Doi. 2002. Synergistic effects on crystalline cellulose degradation between cellulosomal cellulases from Clostridium cellulovorans. J. Bacteriol. 184:5088-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pagès, S., A. Bélaïch, H.-P. Fierobe, C. Tardif, C. Gaudin, and J.-P. Bélaïch. 1999. Sequence analysis of scaffolding protein CipC and ORFXp, a new cohesin-containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localization of ORFXp. J. Bacteriol. 181:1801-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peitsch, M. C. 1996. ProMod and Swiss-Model: Internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans. 24:274-279. [DOI] [PubMed] [Google Scholar]
- 12.Pryor, K. D., and B. Leiting. 1997. High-level expression of soluble protein in Escherichia coli using a His6-tag and maltose-binding-protein double-affinity fusion system. Protein Expr. Purif. 10:309-319. [DOI] [PubMed] [Google Scholar]
- 13.Shimon, L. J., S. Pagès, A. Bélaïch, J.-P. Bélaïch, E. A. Bayer, R. Lamed, Y. Shoham, and F. Frolow. 2000. Structure of a family IIIa scaffoldin CBD from the cellulosome of Clostridium cellulolyticum at 2.2 Å resolution. Acta Crystallogr. Sect. D 56:1560-1568. [DOI] [PubMed] [Google Scholar]
- 14.Shoseyov, O., and R. H. Doi. 1990. Essential 170-kDa subunit for degradation of crystalline cellulose by Clostridium cellulovorans cellulase. Proc. Natl. Acad. Sci. USA 87:2192-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shoseyov, O., T. Hamamoto, F. C. Foong, and R. H. Doi. 1990. Cloning of Clostridium cellulovorans endo-1,4-beta-glucanase genes. Biochem. Biophys. Res. Commun. 169:667-672. [DOI] [PubMed] [Google Scholar]
- 16.Spurlino, J. C., G. Y. Lu, and F. A. Quiocho. 1991. The 2.3- Å resolution structure of the maltose- or maltodextrin-binding protein, a primary receptor of bacterial active transport and chemotaxis. J. Biol. Chem. 266:5202-5219. [DOI] [PubMed] [Google Scholar]
- 17.Tomme, P., A. J. Warren, R. C. Miller, D. G. Kilburn, and N. R. Gilkes. 1995. Cellulose-binding domains: classification and properties, p. 142-163. In J. N. Saddler and M. Penner (ed.), Enzymatic degradation of insoluble carbohydrate. American Chemical Society, Washington, D.C.
- 18.Tormo, J., R. Lamed, A. J. Chirino, E. Morag, E. A. Bayer, Y. Shoham, and T. A. Steitz. 1996. Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J. 15:5739-5751. [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, C., A. F. Castro, D. M. Wilkes, and G. A. Altenberg. 1999. Expression and purification of the first nucleotide-binding domain and linker region of human multidrug resistance gene product: comparison of fusions to glutathione S-transferase, thioredoxin and maltose-binding protein. Biochem. J. 338:77-81. [PMC free article] [PubMed] [Google Scholar]