Abstract
Nucleotide excision repair and translesion DNA synthesis are two processes that operate at arrested replication forks to reduce the frequency of recombination and promote cell survival following UV-induced DNA damage. While nucleotide excision repair is generally considered to be error free, translesion synthesis can result in mutations, making it important to identify the order and conditions that determine when each process is recruited to the arrested fork. We show here that at early times following UV irradiation, the recovery of DNA synthesis occurs through nucleotide excision repair of the lesion. In the absence of repair or when the repair capacity of the cell has been exceeded, translesion synthesis by polymerase V (Pol V) allows DNA synthesis to resume and is required to protect the arrested replication fork from degradation. Pol II and Pol IV do not contribute detectably to survival, mutagenesis, or restoration of DNA synthesis, suggesting that, in vivo, these polymerases are not functionally redundant with Pol V at UV-induced lesions. We discuss a model in which cells first use DNA repair to process replication-arresting UV lesions before resorting to mutagenic pathways such as translesion DNA synthesis to bypass these impediments to replication progression.
Irradiation of cells with 254-nm UV light induces lesions that block DNA polymerases. Lesions that block polymerases are thought to either arrest the progress of the replication machinery or produce nascent-strand gaps depending on which template strand contains the lesion (3, 4, 17, 32, 45, 50, 53, 54). Several studies using plasmid substrates indicate that lesions in the leading-strand template arrest the overall progression of the replication fork, with the nascent lagging strand continuing a short distance beyond the arrested leading strand (17, 30, 50, 53). In contrast, lesions in the lagging-strand template are thought to generate gaps in the nascent DNA strand at sites opposite to the lesion, presumably because discontinuous synthesis of the lagging strand allows the blocked polymerase to reinitiate downstream of the lesion site (17, 30, 50). Events that are consistent with this can also be seen on the chromosome of UV-irradiated Escherichia coli. Following a moderate dose of UV irradiation, the rate of DNA synthesis is transiently inhibited before it efficiently recovers at a time that correlates with lesion removal (8, 45). During this period of inhibition, some limited DNA synthesis is still observed that contains gaps, consistent with replication continuing past a subset of the lesions in the template (13, 42, 43). The repair and restoration of the DNA template in each of these two situations may involve unique enzymatic pathways and are likely to have different consequences for the cell with respect to survival and mutagenesis.
Lesions that arrest the overall progression of the replication machinery would be expected to prevent the replication of the genome and are likely to result in cell lethality if the block to replication cannot be overcome. The ability of E. coli to survive doses of UV irradiation that produce thousands of lesions per genome clearly indicates that efficient mechanisms to deal with replication-arresting lesions exist in the cell. Several proteins associated with the recF pathway, including RecA, RecF, RecO, and RecR, are required to restore replication following arrest by UV-induced DNA lesions (6, 7, 10, 18, 38). In the absence of any of these genes, UV-irradiated cells fail to recover DNA synthesis following arrest, gaps persist in the DNA synthesized postirradiation, and the nascent DNA at the replication fork is extensively degraded (6-8, 10, 19, 38, 41, 47). In vitro, RecA, RecF, RecO, and RecR promote pairing between single-strand DNA and homologous duplex DNA (2, 22, 46, 56, 57), an activity that was originally characterized for its role in bringing together homologous strands of DNA during recombinational processes (5). Cellular assays indicate that the same enzymatic activity is also required during replication to maintain and process the homologous strands of the replication fork when the normal progression of the replication machinery is prevented (reviewed in reference 9). Other recF pathway proteins, RecQ, a 3′→5′ DNA helicase, and RecJ, a 5′→3′ single-strand exonuclease, selectively degrade the nascent lagging strand at blocked replication forks prior to the resumption of DNA synthesis (10). Degradation of nascent DNA by RecJ and RecQ facilitates the timely recovery of DNA synthesis in normal cells and is thought to play a role in suppressing the frequency of illegitimate recombination, perhaps by generating a more extensive substrate for RecA to bind and stabilize at the blocked replication fork (8, 10, 16; also unpublished observations). Consistent with this interpretation, RecQ homologs in yeast, Drosophila melanogaster, and humans have been shown to play critical roles in maintaining processive replication and suppressing the frequency of DNA strand exchanges (reviewed in reference 23). These observations have led to a general model in which RecA and several recF pathway gene products act to maintain and process the arrested replication fork so that repair enzymes or alternative DNA polymerases can gain access to the blocking lesion (6-8, 10). In this way, processive replication is maintained while avoiding strand exchanges that may lead to recombination or rearrangements.
Two mechanisms that operate to reduce the frequency of recombination and promote cell survival following DNA damage are nucleotide excision repair and translesion DNA synthesis (1, 7, 24, 42). Both processes have been proposed to operate at lesion-arrested replication forks to allow DNA synthesis to resume following arrest (7, 34, 37). In E. coli, the uvrA, uvrB, and uvrC gene products form an excinuclease that is required to initiate nucleotide excision repair of UV-induced lesions (reviewed in reference 44). Cells deficient in lesion removal are severely impaired in their ability to resume robust DNA replication and exhibit elevated levels of recombination, genomic rearrangements, and cell lethality (7, 8, 20, 45). In wild-type cells, the time at which robust replication resumes correlates with the removal of the lesions by nucleotide excision repair (8). These observations have been interpreted to support the idea that nucleotide excision repair is a prominent mechanism that operates at replication-arresting DNA lesions (8). However, since nucleotide excision repair is required to remove all lesions throughout the genome, it remains possible that an alternative process, such as translesion DNA synthesis, predominantly operates at lesion-arrested replication forks and that the failure to observe robust replication resumption in uvr mutants occurs due to the rearrest of replication at subsequent downstream lesions.
E. coli encodes three damage-inducible DNA polymerases that have multiple homologs in both prokaryotes and eukaryotes (49). In vitro, polymerase II (Pol II) (polB), Pol IV (dinB), and Pol V (umuDC) are able to incorporate bases opposite to specific lesions in template DNA with higher efficiency than the replicative polymerase, Pol III (34, 39, 52). There are differing reports in the literature as to the contribution of these polymerases to the resumption of DNA synthesis at arrested forks (25, 37, 59). An initial study reported that the recovery of DNA synthesis occurred more slowly in the absence of nucleotide excision repair following UV-induced damage but was not affected by the absence of Pol V (25). However, a subsequent study using repair-deficient mutants found that Pol V was essential and sufficient for DNA synthesis to resume in the absence of repair following exposure to low UV doses in a recA718 background (59). Still a third study reported that the absence of Pol II delayed the recovery of DNA synthesis after UV-induced damage, even when Pol V and nucleotide excision repair were functional (37). However, in an earlier study, this group did not find any contribution of Pol II to lesion bypass following UV irradiation (27). In addition, numerous studies have shown that Pol V, but not Pol II or Pol IV, increases the survival and frequency of mutagenesis in UV-irradiated E. coli (1, 11, 24, 48). These observations suggest that translesion DNA polymerases may operate at lesion-arrested replication forks and raise the possibility that translesion synthesis could be a predominant mechanism that restores DNA synthesis at lesion-arrested replication forks in wild-type cells. While it is apparent that the translesion DNA polymerases play an important role in cellular mutagenesis and genome stability after DNA damage, exactly where and when they operate in the cell are not clear. To date, no study has directly compared the activities of the translesion DNA polymerases and nucleotide excision repair to determine the time and frequency at which each process occurs following UV-induced arrest in wild-type cells.
Utilization of nucleotide excision repair or translesion DNA synthesis at an arrested fork may have very different biological consequences for the cell. Whereas lesion removal by nucleotide excision repair is generally considered to be error free, translesion DNA synthesis by enzymes such as Pol V is responsible for most of the mutagenesis that results from UV-induced DNA damage (24, 26). In this study, we characterized the recovery of replication following arrest in mutants lacking either nucleotide excision repair, the three damage-inducible DNA polymerases, or both to identify when these processes are recruited to the arrested fork in wild-type cells and gain a better understanding of how DNA replication is restored following disruption by UV-induced DNA damage.
MATERIALS AND METHODS
Bacterial strains.
All bacterial strains are in an SR108 background. SR108 is a thyA36 deoC2 derivative of W3110 (31). HL952 (SR108 uvrA::Tn10) and CL579 (SR108 recF6206::Tetr) have been described previously (7, 8). CL575 (SR108 umuC122::Tn5), CL632 (SR108 umuDC595::cat), CL634 (SR108 dinB::Kanr), and CL636 (SR108 polB::Ω Sm-Sp) were constructed by P1 transduction of umuC122::Tn5, umuDC595::cat, dinB::Kanr, and polB::Ω Sm-Sp from GW2100 (11), RW82 (60), MGZdinB (34), and MGZpolB (34), respectively, into SR108. CL637 (SR108 polB::Ω Sm-Sp dinB::Kanr) was constructed by P1 transduction of dinB::Kanr from MGZdinB (34) into CL636. CL646 (SR108 polB::Ω Sm-Sp dinB::Kanr umuDC595::cat) was constructed by P1 transduction of umuDC595::cat from RW82 into CL637. CL681 (SR108 polB::Ω Sm-Sp dinB::Kanr umuDC595::cat uvrA::Tn10) was constructed by P1 transduction of uvrA::Tn10 from HL952 into CL646. Phenotypes were confirmed by antibiotic resistance and, when appropriate, sensitivity to UV. Genotypes for polB, umuDC, and dinB strains were confirmed by PCR and Southern blot analysis.
UV survival.
UV irradiations used a 15-W germicidal lamp (254 nm) at an incident dose of 0.9 J/m2/s (0.2 J/m2/s for doses below 20 J/m2). Cells were grown in Davis medium supplemented with 0.4% glucose, 0.2% Casamino Acids, and 10 μg/ml thymine (DGCthy medium). Fresh overnight cultures were diluted 1:100 and grown to an optical density at 600 nm (OD600) of between 0.4 and 0.5 (approximately 6 × 108 cells/ml). Ten-microliter aliquots of serial 10-fold dilutions were applied as spots in triplicate on Luria-Bertani plates containing 10 μg/ml thymine and UV irradiated at the indicated doses. Viable colonies were counted following overnight incubation at 37°C.
UV-induced mutagenesis.
Mutagenesis induced by UV was measured by the appearance of rifampin-resistant colonies as a result of UV exposure. At least 69 base substitutions within the rpoB gene have been identified that confer resistance to rifampin, allowing one to monitor numerous UV-induced mutation sites in different sequence contexts (15). Overnight cultures were diluted 1:100 and grown in DGCthy medium to an OD600 of 0.4, at which point the culture was split into three equal fractions and irradiated with an incident dose of 0, 2, or 10 J/m2 UV. Following overnight incubation at 37°C, the cultures were plated on Luria-Bertani plates containing 10 μg/ml thymine and 100 μg/ml rifampin. Rifampin-resistant colonies were counted following overnight incubation at 37°C.
DNA synthesis and accumulation.
Overnight cultures were diluted 1:100 and grown in DGCthy medium supplemented with 0.1 μCi/ml of [14C]thymine to an OD600 of precisely 0.3, at which point half of the culture received an incident dose of 27 J/m2 while the other half was mock irradiated. At the times indicated, duplicate 0.5-ml aliquots of culture were pulse-labeled with 1 μCi/ml [3H]thymidine for 2 min at 37°C. Cells were then lysed, and the DNA was precipitated in cold 5% trichloroacetic acid (TCA) and filtered onto Millipore glass fiber filters. The amounts of 3H and 14C on each filter were determined by scintillation counting.
DNA degradation.
Overnight cultures were diluted 1:100 and grown in DGCthy medium supplemented with 0.1 μCi/ml [14C]thymine to an OD600 of 0.4. [3H]thymidine (1 μCi/ml) was then added to the culture. After 5 s, cells were filtered onto a 0.45-μm membrane, rinsed twice with 5 ml of NET buffer (100 mM NaCl, 10 mM Tris, pH 8.0, 10 mM EDTA, pH 8.0), resuspended in prewarmed nonradioactive DGCthy medium, and irradiated with a UV dose of 27 J/m2. At the times indicated, duplicate 0.2-ml aliquots (triplicate for 0 min) of the culture were precipitated in cold 5% TCA and filtered onto Millipore glass fiber filters. The amounts of 3H and 14C were determined as before.
Alkali sucrose gradients.
Overnight cultures were diluted 1:100 and grown in DGC medium supplemented with 0.9 μCi/4 μg/ml [14C]thymine to an OD600 of 0.4. The culture was then UV irradiated with 27 J/m2 before the addition of 9 μCi/ml [3H]thymidine for 5 min at 37°C. Cells were filtered onto a 0.45-μm membrane, rinsed with NET buffer, and resuspended in prewarmed nonradioactive DGCthy medium. At the times indicated, 0.5-ml aliquots of cells were collected into 0.5 ml ice-cold 2× NET, pelleted, and resuspended in 0.1 ml buffered sucrose (10 mM Tris, pH 8.0, 10 mM EDTA, pH 8.0, 110 mM NaCl, 5.1% sucrose), and stored on ice for the duration of the time course. The equivalent of 2.5 × 106 cells was layered on 4.9-ml linear gradients of 5 to 20% (wt/vol) sucrose in 0.1 N NaOH with a 0.1-ml top layer of 5% (wt/vol) Sarkosyl in 0.5 N NaOH. Gradients were centrifuged at 30,000 rpm at 20°C for 120 min in an SW55.1 Ti rotor. Each gradient was dripped onto strips of Whatman no. 17 paper. The strips were washed in 5% TCA and then 95% ethanol, and amounts of 3H and 14C were then determined as before.
RESULTS
The absence of nucleotide excision repair, but not translesion synthesis, impairs recovery of DNA synthesis following UV-induced damage.
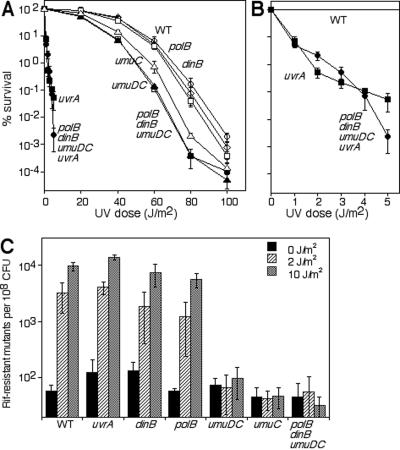
If translesion synthesis were the predominant mechanism by which cells recover replication following arrest, one would predict that the absence of the translesion DNA polymerases would significantly impair cell survival in the presence of DNA damage. Although several studies have examined the survival of individual polymerase mutants following DNA damage, it is possible that these proteins are functionally redundant at UV-induced lesions and that a phenotype would only be revealed in the absence of all three damage-inducible polymerases. To examine this possibility, we constructed isogenic mutants lacking Pol II (polB), Pol IV (dinB), Pol V (umuC and umuDC), or all three gene products. Consistent with previous studies (37), mutations affecting Pol V, but not Pol II or Pol IV, rendered cells modestly hypersensitive to irradiation at higher doses of UV (Fig. 1A). Interestingly, mutants lacking all three DNA polymerases were no more sensitive to UV irradiation than the Pol V single mutant. By comparison, a uvrA mutant was much more sensitive to UV irradiation than the triple-polymerase mutant, and the sensitivity of the quadruple-uvrA-polymerase mutant was similar to that of the uvrA mutant alone (Fig. 1A, B). Similar to previous studies (11, 24, 48, 51), when we measured the frequency of mutagenesis in cultures exposed to UV irradiation as monitored by cells acquiring resistance to the antibiotic rifampin, we found that Pol V, but not Pol II or Pol IV, was responsible for essentially all of the mutagenesis generated by UV-induced DNA damage (Fig. 1C). Thus, the absence of all three translesion DNA polymerases does not severely impair the survival of UV-irradiated E. coli. However, the observation that Pol V contributes to cell survival primarily at high doses of UV could indicate that it plays a critical role in the recovery of replication under conditions when the repair capacity of the cell has been exceeded. Alternatively, the observation that Pol V-dependent mutagenesis increases approximately 100-fold at low doses of UV (2 J/m2) that do not affect cell survival or exceed the repair capacity of the cell suggests that the polymerase is active even at these low doses in wild-type cells. This may imply that its preferred substrate may not relate to lesions that arrest replication forks or impair cell survival. The observation that Pol II and Pol IV do not contribute to survival or mutagenesis and were unable to compensate for the absence of Pol V argues that these polymerases are not functionally redundant with respect to survival or mutagenesis following UV irradiation in vivo.
FIG. 1.
Pol V is required for resistance and mutagenesis following UV irradiation. (A) The survival of parental (▪), polB (▴), dinB (□), umuDC (•), umuC (⋄), uvrA (○), polB dinB umuDC (▵), and polB dinB umuDC uvrA (♦) cultures is shown after UV irradiation at the indicated doses. (B) The survival of parental (▪), uvrA (○), and polB dinB umuDC uvrA (♦) cultures replotted on a different scale. Graphs represent an average of at least three independent experiments. Error bars represent 1 standard deviation. (C) Cultures were irradiated at the indicated doses and examined for the number of rifampin (Rif)-resistant colonies that appeared following an overnight incubation. The number of rifampin-resistant colonies that appeared per 108 cells is plotted. Graphs represent an average of four independent experiments. Error bars represent 1 standard deviation.
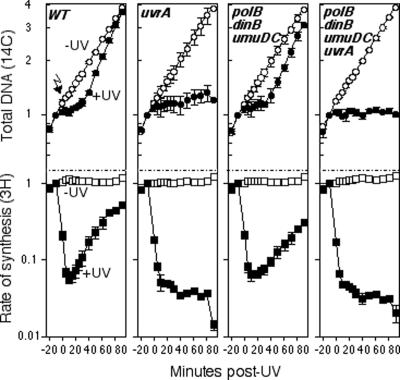
Impaired survival following DNA damage could result from a deficiency in any of several cellular processes and does not directly address the potential role that translesion synthesis may have at sites of replication arrest. Therefore, to examine the recovery of DNA synthesis in these mutants directly, we monitored the overall DNA accumulation and rate that synthesis recovered following UV-induced DNA damage. To this end, duplicate aliquots of [14C]thymine-labeled cultures were pulse-labeled for 2 min with [3H]thymidine at various times after 27 J/m2 UV irradiation. The rate of DNA synthesis (3H incorporation/2 min) could then be determined relative to the total amount of DNA present (14C incorporation) at specific times following treatment. Since the rate of DNA synthesis was found to vary significantly with cell density (data not shown), all experiments included a mock-irradiated control that allowed us to directly compare irradiated and unirradiated cultures and ensure that any observed differences were due to UV treatment rather than culture density. These irradiation conditions generated approximately one cyclobutane-pyrimidine dimer per 9-kb single-strand DNA as measured by T4 endonuclease V-sensitive sites in the DNA (31; data not shown), but did not significantly reduce the survival of wild-type cells (Fig. 1). By this assay, the rate of DNA synthesis in UV-irradiated wild-type cultures initially decreased by more than 90% but began to recover 15 min post-UV irradiation and continued to increase until it approached unirradiated levels, approximately 80 min post-UV irradiation (Fig. 2). At this time, the overall DNA accumulation also approached that of the unirradiated cultures. By comparison, while DNA synthesis was inhibited to a similar extent in uvrA mutants as in wild-type cells, no recovery in the rate of DNA synthesis occurred during the course of the experiment and very little, if any, further DNA accumulation was observed. Similar results were obtained after low doses of UV (5 J/m2). Following a low dose of UV, the rate of DNA synthesis in uvrA mutants was initially inhibited to a lesser extent, but no recovery in DNA synthesis rates was seen during the 90-min time course (data not shown).
FIG. 2.
Nucleotide excision repair, but not translesion DNA synthesis, is required for the recovery of DNA replication after UV irradiation. [3H]thymidine was added to [14C]thymine-prelabeled cultures for 2 min at the indicated times following either 27 J/m2 UV irradiation (filled symbols) or mock irradiation (open symbols) at time zero. The relative amounts of total DNA (14C; ○) and DNA synthesis/2 min (3H; □) are plotted. Graphs represent an average of at least three independent experiments. Error bars represent 1 standard deviation.
In contrast to uvrA, the absence of the damage-inducible polymerases did not affect the time at which DNA synthesis resumed. The overall rate that DNA synthesis increased occurred with a small but reproducible reduction in kinetics in the triple-polymerase mutant compared to wild-type cells; however, DNA synthesis began to resume in UV-irradiated polB dinB umuDC mutants at a similar time to that observed in wild-type cells (Fig. 2). In the absence of both uvrA and the polymerases, no recovery in the rate of DNA synthesis or further accumulation of DNA was observed to occur.
The lack of recovery in uvrA mutants could be interpreted to support a prominent role for nucleotide excision repair operating at lesion-arrested replication forks. However, this result does not exclude the possibility that translesion synthesis can occur at these sites, since the recovery in uvrA mutants may remain below the level of detection due to the persistence of lesions in the uvrA mutant genome. Despite this shortcoming, the recovery of DNA synthesis in the absence of all three damage-inducible polymerases does indicate that these polymerases are not essential for replication to resume following arrest by UV-induced DNA damage. Furthermore, the lack of any significant delay in the recovery argues against the idea that they are used as a predominant mechanism for replication to resume following arrest at UV-induced lesions.
Damage-inducible polymerases act at arrested replication forks in the absence of repair.
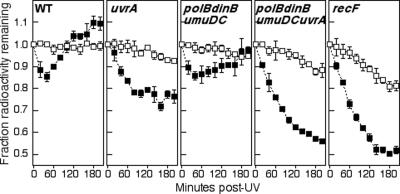
While the previous assay indicated that translesion synthesis is not essential for replication to resume following UV-induced damage, it lacked the sensitivity to determine if the damage-inducible DNA polymerases are capable of acting at sites of replication-arresting DNA lesions. To address this question, we examined the ability of the polymerases to protect the nascent DNA at the arrested replication fork from degradation. In previous studies, this assay has been used to show that the nascent lagging strand of arrested replication forks is subject to degradation by the RecJ nuclease and RecQ helicase at times prior to the resumption of replication (10). In cells that are able to recover replication, the observed degradation of the nascent DNA ceases at the time when replication resumes. However, in mutants that fail to resume DNA synthesis, the nascent DNA degradation continues and is much more extensive (6-8). To examine the degradation that occurs in the polymerase mutants, cultures grown in [14C]thymine were pulse-labeled with [3H]thymidine for 5 s, transferred to nonradioactive medium, and irradiated with 27 J/m2 UV. The amounts of 3H and 14C remaining in the DNA after irradiation were then monitored over time. The 14C label allowed us to compare the degradation that occurred in the overall genome to that which occurred specifically at the 3H-labeled DNA at the arrested fork. As seen in previous studies, the degradation ceased in wild-type cells between 20 to 40 min after irradiation and was limited to less than 20% of the nascent DNA (6, 7, 10) (Fig. 3). The increase in 3H-labeled DNA after 40 min occurs due to the reincorporation of the remaining intracellular pools of [3H]thymidine at the time of replication resumption (10). Comparatively, in recF mutants, which fail to resume replication following arrest (6-8), the nascent DNA degradation continued for more than 100 min until approximately half of the nascent DNA had been degraded (Fig. 3).
FIG. 3.
Increased degradation occurs at the growing fork after irradiation in polB dinB umuDC uvrA cells. [3H]thymidine was added to [14C]thymine-prelabeled cells for 5 s prior to irradiation with 27 J/m2 in nonlabeled medium. The fraction of radioactive nucleotides remaining in the DNA is plotted over time. The initial values for 3H and 14C were between 2,500 to 4,000 and 1,200 to 1,700 cpm, respectively, for all experiments. Graphs represent an average of at least three independent experiments. Error bars represent 1 standard deviation. Results for total DNA (14C; □) and nascent DNA (3H; ▪) are shown.
When we examined cultures of polB dinB umuDC mutants, we found that nascent DNA degradation was limited to the first 40 min and was comparable in extent to that observed in wild-type cells (Fig. 3), consistent with the idea that the time at which replication recovers is not affected by the absence of the polymerases. In contrast, the nascent DNA degradation continued beyond 60 min in the absence of uvrA but then ceased at a point that only moderately exceeded that which occurred in wild-type cells. Interestingly, the extensive nascent DNA degradation seen in recF mutants did not occur in the uvrA mutants despite the fact that robust replication does not detectably resume in either mutant. Surprisingly, when the damage-inducible polymerases were also inactivated, the nascent DNA continued to degrade in uvrA mutants and now exhibited a degradation pattern that was similar in duration and extent to that seen in recF mutants (Fig. 3). The observation that the damage-inducible polymerases are required to protect the nascent DNA from degradation in the absence of nucleotide excision repair suggests that translesion synthesis can act as an alternative to excision repair at sites of blocking DNA lesions. Furthermore, the extensive degradation in polB dinB umuDC uvrA mutants suggests that in the absence of either nucleotide excision repair or translesion DNA synthesis, the ability of the cell to elongate past the arresting lesion is severely compromised, leading to the eventual degradation of the nascent DNA.
Daughter-strand gap repair is delayed in the absence of the damage-inducible polymerases.
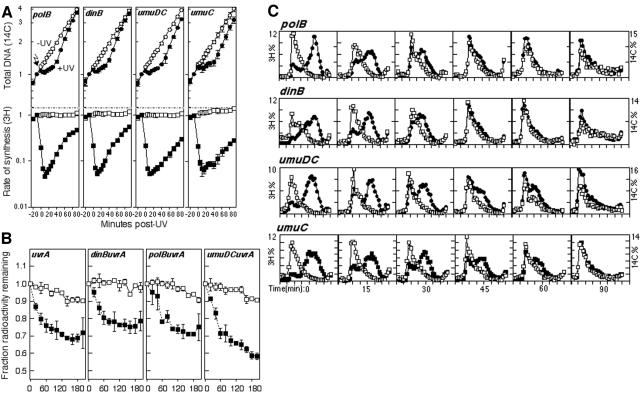
The assays described above would be expected to identify enzymes that specifically participate during the recovery of replication following arrest by a DNA lesion. However, several studies using plasmid substrates have suggested that a subset of DNA lesions, such as those in the lagging-strand template, fail to arrest replication and generate gaps in the daughter-strand DNA. It is reasonable to consider that the processing and repair of these gapped substrates may require a different subset of enzymatic pathways than those that act at arrested replication forks. Consistent with the gapped products observed on plasmids, a number of previous studies have used alkali sucrose gradient analysis to show that immediately after UV irradiation, a limited amount of DNA synthesis can be detected on the chromosome that also contains gaps (13, 42, 43). The repair (or joining) of the chromosomal gapped DNA fragments appears to depend on at least some of the same gene products that are required for the resumption of DNA synthesis, including RecA, RecF, RecO, and RecR (14, 41, 47). Several early studies characterizing the processing and repair of nascent-strand gaps were performed prior to the discovery of the concept that translesion synthesis or nucleotide excision repair may be involved in this pathway. For these reasons, most studies characterizing the repair of nascent-strand gaps were done with uvr mutants and the potential contribution that nucleotide excision repair or the damage-inducible polymerases may have in this process has not been considered or examined.
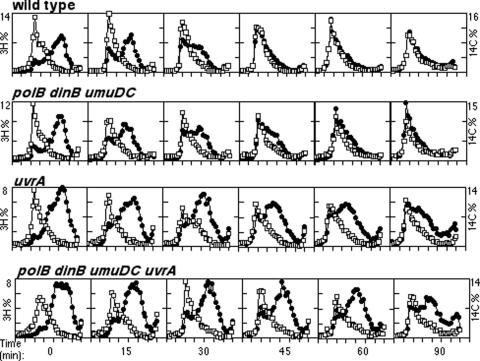
To examine the repair of the postirradiation DNA fragments in these mutants, a 5-min pulse of [3H]thymidine was added to 14C-prelabeled cultures immediately after UV irradiation with 27 J/m2. Cultures were then placed into nonradioactive media, and at the indicated times, cells were lysed and the size of the labeled DNA fragments was analyzed on alkaline sucrose gradients. Whereas the pulse-labeled 3H-DNA cosedimented with the large 14C-labeled genomic DNA in mock-irradiated cultures (data not shown), the DNA synthesized immediately after irradiation consisted of smaller, slower-migrating fragments in UV-irradiated cultures (Fig. 4). The average size of the smaller 3H-labeled fragments was 5 kb, while the size of the 14C-labeled genomic DNA averaged greater than 150 kb (13; data not shown). In wild-type cultures, the size of the postirradiation DNA fragments returned to that of the overall genomic DNA within 45 min of irradiation. However, in polB dinB umuDC mutants, we observed a modest but reproducible delay (of approximately 15 min) before the nascent-strand gaps were repaired, despite the timely recovery of DNA synthesis at the arrested replication forks of these mutants as demonstrated by our previous assays (Fig. 4). We interpret this observation to indicate that translesion synthesis participates in and at least partially contributes to the joining of the observed nascent-strand gaps in wild-type cells.
FIG. 4.
Nucleotide excision repair and translesion DNA synthesis are required for nascent DNA gap filling. The size of the DNA synthesized immediately after irradiation was analyzed by alkali sucrose gradients over time. [14C]thymine-prelabeled cells were irradiated at a dose of 27 J/m2, pulse-labeled with [3H]thymidine for 5 min, and then filtered into nonlabeled medium. Cultures collected immediately after 3H labeling are referred to here as time zero. Amounts of 3H and 14C in each fraction are plotted as a percentage of the total counts in each gradient. 3H and 14C values were between 4,200 to 9,500 and 1,500 to 4,300 cpm per gradient, respectively. Results for DNA synthesized before irradiation (14C; □) and DNA synthesized after treatment (3H; •) are shown. Graphs represent one of at least three independent experiments.
Comparatively, a large portion of the DNA synthesized postirradiation in irradiated uvrA cultures persisted as small fragments throughout the duration of the experiment. Consistent with earlier studies, the size of the postirradiation DNA fragments began to increase at later times, although the average size remained smaller than that of the genomic DNA throughout the 90-min time course in uvrA mutants (13, 43) (Fig. 4). This observation may indicate a potential role for nucleotide excision repair in processing lesions prior to nascent-strand gap repair. However, the delayed gap joining in uvrA mutants could also suggest that the repair of the nascent-strand gaps is coupled to or dependent on the efficient resumption of replication following arrest. In uvrA mutants that also lacked the damage-inducible polymerases, the impaired gap joining was similar in extent to that observed in the uvrA mutants (Fig. 4).
UmuDC participates in the recovery of DNA synthesis and daughter-strand gap repair following UV irradiation.
To identify which of the damage-inducible DNA polymerases acts at arrested replication forks and participates in daughter-strand gap repair following UV-induced DNA damage, we examined mutants that lacked either Pol II (polB), Pol IV (dinB), or Pol V (umuDC) using the assays described above. As shown in Fig. 5A, both polB and dinB mutants recovered DNA synthesis with kinetics that were identical to those of wild-type cultures. By comparison, in umuDC mutants, although the recovery began at a similar time, it occurred with a modest reduction in kinetics that was identical to that in the mutant lacking all three damage-inducible polymerases. Identical results were obtained when we examined a umuC mutant that maintains and expresses a functional copy of the UmuD subunit, but is unable to perform translesion DNA synthesis (11, 35). In addition, we found that following UV irradiation, the presence of Pol V was able to protect the arrested replication fork from degradation when excision repair was absent, whereas the degradation was not affected by the presence or absence of Pol II or Pol IV (Fig. 5B). Similarly, both umuDC and umuC mutants also exhibited a 15-min delay before the postirradiation DNA fragments were fully repaired, whereas no delay was detected in either polB or dinB mutants (Fig. 5C).
FIG. 5.
Pol V contributes to the rate that DNA synthesis resumes, protection of the replication fork in the absence of repair, and daughter-strand gap repair after UV irradiation. (A) Data were obtained and plotted as in Fig. 2. Each graph represents an average of at least three independent experiments. Error bars represent 1 standard deviation. Total DNA (14C) in mock-irradiated cultures (○), total DNA in irradiated cultures (•), the rate of DNA synthesis (3H) in mock-irradiated cultures (□), and the rate of DNA synthesis in irradiated cultures (▪) are shown. (B) Data were obtained and plotted as in Fig. 3. The initial values for 3H and 14C were between 750 to 1,200 and 800 to 1,200 cpm, respectively, for all experiments. Results for total DNA (14C; □) and nascent DNA (3H; ▪) are shown. Each graph represents an average of at least three independent experiments. Error bars represent 1 standard deviation. (C) Data were obtained and plotted as in Fig. 4. 3H and 14C values were between 6,700 to 11,000 and 2,500 to 4,000 cpm per gradient, respectively. Results for DNA synthesized before irradiation (14C; ▪) and DNA synthesized after treatment (3H; •) are shown. Graphs represent one of at least three independent experiments.
We interpret these observations to indicate that following UV-induced DNA damage, Pol V is able to act at lesion-arrested replication forks and promote daughter-strand gap repair. In addition, the observations argue against a functionally redundant role for Pol II or Pol IV with Pol V at sites of UV-induced DNA damage.
DISCUSSION
Both translesion DNA synthesis and nucleotide excision repair have been postulated to act on replication-arresting DNA lesions to promote the recovery of DNA synthesis and reduce the frequency of recombination in wild-type cells. In this study, we show that both processes contribute to replication recovery, but with different efficiencies and kinetics. A role for nucleotide excision repair acting at early times during recovery is supported by the observations that the resumption of DNA synthesis following arrest is severely impaired in the absence of repair enzymes but occurs with nearly wild-type kinetics in the absence of all three damage-inducible DNA polymerases (Fig. 2). Furthermore, the degradation of the DNA at the arrested fork occurs for a longer duration when the recovery process depends solely on translesion synthesis, as in nucleotide excision repair mutants (Fig. 3).
In contrast, several observations are consistent with the idea that translesion synthesis by Pol V can function at the arrested replication fork substrates at later times or as an alternative pathway when the repair capacity of the cell has been exceeded. The hypersensitivity of umuDC mutants is distinct from that of many other UV-sensitive mutants in that it only becomes prominent at higher doses of UV irradiation (Fig. 1A). At lower UV doses, umuDC mutants survive as well as the parental strain and exhibit a similar “shoulder” in their survival curves, consistent with the idea that Pol V becomes essential for survival when the repair capacity of the cell has been exceeded. Furthermore, although robust replication did not resume in the absence of lesion removal, the presence of Pol V reduced the degradation that occurred at the replication fork (Fig. 3 and 5B), suggesting that it is capable of synthesizing past arresting UV lesions, albeit with an efficiency that remains far below that seen during normal replication on undamaged templates.
The data we present indicate that nucleotide excision repair contributes to the recovery of replication at early times after UV irradiation. Although Pol V is not essential for replication to resume, a large induction of Pol V-dependent mutagenesis occurs even after low doses of UV irradiation (Fig. 1C), strongly arguing that Pol V actively participates in some form of DNA synthesis that occurs after UV.
One attractive model to consider is that the prominent replication substrates targeted by nucleotide excision repair and Pol V may be distinct. The observation that the repair of daughter-strand gaps is delayed in Pol V mutants, despite the timely resumption of DNA synthesis, is consistent with the idea that Pol V may predominantly target DNA gaps produced by nonarresting UV-induced lesions. The preferential targeting of Pol V to nonarresting lesions is also more consistent with the modest UV hypersensitivity exhibited in the Pol V mutants. In vitro, efficient translesion synthesis by Pol V requires that the gapped substrate contain a RecA-bound filament and a beta clamp loaded at the site of the lesion (36, 39, 40, 52). Based upon our current understanding of replication fork dynamics, this is similar to the substrate that is expected to be generated following replication through lesions on lagging-strand templates in vivo.
In addition to its role in translesion synthesis, overexpression of UmuD has an inhibitory effect on growth at low temperatures and on exiting stationary phase (28, 33). These observations have led to the proposal that UmuD may function as a cell cycle checkpoint, preventing replication from resuming prematurely in the presence of DNA damage (35). If the protein did act as a damage checkpoint, one would predict that replication would resume more rapidly in the absence of the checkpoint protein than in its presence. In the results presented here, the rate of recovery was modestly hindered in the absence of UmuD (Fig. 5A, B, and C), suggesting that the checkpoint function of UmuD may not be delaying replication recovery from the arrested fork substrate. However, this does not rule out that UmuD may delay synthesis from occurring at other substrates or act during other phases of the culture growth, such as appears to occur during the exit from stationary phase (33).
We observed that Pol V, but not Pol II or Pol IV, detectably contributes to the survival, mutagenesis, resumption of DNA synthesis, and nascent-strand gap repair following UV irradiation in vivo (Fig. 5). These observations are consistent with work by Khidir et al. (25) in which they reported as data not shown that uvrA mutants but not umuC mutants recovered DNA synthesis more slowly than wild-type cells. Using a background that contained a recA718 allele, Witkin et al. (59) found that in the absence of nucleotide excision repair, umuC was essential and sufficient for DNA synthesis to resume. Although it remains difficult to clearly interpret why the phenotype was dependent on the recA718 allele, which has a complex phenotype (29, 58), the observation is consistent with the results presented here, in which Pol V contributes to the recovery of replication when excision repair cannot occur. A previous study observed that mutations in polB delayed the recovery of DNA synthesis by 50 min (37). However in an earlier study, this group did not observe any Pol II-mediated translesion synthesis on UV-damaged templates (27), and in this study, we did not detect a contribution by Pol II at the arrested fork as measured by either the recovery of DNA synthesis, nascent-strand gap repair, or protection of the nascent DNA following arrest. This difference could either be due to experimental conditions or secondary mutations in the strains utilized. The previously described delay in the recovery of polB mutant strain STL1336 was based on a single experiment in which the culture was irradiated early in the growth phase (OD450 of 0.08 equivalent to OD600 of ∼0.06). In our hands, we had difficulty monitoring growth of the culture at this OD and therefore chose to irradiate cultures at an OD600 of 0.3. We also routinely divided cultures at the time of treatment to include an unirradiated control so that the rate of synthesis after irradiation could be directly compared to the rate occurring in an equivalent unirradiated culture. When we irradiated STL1336 at an OD600 of 0.3, DNA synthesis recovered with kinetics similar to those of wild-type cells (data not shown). Previous studies from this group have also reported that the polB strain, STL1336, is prone to accumulate suppressor mutations that alter its response to DNA damage (12). To address the possibility of suppressors in our study, we constructed a new polB::tet deletion and obtained results identical to those shown in Fig. 4 (data not shown). Furthermore, to reduce the potential of suppressor mutations accumulating, strains were frozen immediately after construction and grown the day prior to each experiment. In constructing our polB mutants, we did not observe any reduction in P1 transduction efficiency or growth impairment that may be expected to occur if suppressor mutations arose in culture populations.
The lack of a phenotype for Pol II and Pol IV highlights the need to consider that these polymerases may not be functionally redundant in vivo. The possibilities that they may act during different temporal phases of the cell cycle, on substrates that are unrelated to the recovery of DNA synthesis or specifically on different forms of DNA damage, should not be excluded from consideration. The last possibility is consistent with the observation that the mutations produced by different forms of DNA damage vary, depending on which translesion polymerases are present in the cell (34). Other studies have also suggested that both Pol II and Pol IV are active on undamaged templates in the absence of the replicative polymerase, Pol III (37, 55). The mutagenesis, survival, recovery of replication, and daughter-strand gap repair presented here, as well as previous studies on plasmid substrates (34), are consistent with the idea that Pol V is able to productively bypass lesions generated by 254-nm UV irradiation in vivo.
Although UV-induced lesions appear to specifically require Pol V, there are clear examples, in both E. coli and humans, where there is a functional redundancy in the ability of more than one polymerase to bypass a specific form of DNA damage (34). It is also clear that the competitive or preferential order by which this redundancy occurs can have serious consequences on genomic stability. In humans, Pol η appears to bypass UV-induced DNA damage with relatively high fidelity. However, in its absence, translesion synthesis is achieved by alternative DNA polymerases with much less accuracy and results in the severe cancer-prone phenotype exhibited by patients with the variant form of xeroderma pigmentosum (21).
By analogy, the results presented here suggest that in E. coli, lesions encountered during replication are initially processed with high fidelity through the nucleotide excision repair pathway. At later times, when the cell either cannot or fails to remove the lesions in a timely fashion, these lesions can be bypassed alternatively by Pol V, in a process that is associated with an increased likelihood of mutagenesis. The delayed expression of the active UmuD′ subunit of Pol V following UV irradiation has been previously hypothesized to allow nucleotide excision repair more time to remove DNA lesions (35) and would also be consistent with the results presented here. Thus, in the case of UV-induced damage, in both E. coli and humans, the pathways operating to process lesions encountered during replication appear to be ordered in such a way as to give nonmutagenic pathways priority over those associated with higher mutation frequencies. It will be interesting to see if the same relationship holds true for other forms of DNA damage that are encountered environmentally or therapeutically, which may depend on repair enzymes or alternative polymerases to restore damaged genomic templates.
Acknowledgments
We thank M. Goodman, M. Cox, and A. Ganesan for helpful discussions of the manuscript.
Our work is supported by CAREER award MCB-0448315 from the National Science Foundation and award F32 GM068566 (to C.T.C.) from the NIH-NIGMS.
REFERENCES
- 1.Bagg, A., C. J. Kenyon, and G. C. Walker. 1981. Inducibility of a gene product required for UV and chemical mutagenesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 78:5749-5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bork, J. M., M. M. Cox, and R. B. Inman. 2001. The RecOR proteins modulate RecA protein function at 5′ ends of single-stranded DNA. EMBO J. 20:7313-7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carty, M. P., C. W. Lawrence, and K. Dixon. 1996. Complete replication of plasmid DNA containing a single UV-induced lesion in human cell extracts. J. Biol. Chem. 271:9637-9647. [DOI] [PubMed] [Google Scholar]
- 4.Chan, G. L., P. W. Doetsch, and W. A. Haseltine. 1985. Cyclobutane pyrimidine dimers and (6-4) photoproducts block polymerization by DNA polymerase I. Biochemistry 24:5723-5728. [DOI] [PubMed] [Google Scholar]
- 5.Clark, A. J., and A. D. Margulies. 1965. Isolation and characterization of recombination-deficient mutants of Escherichia coli K12. Proc. Natl. Acad. Sci. USA 53:451-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Courcelle, J., C. Carswell-Crumpton, and P. C. Hanawalt. 1997. recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc. Natl. Acad. Sci. USA 94:3714-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Courcelle, J., D. J. Crowley, and P. C. Hanawalt. 1999. Recovery of DNA replication in UV-irradiated Escherichia coli requires both excision repair and RecF protein function. J. Bacteriol. 181:916-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Courcelle, J., J. R. Donaldson, K. H. Chow, and C. T. Courcelle. 2003. DNA damage-induced replication fork regression and processing in Escherichia coli. Science 299:1064-1067. [DOI] [PubMed] [Google Scholar]
- 9.Courcelle, J., A. K. Ganesan, and P. C. Hanawalt. 2001. Therefore, what are recombination proteins there for? Bioessays 23:463-470. [DOI] [PubMed] [Google Scholar]
- 10.Courcelle, J., and P. C. Hanawalt. 1999. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol. Gen. Genet. 262:543-551. [DOI] [PubMed] [Google Scholar]
- 11.Elledge, S. J., and G. C. Walker. 1983. Proteins required for ultraviolet light and chemical mutagenesis. Identification of the products of the umuC locus of Escherichia coli. J. Mol. Biol. 164:175-192. [DOI] [PubMed] [Google Scholar]
- 12.Escarceller, M., J. Hicks, G. Gudmundsson, G. Trump, D. Touati, S. Lovett, P. L. Foster, K. McEntee, and M. F. Goodman. 1994. Involvement of Escherichia coli DNA polymerase II in response to oxidative damage and adaptive mutation. J. Bacteriol. 176:6221-6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganesan, A. K. 1974. Persistence of pyrimidine dimers during post-replication repair in ultraviolet light-irradiated Escherichia coli. J. Mol. Biol. 87:103-119. [DOI] [PubMed] [Google Scholar]
- 14.Ganesan, A. K., and P. C. Seawell. 1975. The effect of lexA and recF mutations on post-replication repair and DNA synthesis in Escherichia coli K-12. Mol. Gen. Genet. 141:189-205. [DOI] [PubMed] [Google Scholar]
- 15.Garibyan, L., T. Huang, M. Kim, E. Wolff, A. Nguyen, T. Nguyen, A. Diep, K. Hu, A. Iverson, H. Yang, and J. H. Miller. 2003. Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair (Amsterdam) 2:593-608. [DOI] [PubMed] [Google Scholar]
- 16.Hanada, K., T. Ukita, Y. Kohno, K. Saito, J. Kato, and H. Ikeda. 1997. RecQ DNA helicase is a suppressor of illegitimate recombination in Escherichia coli. Proc. Natl. Acad. Sci. USA 94:3860-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higuchi, K., T. Katayama, S. Iwai, M. Hidaka, T. Horiuchi, and H. Maki. 2003. Fate of DNA replication fork encountering a single DNA lesion during oriC plasmid DNA replication in vitro. Genes Cells 8:437-449. [DOI] [PubMed] [Google Scholar]
- 18.Horii, Z., and A. J. Clark. 1973. Genetic analysis of the recF pathway to genetic recombination in Escherichia coli K12: isolation and characterization of mutants. J. Mol. Biol. 80:327-344. [DOI] [PubMed] [Google Scholar]
- 19.Horii, Z., and K. Suzuki. 1968. Degradation of the DNA of Escherichia coli K12 rec− (JC1569b) after irradiation with ultraviolet light. Photochem. Photobiol. 8:93-105. [DOI] [PubMed] [Google Scholar]
- 20.Howard-Flanders, P., L. Theriot, and J. B. Stedeford. 1969. Some properties of excision-defective recombination-deficient mutants of Escherichia coli K-12. J. Bacteriol. 97:1134-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kannouche, P., and A. Stary. 2003. Xeroderma pigmentosum variant and error-prone DNA polymerases. Biochimie 85:1123-1132. [DOI] [PubMed] [Google Scholar]
- 22.Kantake, N., M. V. Madiraju, T. Sugiyama, and S. C. Kowalczykowski. 2002. Escherichia coli RecO protein anneals ssDNA complexed with its cognate ssDNA-binding protein: a common step in genetic recombination. Proc. Natl. Acad. Sci. USA 99:15327-15332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karow, J. K., L. Wu, and I. D. Hickson. 2000. RecQ family helicases: roles in cancer and aging. Curr. Opin. Genet. Dev. 10:32-38. [DOI] [PubMed] [Google Scholar]
- 24.Kato, T., and Y. Shinoura. 1977. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol. Gen. Genet. 156:121-131. [DOI] [PubMed] [Google Scholar]
- 25.Khidhir, M. A., S. Casaregola, and I. B. Holland. 1985. Mechanism of transient inhibition of DNA synthesis in ultraviolet-irradiated E. coli: inhibition is independent of recA whilst recovery requires RecA protein itself and an additional, inducible SOS function. Mol. Gen. Genet. 199:133-140. [DOI] [PubMed] [Google Scholar]
- 26.Kim, S. R., G. Maenhaut-Michel, M. Yamada, Y. Yamamoto, K. Matsui, T. Sofuni, T. Nohmi, and H. Ohmori. 1997. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc. Natl. Acad. Sci. USA 94:13792-13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kow, Y. W., G. Faundez, S. Hays, C. A. Bonner, M. F. Goodman, and S. S. Wallace. 1993. Absence of a role for DNA polymerase II in SOS-induced translesion bypass of φX174. J. Bacteriol. 175:561-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marsh, L., and G. C. Walker. 1985. Cold sensitivity induced by overproduction of UmuDC in Escherichia coli. J. Bacteriol. 162:155-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCall, J. O., E. M. Witkin, T. Kogoma, and V. Roegner-Maniscalco. 1987. Constitutive expression of the SOS response in recA718 mutants of Escherichia coli requires amplification of RecA718 protein. J. Bacteriol. 169:728-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McInerney, P., and M. O'Donnell. 2004. Functional uncoupling of twin polymerases: mechanism of polymerase dissociation from a lagging-strand block. J. Biol. Chem. 279:21543-21551. [DOI] [PubMed] [Google Scholar]
- 31.Mellon, I., and P. C. Hanawalt. 1989. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature 342:95-98. [DOI] [PubMed] [Google Scholar]
- 32.Mitchell, D. L., and R. S. Nairn. 1989. The biology of the (6-4) photoproduct. Photochem. Photobiol. 49:805-819. [DOI] [PubMed] [Google Scholar]
- 33.Murli, S., T. Opperman, B. T. Smith, and G. C. Walker. 2000. A role for the umuDC gene products of Escherichia coli in increasing resistance to DNA damage in stationary phase by inhibiting the transition to exponential growth. J. Bacteriol. 182:1127-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Napolitano, R., R. Janel-Bintz, J. Wagner, and R. P. Fuchs. 2000. All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J. 19:6259-6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Opperman, T., S. Murli, B. T. Smith, and G. C. Walker. 1999. A model for a umuDC-dependent prokaryotic DNA damage checkpoint. Proc. Natl. Acad. Sci. USA 96:9218-9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pham, P., J. G. Bertram, M. O'Donnell, R. Woodgate, and M. F. Goodman. 2001. A model for SOS-lesion-targeted mutations in Escherichia coli. Nature 409:366-370. [DOI] [PubMed] [Google Scholar]
- 37.Rangarajan, S., R. Woodgate, and M. F. Goodman. 1999. A phenotype for enigmatic DNA polymerase II: a pivotal role for pol II in replication restart in UV-irradiated Escherichia coli. Proc. Natl. Acad. Sci. USA 96:9224-9229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rangarajan, S., R. Woodgate, and M. F. Goodman. 2002. Replication restart in UV-irradiated Escherichia coli involving pols II, III, V, PriA, RecA and RecFOR proteins. Mol. Microbiol. 43:617-628. [DOI] [PubMed] [Google Scholar]
- 39.Reuven, N. B., G. Arad, A. Maor-Shoshani, and Z. Livneh. 1999. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD′, RecA, and SSB and is specialized for translesion replication. J. Biol. Chem. 274:31763-31766. [DOI] [PubMed] [Google Scholar]
- 40.Reuven, N. B., G. Tomer, and Z. Livneh. 1998. The mutagenesis proteins UmuD′ and UmuC prevent lethal frameshifts while increasing base substitution mutations. Mol. Cell 2:191-199. [DOI] [PubMed] [Google Scholar]
- 41.Rothman, R. H., and A. J. Clark. 1977. The dependence of postreplication repair on uvrB in a recF mutant of Escherichia coli K-12. Mol. Gen. Genet. 155:279-286. [DOI] [PubMed] [Google Scholar]
- 42.Rupp, W. D., and P. Howard-Flanders. 1968. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J. Mol. Biol. 31:291-304. [DOI] [PubMed] [Google Scholar]
- 43.Rupp, W. D., C. E. I. Wilde, D. L. Reno, and P. Howard-Flanders. 1971. Exchanges between DNA strand in ultraviolet-irradiated Escherichia coli. J. Mol. Biol. 61:25-44. [DOI] [PubMed] [Google Scholar]
- 44.Sancar, A. 1996. DNA excision repair. Annu. Rev. Biochem. 65:43-81. [DOI] [PubMed] [Google Scholar]
- 45.Setlow, R. B., P. A. Swenson, and W. L. Carrier. 1963. Thymine dimers and inhibition of DNA synthesis by ultraviolet irradiation of cells. Science 142:1464-1466. [DOI] [PubMed] [Google Scholar]
- 46.Shan, Q., J. M. Bork, B. L. Webb, R. B. Inman, and M. M. Cox. 1997. RecA protein filaments: end-dependent dissociation from ssDNA stabilization by RecO and RecR proteins. J. Mol. Biol. 265:519-540. [DOI] [PubMed] [Google Scholar]
- 47.Smith, K. C., and D. H. Meun. 1970. Repair of radiation-induced damage in Escherichia coli. I. Effect of rec mutations on post-replication repair of damage due to ultraviolet radiation. J. Mol. Biol. 51:459-472. [DOI] [PubMed] [Google Scholar]
- 48.Steinborn, G. 1978. Uvm mutants of Escherichia coli K12 deficient in UV mutagenesis. I. Isolation of uvm mutants and their phenotypical characterization in DNA repair and mutagenesis. Mol. Gen. Genet. 165:87-93. [DOI] [PubMed] [Google Scholar]
- 49.Sutton, M. D., and G. C. Walker. 2001. Managing DNA polymerases: coordinating DNA replication, DNA repair, and DNA recombination. Proc. Natl. Acad. Sci. USA 98:8342-8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Svoboda, D. L., and J. M. Vos. 1995. Differential replication of a single, UV-induced lesion in the leading or lagging strand by a human cell extract: fork uncoupling or gap formation. Proc. Natl. Acad. Sci. USA 92:11975-11979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang, M., P. Pham, X. Shen, J. S. Taylor, M. O'Donnell, R. Woodgate, and M. F. Goodman. 2000. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature 404:1014-1018. [DOI] [PubMed] [Google Scholar]
- 52.Tang, M., X. Shen, E. G. Frank, M. O'Donnell, R. Woodgate, and M. F. Goodman. 1999. UmuD′(2)C is an error-prone DNA polymerase, Escherichia coli pol V. Proc. Natl. Acad. Sci. USA 96:8919-8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Veaute, X., G. Mari-Giglia, C. W. Lawrence, and A. Sarasin. 2000. UV lesions located on the leading strand inhibit DNA replication but do not inhibit SV40 T-antigen helicase activity. Mutat. Res. 459:19-28. [DOI] [PubMed] [Google Scholar]
- 54.Veaute, X., and A. Sarasin. 1997. Differential replication of a single N-2-acetylaminofluorene lesion in the leading or lagging strand DNA in a human cell extract. J. Biol. Chem. 272:15351-15357. [DOI] [PubMed] [Google Scholar]
- 55.Viguera, E., M. Petranovic, D. Zahradka, K. Germain, D. S. Ehrlich, and B. Michel. 2003. Lethality of bypass polymerases in Escherichia coli cells with a defective clamp loader complex of DNA polymerase III. Mol. Microbiol. 50:193-204. [DOI] [PubMed] [Google Scholar]
- 56.Webb, B. L., M. M. Cox, and R. B. Inman. 1995. An interaction between the Escherichia coli RecF and RecR proteins dependent on ATP and double-stranded DNA. J. Biol. Chem. 270:31397-31404. [DOI] [PubMed] [Google Scholar]
- 57.Webb, B. L., M. M. Cox, and R. B. Inman. 1997. Recombinational DNA repair: the RecF and RecR proteins limit the extension of RecA filaments beyond single-strand DNA gaps. Cell 91:347-356. [DOI] [PubMed] [Google Scholar]
- 58.Witkin, E. M., J. O. McCall, M. R. Volkert, and I. E. Wermundsen. 1982. Constitutive expression of SOS functions and modulation of mutagenesis resulting from resolution of genetic instability at or near the recA locus of Escherichia coli. Mol. Gen. Genet. 185:43-50. [DOI] [PubMed] [Google Scholar]
- 59.Witkin, E. M., V. Roegner-Maniscalco, J. B. Sweasy, and J. O. McCall. 1987. Recovery from ultraviolet light-induced inhibition of DNA synthesis requires umuDC gene products in recA718 mutant strains but not in recA+ strains of Escherichia coli. Proc. Natl. Acad. Sci. USA 84:6805-6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Woodgate, R. 1992. Construction of a umuDC operon substitution mutation in Escherichia coli. Mutat. Res. 281:221-225. [DOI] [PubMed] [Google Scholar]