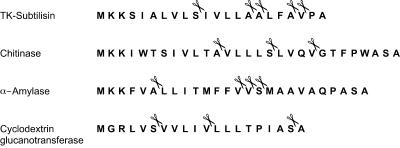Abstract
We have performed the first biochemical characterization of a putative archaeal signal peptide peptidase (SppATk) from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. SppATk, comprised of 334 residues, was much smaller than its counterpart from Escherichia coli (618 residues) and harbored a single predicted transmembrane domain near its N terminus. A truncated mutant protein without the N-terminal 54 amino acid residues (ΔN54SppATk) was found to be stable against autoproteolysis and was examined further. ΔN54SppATk exhibited peptidase activity towards fluorogenic peptide substrates and was found to be highly thermostable. Moreover, the enzyme displayed a remarkable stability and preference for alkaline pH, with optimal activity detected at pH 10. ΔN54SppATk displayed a Km of 240 ± 18 μM and a Vmax of 27.8 ± 0.7 μmol min−1 mg−1 towards Ala-Ala-Phe-4-methyl-coumaryl-7-amide at 80°C and pH 10. The substrate specificity of the enzyme was examined in detail with a FRETS peptide library. By analyzing the cleavage products with liquid chromatography-mass spectrometry, ΔN54SppATk was found to efficiently cleave peptides with a relatively small side chain at the P-1 position and a hydrophobic or aromatic residue at the P-3 position. The positively charged Arg residue was preferred at the P-4 position, while substrates with negatively charged residues at the P-2, P-3, or P-4 position were not cleaved. When predicted signal sequences from the T. kodakaraensis genome sequence were examined, we found that the substrate specificity of ΔN54SppATk was in good agreement with its presumed role as a signal peptide peptidase in this archaeon.
In the domain Archaea, research on protein secretion and the machinery involved are still at an early stage (32). Sequence comparison of genome data have shed light on the features of the archaeal signal peptide (5) and have also indicated the presence or absence of individual components corresponding to eukaryotic or bacterial factors participating in protein secretion (12). Factors that have been analyzed in detail include those involved in flagellum formation in methanogens (8), the signal recognition particle of Archaeoglobus fulgidus (9) and Sulfolobus solfataricus (33), the Sec complex of Methanocaldococcus jannaschii (38), and the secretion ATPase of Sulfolobus spp. (1).
Signal peptide peptidases are enzymes considered to cleave the signal peptide chains of secreted proteins after they are removed from the precursor proteins by signal peptidases (15, 28). Eukaryotic signal peptide peptidases are intramembrane enzymes, with activity dependent on two aspartate residues (21, 39). They have become a center of attention in mammalian cells due to their involvement in immune surveillance. After signal peptide peptidase cleaves signal peptides of the major histocompatibility complex I molecules, the peptide products are presented on the cell surface by a nonclassical major histocompatibility complex class I molecule, HLA-E, indicating to natural killer cells that major histocompatibility complex synthesis is proceeding normally (11, 20).
The bacterial signal peptide peptidase was initially identified in Escherichia coli as a cytoplasmic membrane protein named protease IV (15, 16, 27). The enzyme, encoded by the sppA gene (17, 34), was found to cleave the signal peptide of outer membrane lipoprotein after its release from the precursor protein. Further studies have indicated that protease IV (SppA) carries out only the initial breakdown of the signal peptide into smaller peptide fragments, followed by complete digestion through the functions of cytoplasmic peptidases including oligopeptidase A (25, 26). The gram-positive counterpart of SppA in Bacillus subtilis has also been studied, and has been shown to be involved in signal peptide degradation (10). Furthermore, a cytosolic peptidase, TepA, structurally related to both SppA and ClpP has also been found to actively participate in the degradation of signal peptides in this organism (10).
In terms of signal peptidases and signal peptide peptidases from the Archaea, the type I signal peptidase gene from Methanococcus voltae has been cloned and its product characterized, confirming that the protein exhibits signal peptidase activity (24). Residues critical for the peptidase activity of the protein have been determined (7). FlaK, the signal peptidase for preflagellin signal cleavage, has also been characterized from this organism and has been demonstrated to be an aspartic protease essential for preflagellin cleavage (6). In the Crenarchaeota, the homologue of bacterial type IV prepilin peptidases from S. solfataricus (PibD) has been characterized, and residues on the substrate that are important for recognition by PibD have been examined (2). In contrast to the progress on signal peptidases, experimental examinations of archaeal signal peptide peptidases have not been reported.
Thermococcus kodakaraensis KOD1 is a hyperthermophilic archaeon isolated from a solfatara on Kodakara Island, Kagoshima, Japan (4, 23). The strain is an obligate anaerobe and grows optimally at 85°C. Only heterotrophic growth has been observed, and the strain can efficiently utilize and/or degrade amino acids, pyruvate, tryptone, chitin, and starch. The complete genome sequence of T. kodakaraensis has recently been determined and annotated (13). As expected from the growth characteristics of this strain, the genome sequence revealed the presence of a large number of extracellular enzymes, including chitinase (36), α-amylase (35), and subtilisin-like protease (19). An orthologue search also revealed that T. kodakaraensis harbors a set of factors involved in protein secretion equivalent to those found in various hyperthermophilic archaea (see the Kyoto Encyclopedia of Genes and Genomes; http://www.genome.jp/kegg/).
In this study, we have examined the enzymatic properties of a putative signal peptide peptidase from T. kodakaraensis, revealing that the substrate specificity of the enzyme is consistent with its presumed role as a signal peptide peptidase in this archaeon.
MATERIALS AND METHODS
Strains, media, and plasmids.
T. kodakaraensis KOD1 was cultivated as described elsewhere (4) in order to isolate genomic DNA (29). E. coli DH5α and plasmid pUC18 were used for gene cloning, sequencing, and DNA manipulation. E. coli BL21-CodonPlus(DE3)-RIL (Stratagene, La Jolla, CA) and pET21a(+) (Novagen, Madison, WI) were used for gene expression. E. coli strains were cultivated in Luria-Bertani medium (10 g liter−1 of tryptone, 5 g liter−1 of yeast extract, and 10 g liter−1 of NaCl) with 100 μg ml−1 ampicillin at 37°C.
DNA manipulation and sequence analysis.
Restriction and modification enzymes were purchased from Toyobo (Osaka, Japan). Plasmid DNA was isolated with the Plasmid mini-kit from QIAGEN (Hilden, Germany). KOD Plus (Toyobo) was used as a polymerase for PCR, and a GFX PCR DNA and gel band purification kit (Amersham Biosciences, Buckinghamshire, United Kingdom) was used to recover DNA fragments from agarose gels after electrophoresis. DNA sequencing was performed using BigDye terminator cycle sequencing kit v.3.0 and a model 3100 capillary DNA sequencer (Applied Biosystems, Foster City, CA). Sequence alignments and construction of the phylogenetic tree with the neighbor-joining method were performed with the ClustalW program available at the DNA Data Bank of Japan. Bootstrap resampling was performed 1,000 times with the BSTRAP program.
Expression of the sppATk gene in E. coli.
The sppATk gene initiating with a Met residue preceding Gln30, omitting the transmembrane domain, was amplified from the genomic DNA of T. kodakaraensis using the primer set sppN1 and sppC1 (sppN1, 5′-GTTCTCCATATGCAGGTCAATCCCCCCGCTGT-3′; sppC1, 5′-CAGAATTCAACCACCCCCAATGAGGG-3′). The gene for the truncated protein initiating with a Met residue preceding Cys55 was amplified with sppN2 and sppC1 (sppN2, 5′-ACTTTACGCATATGTGTGAAGGCAGTGTTAAC-3′). After confirming the sequences of the DNA fragments, they were inserted into pET21a(+) at the NdeI and EcoRI sites. After introduction into E. coli BL21-CodonPlus(DE3)-RIL cells, gene expression was induced with 0.1 mM isopropylthiogalactopyranoside (IPTG) at the mid-exponential growth phase with further incubation for 6 h at 37°C.
Purification of recombinant SppATk.
After inducing gene expression, cells were washed with 50 mM Tris-HCl (pH 8) and resuspended in the same buffer. Cells were sonicated on ice, and the supernatant after centrifugation (20,000 × g, 30 min at 4°C) was applied to heat treatment at 85°C for 15 min, immediately cooled on ice, and then centrifuged (20,000 × g, 30 min at 4°C). The soluble protein sample was brought to 35% saturation with ammonium sulfate and the precipitate which included SppATk was dissolved in 50 mM Tris-HCl (pH 8) at a concentration of 1 mg ml−1. This was applied to anion exchange chromatography (ResourceQ, Amersham Biosciences) equilibrated with 50 mM Tris-HCl (pH 8), 0.2 M NaCl, and proteins were eluted with a linear gradient (0.2 to 1.0 M, 42 ml) of NaCl. After desalting with a HiPrep26/10 column (Amersham Biosciences), the sample was applied to gel filtration chromatography (Superdex 200 HR 10/30, Amersham Biosciences) equilibrated with 50 mM Tris-HCl (pH 8), 0.15 M NaCl, and the fractions obtained were used for enzyme analysis. Approximately 3 to 7 mg of purified octameric protein was obtained per liter of culture.
Protein analysis of purified recombinant SppATk.
The native molecular mass of the purified protein was examined by gel-filtration chromatography using Superdex 200 HR 10/30 in 50 mM Tris-HCl (pH 8), 0.15 M NaCl. The retention time was calibrated with those of the standard proteins thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), aldolase (158 kDa), albumin (67 kDa), ovalbumin (43 kDa), chymotrypsinogen A (25 kDa), and RNase A (13.7 kDa). Protein concentration was determined with the protein assay kit (Bio-Rad, Hercules, CA) using bovine serum albumin as a standard. Determination of N-terminal amino acid sequences of proteins were performed with a protein sequencer (model 491 cLC, Applied Biosystems) after separation by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and electroblotting onto a polyvinylidene difluoride membrane (Millipore, Bedford, MA).
Enzyme activity measurements.
Most activity measurements were performed with peptidyl-MCA substrates [peptidyl-α-(4-methylcoumaryl-7-amide) substrates] available from Peptide Institute (Osaka, Japan). Release of 7-amino-4-methylcoumarin was monitored consecutively with a fluorescence spectrophotometer capable of maintaining the cuvette at desired temperatures between 30 and 100°C. Excitation and emission wavelengths were 380 nm and 460 nm, respectively. Standard activity measurements were performed at 60°C in a final volume of 1 ml with 0.1 μg of purified protein and Ala-Ala-Phe-MCA (200 μM) in 50 mM CHES (N-cyclohexyl-2-aminoethanesulfonic acid; pH 10.0). The final concentration of dimethyl sulfoxide used to dissolve the substrate was constant at 3% of the reaction mixture. Kinetic parameters were calculated with SigmaPlot (SPSS Science, Chicago, IL).
Effects of temperature and pH on enzyme activity.
All buffers were prepared so that they would reflect accurate values at the applied temperatures. In examining the effect of temperature, the standard assay method was applied at each temperature. The effect of pH was examined in the presence of 50 mM of morpholineethanesulfonic acid (MES)-NaOH (pH 6.0 to 7.0), HEPES-NaOH (pH 7.0 to 8.0), Bicine-NaOH (pH 8.0 to 9.0), CHES-NaOH (pH 9.0 to 10.0), and CAPS (N-cyclohexyl-3-aminopropanesulfonic acid)-NaOH (pH 10.0 to 12.0), respectively. Thermostability of the protein was analyzed by measuring the residual activity of the protein after incubation at various temperatures in 50 mM CHES-NaOH (pH 10.0). The initial activity of the enzyme incubated at 60°C was designated as 100%. Alkaline stability was analyzed by measuring the residual activity of the protein after incubation at various pH in 50 mM CAPS-NaOH at 60°C. The initial activity of the enzyme incubated in 50 mM CAPS-NaOH (pH 10.0) was designated as 100%. In measuring thermostability and alkaline stability, the protein concentration during incubation was 1 μg ml−1. Residual activities were measured with the standard assay method described above.
Effect of protease inhibitors.
The effects of various protease inhibitors at concentrations of 200 μM, 1 mM, or 10 mM were examined at 60°C and pH 10. The substrate Ala-Ala-Phe-MCA was present at a concentration of 200 μM. Activity in the absence of inhibitors was defined as 100%.
Determination of substrate specificity.
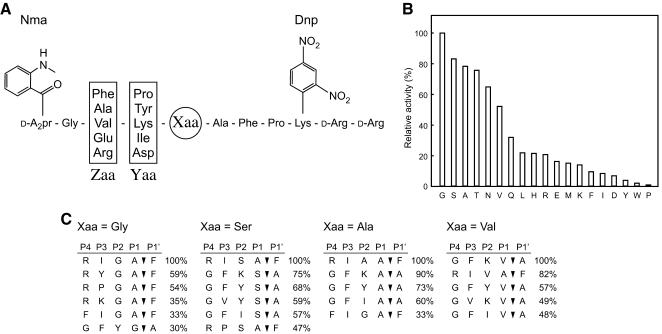
Substrate specificity was examined with a FRETS peptide library (25Xaa series, Peptide Institute) (37). These peptide substrates harbor a highly fluorescent 2-(N-methylamino)benzoyl group linked to the side chain of the amino terminal d-2,3-diaminopropionic acid residue (d-A2pr), along with a 2,4-dinitrophenyl group (quencher) linked to the ε-amino group of a Lys residue. In between the d-A2pr and Lys residue lies the peptide Gly-Zaa-Yaa-Xaa-Ala-Phe-Pro, where Zaa is a mixture of Phe, Ala, Val, Glu, and Arg, Yaa is a mixture of Pro, Tyr, Lys, Ile, and Asp, and the Xaa residue is a defined single amino acid of choice (see below). Excitation and emission wavelengths were 340 nm and 440 nm, respectively.
In the initial assay to examine the preference for residues at the P-1 position, 1 μg of purified enzyme was added to the reaction mixture with a final volume of 1 ml containing 30 μM substrate in 50 mM CHES (pH 10). The final concentration of dimethyl sulfoxide used to dissolve the substrate was constant at 3% of the reaction mixture. A second assay to identify the cleavage sites and the preference towards residues at the P-2 to P-4 positions was performed on selected substrates. Aliquots (100 μl) from the cleavage reactions were taken at various time intervals that corresponded to 15 to 30% cleavage of the substrates, and subjected to liquid chromatography (LC)-mass spectrometry analysis. An ODS A-302 column (YMC, Kyoto, Japan) was used for separation with 0.05% trifluoroacetic acid in H2O as eluant A and 0.05% trifluoroacetic acid in CH3CN as eluant B. The gradient was 5-40% of eluant B in A at a flow rate of 1.0 ml min−1 over a time span of 55 min. Aliquots taken from the cleavage reactions were injected and the cleaved products were monitored with absorbance at 220 nm, as well as fluorescence intensity in order to identify the N-terminal segments. The structures of the cleaved products were deduced from the theoretical molecular weights.
RESULTS
Putative signal peptide peptidase gene on the genome of T. kodakaraensis.
Using the primary structure of the signal peptide peptidase from E. coli (SppAEc), we performed a BLAST search against the protein sequences of T. kodakaraensis. Although significantly smaller in size than SppAEc (618 amino acid residues), one open reading frame, encoding TK1164 (334 residues), displayed significant similarity (27% identical) to SppAEc, and was therefore designated sppATk (Fig. 1). The deduced molecular mass of the protein was 36,211 Da. A BLAST search against the complete genome sequences of various archaeal strains was performed with the SppAEc and SppATk sequences. Similar orthologues were found in many genera of the Euryarchaeota, including Picrophilus, Pyrococcus, and Thermoplasma, as well as the methanogens and the haloarchaea. We also found orthologues in Nanoarchaeum and the crenarchaeon Pyrobaculum.
FIG. 1.
Sequence alignment of putative signal peptide peptidase proteins from E. coli, B. subtilis, and representative archaeal strains. The 19 archaeal SppA sequences described in Fig. 2 were aligned with the sequences of SppA from E. coli and B. subtilis. After alignment, representative sequences were selected. Asterisks indicate highly conserved residues that were present in at least 18 of the 21 sequences aligned. Among these, residues that were conserved in all sequences examined are indicated by stars. The bar above the alignment indicates the putative transmembrane domain of signal peptide peptidase from T. kodakaraensis. Arrowheads indicate the residues that immediately follow the artificial Met residue incorporated in ΔN29SppATk and ΔN54SppATk. Tko, T. kodakaraensis; Mja, Methanocaldococcus jannaschii; Tac, Thermoplasma acidophilum; PaI, Pyrobaculum aerophilum I; Neq, Nanoarchaeum equitans; Hba, Halobacterium sp. strain NRC-1; Bsp, B. subtilis; Eco, E. coli. Due to their lengths, not all residues are shown for Pyrobaculum aerophilum I (608 residues), Halobacterium sp. strain NRC-1 (300 residues), and E. coli (618 residues).
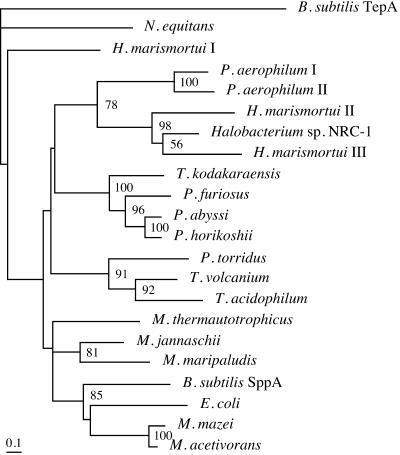
A phylogenetic analysis of these sequences along with several selected bacterial sequences is shown in Fig. 2. Although the catalytic residues have not been experimentally verified, effects of various inhibitors have suggested that SppAEc is a serine protease (16). We indeed observed multiple serine residues that were highly conserved among the archaeal and bacterial SppA sequences (Fig. 1). As in the case of SppAEc, SppATk is structurally categorized in the S49 family of the SK clan of serine proteases (MEROPS, the peptidase database, http://merops.sanger.ac.uk/) (31). Another common feature was that both proteins harbored a putative transmembrane region(s) near their N termini. Based on these similar features, we set out to express the sppATk gene and examine the enzymatic properties of the recombinant protein.
FIG. 2.
Phylogenetic tree of putative archaeal SppA sequences. SppA sequences from archaea that displayed high similarity to SppAEc and SppATk were analyzed along with the sequences of SppAEc and the SppA and TepA from B. subtilis. The proteins used (accession numbers) were B. subtilis SppA (CAB14931) and TepA (CAB13552), E. coli (BAA15557), Haloarcula marismortui I (AAV45638), II (AAV46904), and III (AAV47811), Halobacterium sp. strain NRC-1 (AAG19125), Methanocaldococcus jannaschii (AAB98642), Methanococcus maripaludis (CAF30625), Methanosarcina acetivorans (AAM07395), Methanosarcina mazei (AAM30562), Methanothermobacter thermautotrophicus (AAB85306), Nanoarchaeum equitans (AAR39164), Picrophilus torridus (AAT42796), Pyrobaculum aerophilum I (AAL65089) and II (AAL64441), Pyrococcus abyssi (CAB49512), Pyrococcus furiosus (AAL81707), Pyrococcus horikoshii (BAA30681), Thermococcus kodakaraensis (BAD85353), Thermoplasma acidophilum (CAC11222), and Thermoplasma volcanium (BAB59171). Only bootstrap values above 50 are indicated.
Expression of the sppATk gene in E. coli and purification of the recombinant protein.
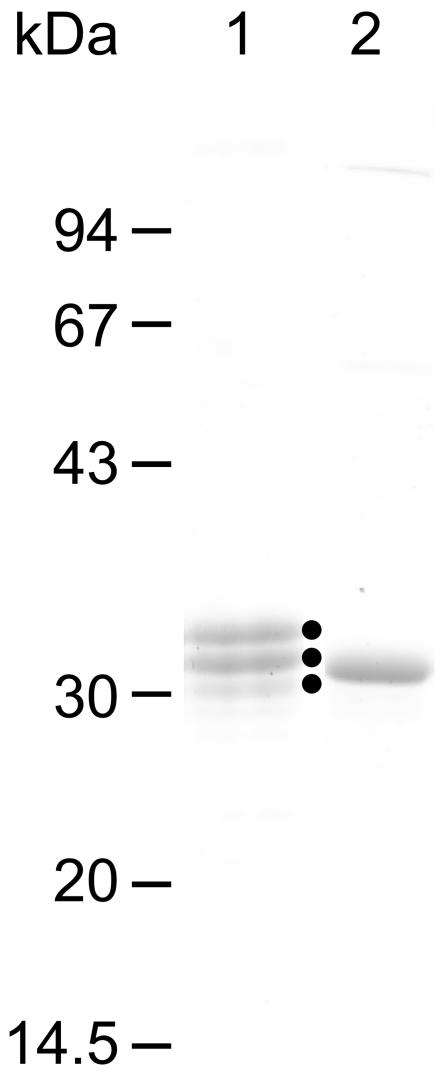
The putative transmembrane domain of SppATk corresponds to residues Lys7 to Tyr29 (Fig. 1). In order to characterize the catalytic domain of the protein, we omitted this region when constructing the expression vector. An artificial Met residue was incorporated in the place of Tyr29, and this gene was expressed in E. coli. As expected, we obtained a soluble protein (ΔN29SppATk) which was resistant to heat treatment at 85°C for 15 min. We found that the thermostable protein exhibited peptidase activity towards peptide substrates such as Ala-Ala-Phe-MCA. The recombinant protein was purified with ammonium sulfate fractionation, anion exchange chromatography, and gel filtration chromatography. During these procedures, we observed a gradual decrease in the molecular weight of the protein as judged by SDS-PAGE, leading to three major molecular species (Fig. 3).
FIG. 3.
SDS-PAGE analysis of purified ΔN29SppATk (lane 1) and ΔN54SppATk (lane 2). Solid dots to the right of lane 1 indicate the purified ΔN29SppATk and the major degradation products.
We determined the N-terminal amino acid sequences of each species and found that in the smaller proteins, degradation at the N-terminal region had occurred, probably due to autoproteolysis. As the smallest species harbored the Cys55 at its extreme N terminus, we reconstructed an expression plasmid so that translation initiated at a Met residue adjacent to Cys55. The protein produced (ΔN54SppATk) was purified with the methods mentioned above (Fig. 3), and was found to be relatively stable in terms of proteolytic degradation. This protein, ΔN54SppATk, was therefore used for further biochemical examination. Although not shown in the results described below, ΔN29SppATk and ΔN54SppATk displayed similar tendencies in terms of specific activity, pH dependency, and substrate preference using the FRETS peptide library.
Oligomeric form of ΔN54SppATk.
The molecular mass of the purified ΔN54SppATk using gel filtration chromatography was estimated to be slightly larger than that of catalase (232 kDa). The molecular mass of a single subunit of ΔN54SppATk is 30,421 Da, suggesting that the protein formed an octamer. Besides this major peak, we observed a second peak at 460 to 500 kDa. The octameric form of the protein was used for further examination. It should be noted that the results obtained here are those of a truncated protein, and may not accurately reflect the oligomeric state of the native protein, which can be assumed to be associated with the membrane. It has previously been reported that the oligomeric form of the native SppAEc was suggested to be tetrameric through cross-linking experiments, while the results of native PAGE raised the possibility of an even higher oligomeric form (17).
Optimal pH and temperature.
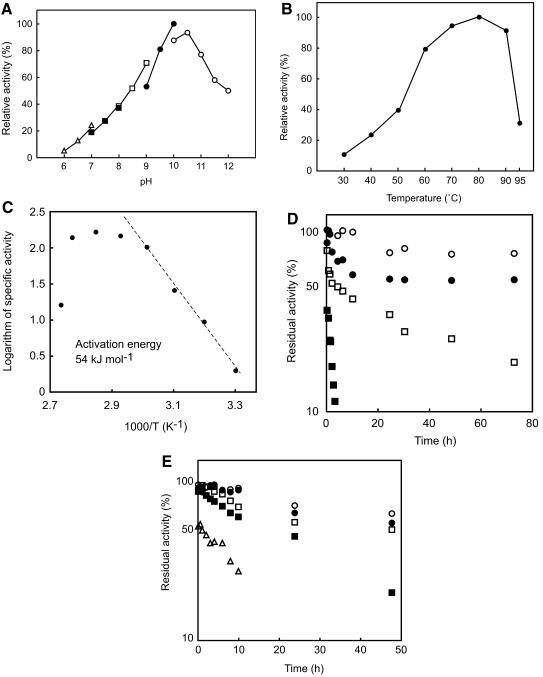
The effects of pH and temperature on the activity of ΔN54SppATk were examined with the substrate Ala-Ala-Phe-MCA. As shown in Fig. 4A, the protein exhibited maximal activity at an unexpectedly high pH range of 10 to 10.5. High levels of activity were maintained at even higher pH values of 11.5 (58%) and 12 (50%). The effects of temperature on activity were analyzed with the same substrate at pH 10. Maximum activity under the conditions examined was observed at approximately 80°C (Fig. 4B). The Arrhenius plot gave a constant slope from 30°C to 60°C, and the activation energy of the reaction was calculated to be 54 kJ mol−1 (Fig. 4C). The thermostability of the enzyme was examined for 72 h at pH 10 (Fig. 4D), and alkaline stability was measured at 60°C for 48 h (Fig. 4E). In terms of temperature, we observed extremely high stability at 60 and 70°C, with over 60% of the initial activity remaining after 72 h. At 80°C, we detected a relatively greater decrease in activity at the initial phases of incubation, which was consistently reproduced in multiple experiments. However after 30 min, the enzyme seemed to stabilize and follow the usual deactivation kinetics, where the deactivation rate (−dNE/dt) is proportional to the amount of enzyme (NE), or −dNE/dt = kNE. We still observed over 30% residual activity after 48 h at 80°C and pH 10. ΔN54SppATk also exhibited high alkaline stability. Nearly 50% residual activity was observed after 24 h at pH 11.5 and 60°C, and the calculated half-life at pH 12 was approximately 10 h.
FIG. 4.
(A) Effect of pH on the activity of ΔN54SppATk. Measurements were performed at 60°C in the following buffers at a concentration of 50 mM: MES-NaOH (open triangles), HEPES-NaOH (solid squares), Bicine-NaOH (open squares), CHES-NaOH (solid circles), and CAPS-NaOH (open circles). (B) Effect of temperature on the activity of ΔN54SppATk. Reactions were carried out in 50 mM CHES-NaOH (pH 10.0). (C) Arrhenius plot of B, indicating the activation energy of substrate hydrolysis catalyzed by ΔN54SppATk. (D) Thermostability of ΔN54SppATk at various temperatures. Incubation of the enzyme was carried out in 50 mM CHES-NaOH (pH 10.0). Symbols: 60°C, open circles; 70°C, solid circles; 80°C, open squares; 90°C, solid squares. (E) Stability of ΔN54SppATk at various pHs. Enzyme incubation was carried out at 60°C in 50 mM CAPS-NaOH (pH 10.0, open circles; pH 10.5, solid circles; pH 11.0, open squares; pH 11.5, solid squares; pH 12.0, open triangles). All activity measurements (A to E) were carried out with 200 μM Ala-Ala-Phe-MCA.
Effects of various inhibitors.
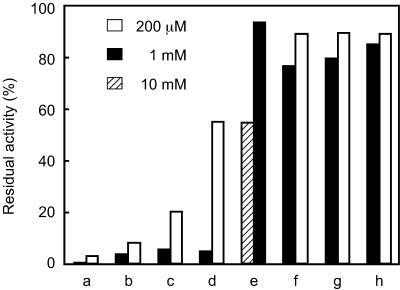
The effects of various protease inhibitors were examined at 60°C and pH 10 (Fig. 5). Leupeptin, chymostatin, and antipain exhibited relatively strong inhibition, leading to an 80% or higher decrease in activity at 200 μM. Diisopropyl fluorophosphate, which specifically reacts with serine residues, also strongly inhibited the activity of SppATk at 1 mM. In contrast, addition of EDTA, a typical inhibitor of metalloproteases, and pepstatin, an inhibitor of aspartate proteases, did not result in significant decreases in activity. Although the effects of phenylmethylsulfonyl fluoride were lower than expected, the results as a whole agree with the assumption that SppATk is a serine protease.
FIG. 5.
Effect of various protease inhibitors on the activity of ΔN54SppATk. The inhibitors examined were leupeptin (a), antipain (b), chymostatin (c), diisopropyl fluorophosphate (d), phenylmethylsulfonyl fluoride (e), pepstatin (f), elastatinal (g), and EDTA (h). Activity measurements were performed at 60°C with the indicated inhibitor concentrations.
Preference of ΔN54SppATk for various peptidyl-MCA substrates.
We examined the activity of ΔN54SppATk on various peptidyl-MCA substrates shown in Table 1. At pH 10, we found that Ala-Ala-Phe-MCA was by far the preferred substrate, followed by moderate activities towards N-glutaryl (Glt)-Ala-Ala-Phe-MCA and N-benzyloxycarbonyl (Z)-Val-Lys-Met-MCA. At pH 8, the preference became stricter, with Ala-Ala-Phe-MCA and Glt-Ala-Ala-Phe-MCA the only substrates leading to significant cleavage.
TABLE 1.
Activity of ΔN54SppATk towards various peptidyl-MCA substratesa
| Substrateb | Sp act (μmol mg protein−1 min−1)
|
|
|---|---|---|
| pH 10.0 (%) | pH 8.0 | |
| Ala-Ala-Phe-MCA | 9.80 (100) | 4.70 |
| Glt-Ala-Ala-Phe-MCA | 2.85 (29) | 0.98 |
| Z-Val-Lys-Met-MCA | 1.05 (11) | 0.05 |
| Phe-MCA | 0.15 (1.5) | 0.04 |
| Suc-Ala-Ala-Ala-MCA | 0.06 (0.6) | 0.04 |
| Suc-Ile-Ile-Trp-MCA | 0.03 (0.3) | n.d. |
| Suc-Leu-Leu-Val-Tyr-MCA | 0.02 (0.2) | 0.03 |
| Ala-MCA | n.d. | 0.04 |
| Suc(OMe)-Ala-Ala-Pro-Val-MCA | n.d. | 0.01 |
| Suc-Ala-Ala-Pro-Phe-MCA | n.d. | 0.01 |
| Z-Leu-Leu-Glu-MCA | n.d. | n.d. |
| Z-Ala-Ala-Asn-MCA | n.d. | n.d. |
| Z-Leu-Leu-Leu-MCA | n.d. | n.d. |
| Z-Leu-Arg-Gly-Gly-MCA | n.d. | n.d. |
Activities were examined with a substrate concentration of 300 μM in 50 mM CHES (pH 10.0) or 50 mM HEPES (pH 8.0) at 60°C. n.d., activity not detected.
Suc, N-succinyl; Suc(OMe), N-methoxysuccinyl.
Kinetic analysis.
A kinetic analysis of the reaction catalyzed by ΔN54SppATk was carried out at 80°C and pH 10 with the substrate Ala-Ala-Phe-MCA. The reaction followed Michaelis-Menten kinetics with a Km of 240 ± 18 μM and a Vmax of 27.8 ± 0.7 μmol min−1 mg protein−1. The kcat of the enzyme was calculated to be 14.1 s−1.
Examining the substrate preference of ΔN54SppATk with a FRETS peptide library.
We next evaluated the peptidase activity of ΔN54SppATk against a FRETS peptide library described in Materials and Methods. This analysis provides detailed information on the preference of a peptidase towards substrate residues at the P-1, P-2, P-3, and, in some cases, the P-4 position. An initial analysis was carried out with 19 substrates corresponding to all amino acids at the Xaa site with the exception of cysteine (Fig. 6A). We detected a preference of ΔN54SppATk for substrates with rather small residues (Gly, Ser, Ala, and Thr) at the Xaa position (Fig. 6B). Cleavage rates of substrates with charged or aromatic residues were low.
FIG. 6.
(A) Structure of the peptide substrates from a FRETS peptide library utilized in B and C. (B) Peptidase activity towards various substrates from the FRETS peptide library. Amino acid residues at the Xaa position are indicated. Each substrate was examined at a concentration of 30 μM. (C) The major cleaved products of four substrates (Xaa = Gly, Ser, Ala, and Val) were detected by LC-mass spectrometry. Relative quantities are indicated to the right of the sequences.
We next selected four substrates with high cleavage rates (Xaa = Gly, Ser, Ala, and Val), and subjected the cleaved products to LC-mass spectrometry analysis. In order to identify products generated from the initial cleavage reaction of the substrate, reactions were stopped at various intervals corresponding to 15 to 30% substrate cleavage. The most abundant cleavage products for each of the four substrates are shown in Fig. 6C. In the case of Xaa = Gly, we found that cleavage predominantly occurred at the C-terminal amide bond of Ala, and not the expected Gly residue. In this case, we can obtain insight on the preferred residues at the P-3 and P-4 sites. At the P-3 site, Ile led to the highest levels of cleavage, followed by Tyr and Pro. Products with Asp at the P-3 position could not be found. Concerning the P-4 position, we found a high preference for Arg. We also detected products with Phe or Val at the P-4 site. As in the case of the P-3 residue, we could not find products with the negatively charged Glu residue at the P-4 site. Among the products that were actually cleaved at the Gly residue, Tyr and Lys were found at the P-2 site, and Phe and Val at the P-3 site.
With Xaa = Ala, Ser, or Val, we found a common tendency with the results described above for the Xaa = Gly substrate. In all three cases, the most preferred P-3 residue was Ile or Phe, followed by Val, while cleavage products with negatively charged residues at this site were not found. A preference for the positively charged Arg at the P-4 site was also observed for all three substrates. The results also indicated a broad substrate specificity of ΔN54SppATk towards the P-2 residue.
DISCUSSION
We have described the biochemical properties of ΔN54SppATk, a truncated form of the putative signal peptide peptidase from T. kodakaraensis. One remarkable feature of the enzyme was its high activity at high alkaline pH. Alkaliphilic proteases have attracted much attention due to their high demand in application, particularly in the detergent industry (3, 14, 18, 22). A number of enzymes are commercially available, including Savinase, subtilisin Carlsberg, and subtilisin BPN′. Various strategies in the protein engineering field have been applied to alkaliphilic proteases with the aim to improve their (thermo)stability, and this has led to significant improvements in terms of catalytic efficiency and stability (14). However, as ΔN54SppATk was obtained from a hyperthermophile, the stability of the enzyme can be regarded as exceptionally high in comparison with previously known alkaline proteases/peptidases. We have detected over 50% residual activity after incubation at 70°C and pH 10 for 3 days.
As the enzyme exhibits intrinsic thermophilic and alkaliphilic properties, ΔN54SppATk should be an attractive target for future protein engineering focusing on the modification of its substrate specificity. We have examined whether ΔN54SppATk exhibits hydrolase activity towards proteins at 60°C and pH 10. Using bovine serum albumin, ovalbumin, α-casein, hemoglobin, and lysozyme, we found that protease activity of ΔN54SppATk was dependent on the particular protein substrate. Degradation of bovine serum albumin and ovalbumin was not observed, while α-casein, hemoglobin, and lysozyme were degraded to various extents (data not shown).
SppATk is structurally categorized in the S49 family of serine proteases, a prokaryotic family of proteases of which still little is understood. There are no three-dimensional structures available, and moreover, the catalytic mechanism of signal peptide peptidases and the residues involved have not yet been experimentally determined. Information on the enzymatic properties of previously identified signal peptide peptidases is also limited. The specific activity of SppAEc has been measured in several studies and ranges between 0.733 μmol mg−1 min−1 at 25°C against Z-valine p-nitrophenyl ester (27) and 13.6 μmol mg−1 min−1 at 37°C against Z-valine β-naphthyl ester (17). Taking into account the Vmax at 80°C and the effects of temperature, the specific activity of the archaeal SppATk was calculated to be 6.6 μmol mg−1 min−1 at 40°C against Ala-Ala-Phe-MCA.
Residues preferred at the cleavage site have also been examined for SppAEc with both synthetic substrates and signal peptides. The former revealed a preference of cleavage at the C-terminal side of Ala, Leu, Val, Gly, and Phe (27), while the latter indicated a preference for Val, Leu, Ile, Gly, Thr, and Ala (25). As in the case of the specific activity described above, the substrates used and the conditions applied in these experiments vary greatly, making it difficult to accurately compare the two enzymes. One point that is worthy of note is that many residues (Ala, Val, Thr, and Gly) are commonly preferred at the P-1 site by both SppAEc and SppATk.
By performing a BLAST search against the genome sequences of archaea, we were able to identify SppA orthologues in most strains of Euryarchaeota (Fig. 2). As exceptions, we could not identify highly similar orthologues on the A. fulgidus and Methanopyrus kandleri genomes. Interestingly, most members of the Crenarchaeota do not seem to utilize structurally related signal peptide peptidases. The two protein sequences from Pyrobaculum aerophilum, the only SppA orthologues from Crenarchaeota, contained exceptionally long extensions in their C-terminal domains. Residues that are highly conserved among bacterial and archaeal SppA sequences are indicated in Fig. 1 and may be involved in catalysis and/or substrate specificity. In particular, Ser162 in SppATk is conserved in all SppA sequences and is also conserved in the functionally and structurally related B. subtilis TepA protease (10). Although this serine residue of TepA has not been experimentally analyzed, it is included in a region which displays similarity to the region containing the active site serine of E. coli ClpP, raising the possibilities that this residue is the nucleophile in all of these proteins. However, the His and Asp/Glu residues that constitute the conventional catalytic triad found in ClpP are not conserved among the archaeal proteins and are not even present in the SppA proteins from E. coli and B. subtilis. In order to accurately determine the active-site residues of SppA proteins, including the nucleophilic serine, site-directed mutagenesis studies will be necessary.
Using the FRET peptide library, we have been able to clarify some of the preferences of ΔN54SppATk towards residues at the P-1, P-2, P-3, and P-4 sites. A relatively small side chain seems to be preferred at the P-1 position. The specificity at the P-2 position can be regarded as broad. Hydrophobic and/or aromatic residues are recognized most at the P-3 site, while the positively charged Arg enhances the activity of ΔN54SppATk when present at the P-4 position. Our results also indicate that the presence of acidic residues at any one of the sites from P-2 to P-4 has a negative effect on the substrate recognition of ΔN54SppATk. With the MCA substrates, a direct comparison among substrates to determine the residue preference of ΔN54SppATk was difficult, as multiple factors differ even between two given substrates. One point that can be noted is that a negative charge at the P-4 position has a large negative effect on substrate recognition (Ala-Ala-Phe-MCA > Glt-Ala-Ala-Phe-MCA).
Taking into account the specificity of ΔN54SppATk, we examined a vast number of putative signal sequences that were identified on the T. kodakaraensis genome using the SOSUI program. Some representative signal sequences, those from four proteins that have been experimentally proven to be secreted from T. kodakaraensis (19, 30, 35, 36; unpublished data), are shown in Fig. 7. As in the case of most putative signal sequences on the T. kodakaraensis genome, acidic residues are not found, consistent with the fact that ΔN54SppATk does not cleave peptides with acidic residues in the P-2 to P-4 sites. Further, we found a number of candidate sequences in each signal sequence that can be presumed to be efficiently recognized and cleaved by ΔN54SppATk.
FIG. 7.
Predicted signal sequences of four T. kodakaraensis proteins that have been biochemically characterized. Prediction was carried out with the SOSUI program. The signal sequence of α-amylase has been confirmed experimentally. Scissors indicate sites that can be presumed to be cleaved by SppATk when its substrate specificity is taken into account.
Although future gene disruption studies will be necessary to confirm the physiological role of SppATk, the similarity in primary structure to SppAEc as well as the enzymatic properties of the enzyme are in good agreement with the assumption that SppATk functions as a signal peptide peptidase in T. kodakaraensis.
Acknowledgments
This work was supported by a grant of the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Albers, S.-V., and A. J. M. Driessen. 2005. Analysis of ATPases of putative secretion operons in the thermoacidophilic archaeon Sulfolobus solfataricus. Microbiology 151:763-773. [DOI] [PubMed] [Google Scholar]
- 2.Albers, S.-V., Z. Szabó, and A. J. M. Driessen. 2003. Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. J. Bacteriol. 185:3918-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atomi, H. 2005. Recent progress towards the application of hyperthermophiles and their enzymes. Curr. Opin. Chem. Biol. 9:166-173. [DOI] [PubMed] [Google Scholar]
- 4.Atomi, H., T. Fukui, T. Kanai, M. Morikawa, and T. Imanaka. 2004. Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1:263-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bardy, S. L., J. Eichler, and K. F. Jarrell. 2003. Archaeal signal peptides-a comparative survey at the genome level. Protein Sci. 12:1833-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bardy, S. L., and K. F. Jarrell. 2003. Cleavage of preflagellins by an aspartic acid signal peptidase is essential for flagellation in the archaeon Methanococcus voltae. Mol. Microbiol. 50:1339-1347. [DOI] [PubMed] [Google Scholar]
- 7.Bardy, S. L., S. Y. M. Ng, D. S. Carnegie, and K. F. Jarrell. 2005. Site-directed mutagenesis analysis of amino acids critical for activity of the type I signal peptidase of the archaeon Methanococcus voltae. J. Bacteriol. 187:1188-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bardy, S. L., S. Y. M. Ng, and K. F. Jarrell. 2004. Recent advances in the structure and assembly of the archaeal flagellum. J. Mol. Microbiol. Biotechnol. 7:41-51. [DOI] [PubMed] [Google Scholar]
- 9.Bhuiyan, S. H., K. Gowda, H. Hotokezaka, and C. Zwieb. 2000. Assembly of archaeal signal recognition particle from recombinant components. Nucleic Acids Res. 28:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolhuis, A., A. Matzen, H.-L. Hyyryläinen, V. P. Kontinen, R. Meima, J. Chapuis, G. Venema, S. Bron, R. Freudl, and J. M. van Dijl. 1999. Signal peptide peptidase- and ClpP-like proteins of Bacillus subtilis required for efficient translocation and processing of secretory proteins. J. Biol. Chem. 274:24585-24592. [DOI] [PubMed] [Google Scholar]
- 11.Braud, V. M., D. S. J. Allan, C. A. O'Callaghan, K. Söderström, A. D'Andrea, G. S. Ogg, S. Lazetic, N. T. Young, J. I. Bell, J. H. Phillips, L. L. Lanier, and A. J. McMichael. 1998. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 391:795-799. [DOI] [PubMed] [Google Scholar]
- 12.Cao, T. B., and M. H. Saier, Jr. 2003. The general protein secretory pathway: phylogenetic analyses leading to evolutionary conclusions. Biochim. Biophys. Acta 1609:115-125. [DOI] [PubMed] [Google Scholar]
- 13.Fukui, T., H. Atomi, T. Kanai, R. Matsumi, S. Fujiwara, and T. Imanaka. 2005. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 15:352-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta, R., Q. K. Beg, and P. Lorenz. 2002. Bacterial alkaline proteases: molecular approaches and industrial applications. Appl. Microbiol. Biotechnol. 59:15-32. [DOI] [PubMed] [Google Scholar]
- 15.Hussain, M., S. Ichihara, and S. Mizushima. 1982. Mechanism of signal peptide cleavage in the biosynthesis of the major lipoprotein of the Escherichia coli outer membrane. J. Biol. Chem. 257:5177-5182. [PubMed] [Google Scholar]
- 16.Ichihara, S., N. Beppu, and S. Mizushima. 1984. Protease IV, a cytoplasmic membrane protein of Escherichia coli, has signal peptide peptidase activity. J. Biol. Chem. 259:9853-9857. [PubMed] [Google Scholar]
- 17.Ichihara, S., T. Suzuki, M. Suzuki, and S. Mizushima. 1986. Molecular cloning and sequencing of the sppA gene and characterization of the encoded protease IV, a signal peptide peptidase, of Escherichia coli. J. Biol. Chem. 261:9405-9411. [PubMed] [Google Scholar]
- 18.Ito, S., T. Kobayashi, K. Ara, K. Ozaki, S. Kawai, and Y. Hatada. 1998. Alkaline detergent enzymes from alkaliphiles: enzymatic properties, genetics, and structures. Extremophiles 2:185-190. [DOI] [PubMed] [Google Scholar]
- 19.Kannan, Y., Y. Koga, Y. Inoue, M. Haruki, M. Takagi, T. Imanaka, M. Morikawa, and S. Kanaya. 2001. Active subtilisin-like protease from a hyperthermophilic archaeon in a form with a putative prosequence. Appl. Environ. Microbiol. 67:2445-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemberg, M. K., F. A. Bland, A. Weihofen, V. M. Braud, and B. Martoglio. 2001. Intramembrane proteolysis of signal peptides: an essential step in the generation of HLA-E epitopes. J. Immunol. 167:6441-6446. [DOI] [PubMed] [Google Scholar]
- 21.Lemberg, M. K., and B. Martoglio. 2004. On the mechanism of SPP-catalysed intramembrane proteolysis; conformational control of peptide bond hydrolysis in the plane of the membrane. FEBS Lett. 564:213-218. [DOI] [PubMed] [Google Scholar]
- 22.Maurer, K.-H. 2004. Detergent proteases. Curr. Opin. Biotechnol. 15:330-334. [DOI] [PubMed] [Google Scholar]
- 23.Morikawa, M., Y. Izawa, N. Rashid, T. Hoaki, and T. Imanaka. 1994. Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl. Environ. Microbiol. 60:4559-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng, S. Y. M., and K. F. Jarrell. 2003. Cloning and characterization of archaeal type I signal peptidase from Methanococcus voltae. J. Bacteriol. 185:5936-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novak, P., and I. K. Dev. 1988. Degradation of a signal peptide by protease IV and oligopeptidase A. J. Bacteriol. 170:5067-5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novak, P., P. H. Ray, and I. K. Dev. 1986. Localization and purification of two enzymes from Escherichia coli capable of hydrolyzing a signal peptide. J. Biol. Chem. 261:420-427. [PubMed] [Google Scholar]
- 27.Pacaud, M. 1982. Purification and characterization of two novel proteolytic enzymes in membranes of Escherichia coli. Protease IV and protease V. J. Biol. Chem. 257:4333-4339. [PubMed] [Google Scholar]
- 28.Paetzel, M., A. Karla, N. C. J. Strynadka, and R. E. Dalbey. 2002. Signal peptidases. Chem. Rev. 102:4549-4579. [DOI] [PubMed] [Google Scholar]
- 29.Ramakrishnan, V., and M. W. W. Adams. 1995. Preparation of genomic DNA from sulfur-dependent hyperthermophilic archaea, p. 95-96. In F. T. Robb and A. R. Place (ed.), Archaea—a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 30.Rashid, N., J. Cornista, S. Ezaki, T. Fukui, H. Atomi, and T. Imanaka. 2002. Characterization of an archaeal cyclodextrin glucanotransferase with a novel C-terminal domain. J. Bacteriol. 184:777-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rawlings, N. D., D. P. Tolle, and A. J. Barrett. 2004. MEROPS: the peptidase database. Nucleic Acids Res. 32:D160-D164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ring, G., and J. Eichler. 2004. Extreme secretion: protein translocation across the archaeal plasma membrane. J. Bioenerg. Biomembr. 36:35-45. [DOI] [PubMed] [Google Scholar]
- 33.Rosendal, K. R., K. Wild, G. Montoya, and I. Sinning. 2003. Crystal structure of the complete core of archaeal signal recognition particle and implications for interdomain communication. Proc. Natl. Acad. Sci. USA 100:14701-14706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki, T., A. Itoh, S. Ichihara, and S. Mizushima. 1987. Characterization of the sppA gene coding for protease IV, a signal peptide peptidase of Escherichia coli. J. Bacteriol. 169:2523-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tachibana, Y., M. M. Leclere, S. Fujiwara, M. Takagi, and T. Imanaka. 1996. Cloning and expression of the α-amylase gene from the hyperthermophilic archaeon Pyrococcus sp. KOD1, and characterization of the enzyme. J. Ferment. Bioeng. 82:224-232. [Google Scholar]
- 36.Tanaka, T., S. Fujiwara, S. Nishikori, T. Fukui, M. Takagi, and T. Imanaka. 1999. A unique chitinase with dual active sites and triple substrate binding sites from the hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1. Appl. Environ. Microbiol. 65:5338-5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanskul, S., K. Oda, H. Oyama, N. Noparatnaraporn, M. Tsunemi, and K. Takada. 2003. Substrate specificity of alkaline serine proteinase isolated from photosynthetic bacterium, Rubrivivax gelatinosus KDDS1. Biochem. Biophys. Res. Commun. 309:547-551. [DOI] [PubMed] [Google Scholar]
- 38.Van den Berg, B., W. M. Clemons, Jr., I. Collinson, Y. Modis, E. Hartmann, S. C. Harrison, and T. A. Rapoport. 2004. X-ray structure of a protein-conducting channel. Nature 427:36-44. [DOI] [PubMed] [Google Scholar]
- 39.Wolfe, M. S., and R. Kopan. 2004. Intramembrane proteolysis: theme and variations. Science 305:1119-1123. [DOI] [PubMed] [Google Scholar]