Abstract
Archaea encode a DNA ligase composed of a C-terminal catalytic domain typical of ATP-dependent ligases plus an N-terminal domain similar to that found in eukaryotic cellular and poxvirus DNA ligases. All archaeal DNA ligases characterized to date have ATP-dependent adenylyltransferase and nick-joining activities. However, recent reports of dual-specificity ATP/NAD+ ligases in two Thermococcus species and Pyrococcus abyssi and an ATP/ADP ligase in Aeropyrum pernix raise the prospect that certain archaeal enzymes might exemplify an undifferentiated ancestral stage in the evolution of ligase substrate specificity. Here we analyze the biochemical properties of Pyrococcus horikoshii DNA ligase. P. horikoshii ligase catalyzes autoadenylylation and nick sealing in the presence of a divalent cation and ATP; it is unable to utilize NAD+ or ADP to promote ligation in lieu of ATP. P. horikoshii ligase is thermophilic in vitro, with optimal adenylyltransferase activity at 90°C and nick-joining activity at 70 to 90°C. P. horikoshii ligase resembles the ligases of Methanobacterium thermautotrophicum and Sulfolobus shibatae in its strict specificity for ATP.
DNA ligases are essential components of the DNA replication, repair, and recombination machinery in all domains of the phylogenetic tree: eucarya, archaea, and bacteria. Ligases seal 3′ OH and 5′ PO4 ends via three nucleotidyl transfer reactions (10). First, a lysine nucleophile on the enzyme attacks the α-phosphorus of ATP or NAD+, which results in the formation of a covalent ligase-adenylate intermediate and the release of pyrophosphate or nicotinamide mononucleotide. Second, attack by the 5′ PO4 on ligase-adenylate results in expulsion of the active-site lysine and formation of an activated DNA-adenylate intermediate. Third, ligase catalyzes the attack of a 3′ OH on the DNA-adenylate, resulting in the release of AMP and formation of a phosphodiester.
DNA ligases are grouped into two families, depending on their requirement for ATP or NAD+ in the ligase adenylylation reaction. Whereas NAD+-dependent ligases are found only in bacteria and eukaryotic viruses, ATP-dependent DNA ligases are found in bacteria (and bacteriophages), eucarya (and eukaryotic viruses), and archaea (8, 10, 21, 27). A core ligase module composed of nucleotidyltransferase and OB-fold domains is shared by all known DNA ligases (3, 15, 16, 24). The adenylate-binding pocket is located within the nucleotidyltransferase domain and is composed of five conserved peptide motifs (19). The substrate specificity of NAD+-dependent ligase is dictated by a unique domain that binds the nicotinamide mononucleotide component of the nucleotide substrate (3, 23); the specificity of ATP-dependent ligases is determined, at least in part, by a distinctive motif within the OB-fold domain that coordinates the beta and gamma phosphates of the nucleotide (5, 19, 22).
The archaea are unicellular anucleate organisms with distinctive biosynthetic capacities and an ability to thrive under extreme environmental conditions. The domain Archaea is subdivided into three kingdoms: Euryarchaeota (encompassing the methanogens, halobacteria, thermococci, archeaoglobi, and others), Crenarchaeota (embracing many hyperthermophilic species), and Nanoarchaeota (13, 25). Sequencing of archaeal genomes has illuminated evolutionary relationships among the three domains of life. A common ancestry for archaea and eucarya is supported by the similarities in their DNA replication machineries, which differ from those of bacteria (2, 11). Indeed, the first identification of a DNA ligase gene from an archaeon (Desulfurolobus ambivalens) highlighted the similarity of its encoded polypeptide to the ATP-dependent DNA ligases of eucarya and poxviruses and the absence of overt similarity to the NAD+-dependent ligases of bacteria (8). The D. ambivalens ligase polypeptide includes all of the essential motifs found in ATP-dependent ligases and lacks the unique nicotinamide mononucleotide-binding domain found in the NAD+-dependent ligase family (19). The biochemical characterization of DNA ligase from the euryarchaeon Methanobacterium thermautotrophicum revealed that its sealing reaction depended on ATP and that NAD+ could not substitute for ATP (20). Utilization of ATP but not NAD+ was also reported for the DNA ligase encoded by the crenarchaeon Sulfolobus shibatae (9). Given that all known archaeal proteomes contain a single homolog of the M. thermautotrophicum ligase enzyme and none contain an obvious homolog of bacterial NAD+-dependent ligase, it was presumed that the archaeal ligases would rely exclusively on ATP as their nucleotide substrate.
This notion was challenged by two reports that the DNA ligases of the hyperthermophilic euryarchaea Thermococcus kodakaraensis and Thermococcus fumicolans could utilize either ATP or NAD+ in the ligase adenylylation and nick-sealing reactions (14, 17). Nakatani et al. (14) found that nick sealing by T. kodakaraensis ligase depended on ATP (and ADP could not substitute), but they could also detect low adenylyltransferase and nick-joining activities in the presence of NAD+. Rolland et al. (17) showed that T. fumicolans ligase could use ATP and NAD+ with comparable efficacies in nick sealing (similar Km and Vmax values) and that T. fumicolans ligase reacted with either ATP or NAD+ to form the ligase-AMP intermediate. Rolland et al. cited unpublished findings that Pyrococcus abyssi DNA ligase could also utilize either ATP or NAD+. These results raised the prospect that some of the archaeal DNA ligases might exemplify an undifferentiated ancestral stage in the evolution of ligase substrate specificity.
The ability of certain ligases from the order Thermococcales to utilize either ATP or NAD+ as a substrate might be explained if they recognized only the ADP component common to both nucleotide substrates. From an evolutionary standpoint, this suggests that ATP-dependent and NAD+-dependent DNA ligases might have evolved from a common ancestral enzyme by the acquisition of protein structural elements that interact with the gamma phosphate of ATP or the nicotinamide ribonucleoside of NAD+, in which case the aboriginal ligase might have utilized ADP as the immediate substrate. In this light, it is remarkable that Jeon and Ishikawa (6) found the DNA ligase from the crenarchaeon Aeropyrum pernix catalyzed nick sealing in the presence of ADP or ATP, but not NAD+ or AMP.
Here, we query the biochemical properties and nucleotide specificity of the DNA ligase encoded by the euryarchaeon Pyrococcus horikoshii (7). We produced the 559-amino-acid (aa) P. horikoshii ligase in Escherichia coli and purified the recombinant protein. P. horikoshii ligase displays optimal ATP-dependent nick-joining and adenylyltransferase activities at 90°C and is unable to utilize NAD+ or ADP to promote ligation in lieu of ATP. Moreover, ADP inhibits the catalysis of nick sealing by the ligase-AMP intermediate.
MATERIALS AND METHODS
T7-based vector for expression of P. horikoshii DNA ligase.
The P. horikoshii ligase open reading frame was amplified by PCR. P. horikoshii genomic DNA (obtained from ATCC) was used as the template. The primers were designed to introduce an NdeI site at the translation start codon and a BamHI restriction site 3′ of the stop codon. The PCR product was digested with NdeI and BamHI and then inserted into the NdeI and BamHI sites of T7-based expression plasmid pET16b (Novagen) to yield pET-PhoLig. Dideoxy sequencing of the entire insert of pET-PhoLig confirmed that no alterations of the genomic DNA sequence were introduced during PCR amplification and cloning.
Recombinant P. horikoshii DNA ligase.
The pET-PhoLig plasmid was transformed into E. coli BL21(DE3). A 1-liter bacterial culture was grown at 37°C in Luria-Bertani medium containing 0.2 mg/ml ampicillin until the A600 reached ∼0.6. The culture was adjusted to 0.4 mM isopropyl-β-d-thiogalactopyranoside and incubated at 37°C for 6 h with constant shaking. Cells were harvested by centrifugation, and the pellet was stored at −80°C. All subsequent procedures were performed at 4°C. Thawed bacteria were resuspended in 50 ml of buffer A (50 mM Tris-HCl [pH 7.5], 0.25 M NaCl, 10% sucrose). Cell lysis was achieved by the addition of lysozyme and Triton X-100 to final concentrations of 0.1 mg/ml and 0.1%, respectively. The lysate was sonicated to reduce viscosity, and insoluble material was removed by centrifugation. The soluble extract was applied to a 5-ml column of Ni-nitrilotriacetic acid-agarose (QIAGEN) that had been equilibrated with buffer A containing 0.1% Triton X-100. The column was washed with 20 ml of the same buffer and then eluted stepwise with 10-ml aliquots of buffer B (50 mM Tris-HCl [pH 8.0], 0.25 M NaCl, 0.05% Triton X-100, 10% glycerol) containing 50, 100, 200, and 500 mM imidazole. The polypeptide compositions of the column fractions were monitored by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). The His-tagged P. horikoshii ligase was recovered predominantly in the 200 mM imidazole eluate. The preparation was dialyzed against a buffer containing 50 mM Tris-HCl (pH 7.5), 0.2 M NaCl, 5 mM EDTA, 2 mM dithiothreitol (DTT), and 10% glycerol and then stored at −80°C. The protein concentration of the enzyme preparation was determined with the Bio-Rad dye reagent with bovine serum albumin as the standard. Approximately 30 mg of P. horikoshii ligase was recovered from a 1-liter bacterial culture.
Adenylyltransferase assay.
Reaction mixtures (20 μl) containing 50 mM Tris-HCl (pH 7.0), 5 mM DTT, 1 mM MgCl2, 20 μM [α-32P]ATP, and P. horikoshii ligase as specified were incubated for 15 min at 90°C. The reactions were quenched by adding SDS to 1.2%. The products were analyzed by SDS-PAGE (12% acrylamide). The ligase-[32P]AMP adduct was visualized by autoradiography of the dried gel and quantified by scanning the gel with a Fujifilm FLA5000 imaging apparatus.
Ligation assay.
A 30-mer oligodeoxyribonucleotide was 5′ 32P labeled by using T4 polynucleotide kinase and [γ-32P]ATP and then purified by electrophoresis through a nondenaturing 17% polyacrylamide gel. The labeled 30-mer was annealed to a complementary 66-mer to form the nicked hairpin DNA substrate shown in Fig. 4. Ligase reaction mixtures (20 μl) containing 50 mM Tris-HCl (pH 8.0), 5 mM DTT, 5 mM MgCl2, 10 μM ATP, 1 pmol radiolabeled nicked hairpin DNA, and P. horikoshii ligase as specified were incubated at 90°C for 60 min. The reactions were quenched by adding 40 μl of 95% formamide-20 mM EDTA. The samples were heated at 95°C for 5 min and then analyzed by electrophoresis through a 12% polyacrylamide gel containing 7 M urea in 45 mM Tris-borate-1 mM EDTA. The extent of ligation (96-mer/[30-mer + 96-mer]) was determined by scanning the gel with a Fujifilm FLA5000 imaging apparatus.
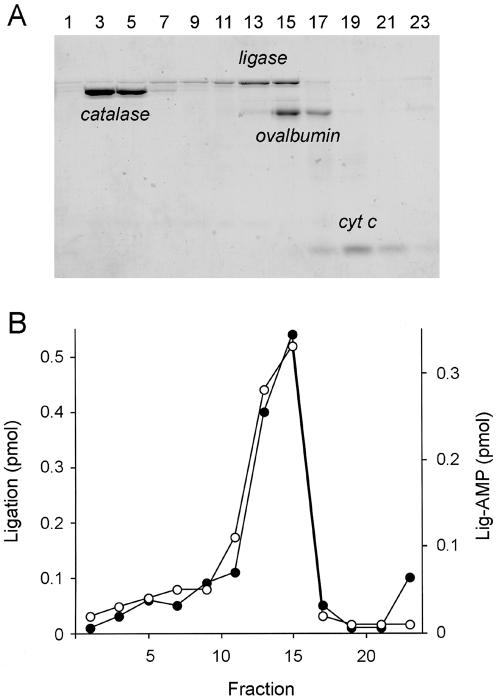
FIG. 4.
Glycerol gradient sedimentation. (A) Aliquots (18 μl) of the odd-numbered gradient fractions were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The positions of the ligase polypeptide and the internal standards catalase, ovalbumin, and cytochrome (cyt) c are indicated. (B) Adenylyltransferase (○) reaction mixtures (20 μl) containing 50 mM Tris-HCl (pH 7.0), 5 mM DTT, 1 mM MgCl2, 2 μM [α-32P]ATP, and 4 μl of the indicated gradient fractions were incubated for 15 min at 90°C. Nick-joining (•) reaction mixtures (20 μl) containing 50 mM Tris-HCl (pH 8.0), 5 mM DTT, 5 mM MgCl2, 20 μM ATP, 1 pmol 32P-labeled nicked DNA, and 2 μl of a 1:10 dilution of the indicated gradient fractions were incubated for 1 h at 90°C.
Glycerol gradient sedimentation.
An aliquot (50 μg) of P. horikoshii ligase was mixed with catalase (50 μg), ovalbumin (50 μg), and cytochrome c (50 μg). The mixture was applied to a 4.8-ml 15 to 30% glycerol gradient containing 50 mM Tris-HCl (pH 7.5), 0.2 M NaCl, 1 mM EDTA, 0.5 mM DTT, and 0.025% Triton X-100. The gradient was centrifuged for 18 h at 4°C in a Beckman SW55Ti rotor at 50,000 rpm. Fractions (∼0.2 ml) were collected from the bottom of the tube. Aliquots were analyzed by SDS-PAGE and assayed for adenylyltransferase and nick-joining activities.
Materials.
Oligodeoxyribonucleotides were purchased from Biosource International. [α-32P]ATP was purchased from Perkin-Elmer Life Sciences. ATP was from Sigma, NAD+ was from Roche Diagnostics, and ADP was from MP Biomedicals Inc. Concentrations of nucleotide stock solutions were determined by UV absorbance at 260 nm.
RESULTS
Purification of P. horikoshii ligase and demonstration of adenylyltransferase activity.
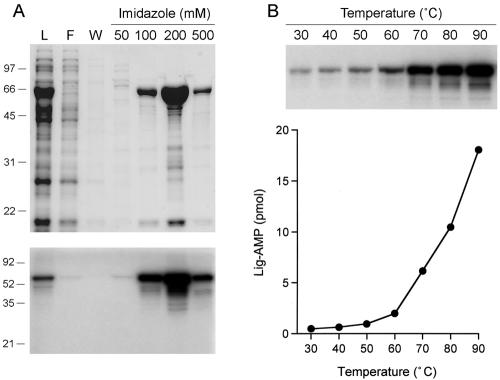
The P. horikoshii open reading frame encoding a 559-aa ligase-like polypeptide (National Center for Biotechnology Information accession no. NP_143476) was PCR amplified from genomic DNA and cloned into a T7 RNA polymerase-based bacterial expression vector so as to fuse the P. horikoshii ligase protein to a 20-aa N-terminal leader peptide containing 10 tandem histidines. We produced P. horikoshii ligase in E. coli and purified the 66-kDa recombinant protein from a soluble bacterial extract by adsorption to Ni-agarose and step elution with imidazole (Fig. 1A).
FIG. 1.
Purification and adenylyltransferase activity of P. horikoshii ligase. (A) Aliquots (10 μl) of the soluble lysate (L), the nickel agarose flowthrough (F), wash (W), and the indicated imidazole elute fractions were analyzed by SDS-PAGE. The gel was stained with Coomassie blue dye. The positions and sizes (kilodaltons) of marker polypeptides are shown on the left. Reaction mixtures (20 μl) containing 50 mM Tris-HCl (pH 7.0), 5 mM DTT, 1 mM MgCl2, 20 μM [α-32P]ATP, and 1 μl of each of the indicated fractions were incubated at 80°C for 10 min. Reactions were quenched by adding SDS to 1.2%. The reaction products were resolved by SDS-PAGE. An autoradiogram of the dried gel is shown. (B) Reaction mixtures (20 μl) containing 50 mM Tris-HCl (pH 7.0), 5 mM DTT, 1 mM MgCl2, 20 μM [α-32P]ATP, and 50 pmol P. horikoshii ligase were incubated for 10 min at the indicated temperatures. The reaction products were analyzed by SDS-PAGE and visualized by autoradiography (top). Ligase-adenylate (Lig-AMP) formation is plotted as a function of temperature (bottom).
The initial step in DNA ligation involves the formation of a covalent enzyme-adenylate intermediate (10). The adenylyltransferase activity of P. horikoshii ligase was evinced by label transfer from [α-32P]ATP to the P. horikoshii ligase to form a covalent enzyme-adenylate adduct that migrated as a 66-kDa species during SDS-PAGE. Adenylyltransferase activity paralleled the elution profile of the P. horikoshii ligase protein during Ni-agarose chromatography (Fig. 1A). We conclude that recombinant P. horikoshii ligase is active in nucleotidyl transfer. Further characterization of P. horikoshii ligase was performed with the 200 mM imidazole fraction.
Characterization of the adenylyltransferase activity.
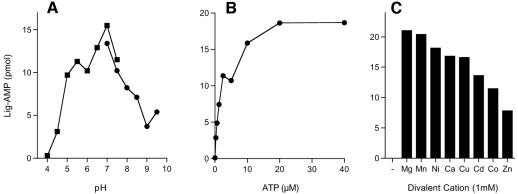
The extent of ligase-AMP formation was extremely sensitive to the reaction temperature. Adenylyltransferase activity was maximal at 90°C and declined sharply as the temperature was lowered to ≤60°C (Fig. 1B). Activity was optimal in either Tris-acetate or Tris-HCl buffer at pH 7.0 (Fig. 2A). (The stated pH values were determined at 22°C and do not necessarily reflect the pH at a reaction temperature of 90°C.) The extent of adenylylation increased with the ATP concentration over a range of 0.2 to 10 μM and reached saturation at 20 μM (Fig. 2B). We calculated that ∼40% of the P. horikoshii ligase protein was adenylated with [32P]AMP at a saturating ATP concentration. The remainder of the P. horikoshii ligase preparation likely consists of preformed ligase-AMP intermediate (see below). A divalent-cation cofactor was required for ligase adenylylation (Fig. 2C). MgCl2 and MnCl2 supported approximately equivalent activities over an optimal concentration range of 0.5 to 1 mM (data not shown). P. horikoshii ligase displayed a distinctively broad metal cofactor specificity, insofar as Mg2+, Mn2+, Ni2+, Ca2+, Cu2+, Cd2+ Co2+, and Zn2+ at 1 mM all supported the autoadenylylation reaction (Fig. 2C).
FIG. 2.
Characterization of the adenylyltransferase reaction. (A) pH dependence. Reaction mixtures (20 μl) containing 50 mM buffer (either Tris-acetate at pH 4.0, 4.5, 5.0, 5.5, 6.0, 6.5, 7.0, or 7.5 or Tris-HCl at pH 7.0, 7.5, 8.0, 8.5, 9.0, or 9.5; the pH values of the buffers were determined at 22°C for 1 M stock solutions), 5 mM DTT, 1 mM MgCl2, 20 μM [α-32P]ATP, and 50 pmol ligase were incubated for 15 min at 90°C. The extent of ligase (Lig)-AMP formation is plotted as a function of the pH. (B) ATP dependence. Reaction mixtures (20 μl) containing 50 mM Tris-HCl (pH 7.0), 5 mM DTT, 1 mM MgCl2, 50 pmol P. horikoshii ligase, and [α-32P]ATP as specified were incubated for 15 min at 90°C. Ligase-AMP formation is plotted as a function of the ATP concentration. (C) Divalent-cation specificity. Reaction mixtures (20 μl) containing 50 mM Tris-HCl (pH 7.0), 20 μM [α-32P]ATP, 50 pmol ligase, and either no divalent cation (lane −) or 1 mM MgCl2, MnCl2, NiCl2, CaCl2, CuCl2, CdCl2, CoCl2, or ZnCl2 were incubated for 15 min at 90°C.
Nick-joining activity of P. horikoshii ligase.
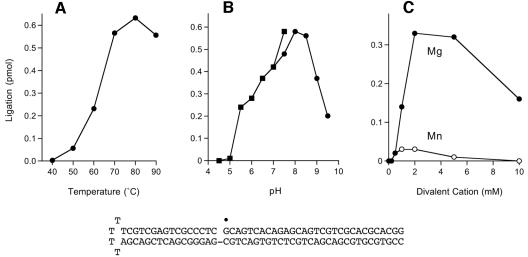
The ligation reaction of P. horikoshii ligase was assayed with a singly nicked DNA substrate composed of a 5′-32P-labeled 30-mer annealed to an unlabeled 66-mer 5′-tailed hairpin strand to form the nicked hairpin structure shown in Fig. 3. The reaction of P. horikoshii ligase with this substrate in the presence of ATP and magnesium generated a 32P-labeled 96-mer product that was resolved from the radiolabeled 30-mer substrate strand by denaturing PAGE (not shown). The extent of nick joining was optimal at 70 to 90°C and declined sharply at ≤60°C (Fig. 3A). Ligation was optimal in Tris-acetate or Tris-HCl buffer at pH 7.5 to 8.5 (Fig. 3B). An exogenous divalent-cation cofactor was required for nick joining. The metal cofactor requirement was satisfied by Mg2+ and to a much lesser extent by Mn2+ (Fig. 3C). Ligation was optimal at 2 to 5 mM MgCl2. Neither Ni2+, Ca2+, Cu2+, Cd2, Co2+, nor Zn2+ was able to support the nick-joining activity at 2 or 5 mM concentration (data not shown). Thus, P. horikoshii ligase displays a more stringent metal requirement for overall ligation than for the isolated step 1 adenylyltransferase reaction. A mixing experiment was performed in which ligation reaction mixtures containing 5 mM magnesium were supplemented with each of the other divalent cations at 5 mM. We found that Ni2+, Mn2+, Cd2, Co2+, and Zn2+ abolished nick-joining activity, whereas Cu2+ and Ca2+ were partially inhibitory (about fourfold for Cu2+ and about eightfold for Ca2+; data not shown).
FIG. 3.
Characterization of the ligation reaction. (A) Temperature dependence. Reaction mixtures (20 μl) containing 50 mM Tris-HCl (pH 8.0), 5 mM DTT, 5 mM MgCl2, 10 μM ATP, 1 pmol radiolabeled nicked hairpin DNA, and 0.1 pmol ligase were incubated at the indicated temperatures for 1 h. (B) pH dependence. Reaction mixtures (20 μl) containing 50 mM buffer (either Tris-acetate at pH 4.0, 4.5, 5.0, 5.5, 6.0, 6.5, 7.0, or 7.5 or Tris-HCl at pH 7.0, 7.5, 8.0, 8.5, 9.0, or 9.5; the pH values of the buffers were determined at 22°C for 1 M stock solutions), 5 mM DTT, 5 mM MgCl2, 10 μM ATP, 1 pmol radiolabeled nicked hairpin DNA, and 0.1 pmol ligase were incubated at 90°C for 1 h. (C) Divalent-cation dependence. Reaction mixtures (20 μl) containing 50 mM Tris-HCl (pH 8.0), 10 μM ATP, 1 pmol radiolabeled nicked hairpin DNA, 0.1 pmol ligase, and either 0, 0.2, 0.5, 1, 2, 5, or 10 mM MgCl2 or MnCl2 were incubated for 1 h at 90°C. The nicked hairpin DNA substrate is depicted at the bottom with the 5′ 32P label of the 30-mer strand denoted by the symbol •.
Velocity sedimentation of P. horikoshii ligase.
The native size of the P. horikoshii ligase was gauged by zonal velocity sedimentation through a 15 to 30% glycerol gradient containing 0.2 M NaCl. The marker proteins catalase (248 kDa), ovalbumin (45 kDa), and cytochrome c (13 kDa) were included as internal standards. The majority of the applied P. horikoshii ligase sedimented as a discrete peak in fractions 13 to 15 on the “heavy” side of ovalbumin (Fig. 4A). A minor fraction was present in a more rapidly sedimenting shoulder. The adenylyltransferase and nick-joining activity profiles coincided with those of the P. horikoshii ligase protein, peaking in fractions 13 to 15 but trailing off as a “heavy” shoulder (Fig. 4B). A plot of the S values of the three standards versus the fraction number yielded a straight line (not shown). An S value of 4.2 was determined for the major peak component of P. horikoshii ligase by interpolation to the internal standard curve. We surmise that the major component of P. horikoshii ligase is a monomer.
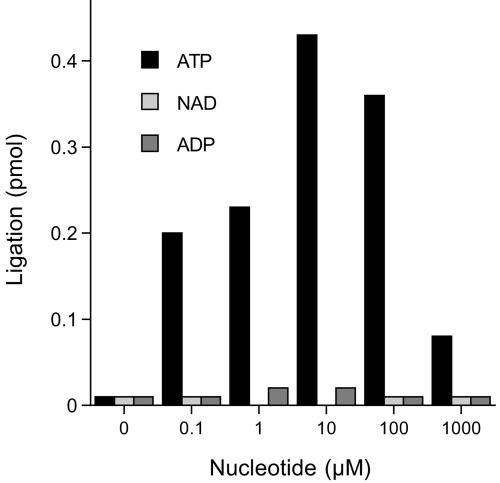
Nucleotide substrate specificity in nick joining.
Nick-joining activity was assayed in DNA substrate excess in the presence of 0, 0.1, 1, 10, 100, or 1,000 μM ATP, NAD, and ADP (Fig. 5). A low level of sealing was detected in the absence of nucleotide that was attributed to preadenylated ligase in the enzyme preparation. Ligation was specifically stimulated by ATP, with optimal activity at 10 μM ATP. Neither NAD+ nor ADP was able to satisfy the nucleotide substrate requirement for nick joining. Note that low concentrations of ATP (0.1 to 1 μM) were sufficient to sustain about half of the optimum sealing activity and that increasing the ATP concentration to 1 mM inhibited nick joining by ∼80% compared to the peak value (Fig. 5). This inhibition cannot be explained by ATP sequestration of the divalent cation, because the reaction mixtures contained 5 mM Mg2+ and we have demonstrated that 2 mM Mg2+ suffices for optimal ligation (Fig. 3C).
FIG. 5.
Nucleotide specificity. Reaction mixtures (20 μl) containing 50 mM Tris-HCl (pH 7.0), 5 mM DTT, 5 mM MgCl2, 1 pmol radiolabeled nicked hairpin DNA, 0.05 pmol ligase, and ATP, NAD+, or ADP as specified were incubated for 1 h at 90°C.
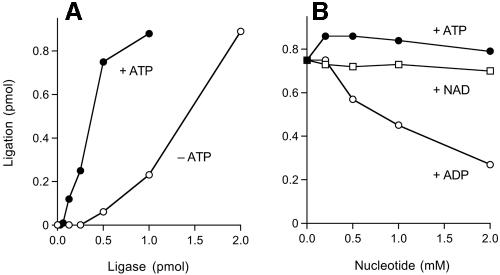
The extent of nick sealing in the presence of 10 μM ATP was proportional to the input P. horikoshii ligase protein (Fig. 6A). In the linear range of the titration curve, about 25 pmol of nicks was sealed per pmol of P. horikoshii ligase. At a saturating enzyme concentration, 90% of the input 30-mer strand was ligated to the 66-mer hairpin (Fig. 6A). This upper limit value likely reflects incomplete annealing of the component strands. There was a significant extent of ligation in the absence of added nucleotide (Fig. 6A); the titration curve was sigmoidal, perhaps because the ligase was unstable at low protein concentrations at 90°C in the ligation reaction buffer. Simple comparison of the plus-ATP and minus-ATP activities at 0.5 pmol of ligase indicated that ATP stimulated activity by 12-fold. The extent of ATP-independent sealing was substoichiometric with respect to the input P. horikoshii ligase, as expected for a single-turnover reaction by the fraction of preformed ligase-AMP in the recombinant protein preparation. From the extent of ATP-independent sealing by 2 pmol of enzyme, we calculated that at least 45% of the P. horikoshii ligase was ligase-AMP.
FIG. 6.
Nucleotide stimulation and inhibition. (A) Reaction mixtures (20 μl) containing 50 mM Tris-HCl (pH 8.0), 5 mM DTT, 5 mM MgCl2, 1 pmol radiolabeled nicked hairpin DNA, ligase as specified, and either 10 μM ATP (+ATP) or no ATP (−ATP) were incubated for 1 h at 90°C. (B) Reaction mixtures (20 μl) containing 50 mM Tris-HCl (pH 8.0), 5 mM DTT, 5 mM MgCl2, 1 pmol radiolabeled nicked hairpin DNA, 2 pmol ligase, and ATP, NAD+, or ADP as specified were incubated for 1 h at 90°C. The extent of ligation is plotted as a function of the nucleotide concentration.
ADP inhibits single-turnover nick joining by preformed ligase-adenylate.
The effects of exogenous NAD+ and ADP on single-turnover nick sealing in the absence of ATP are shown in Fig. 6B. The amount of input enzyme was adjusted to achieve ligation of ∼75% of the input substrate. NAD+ at concentrations of up to 2 mM had no effect on the extent of single-turnover ligation. In contrast, ADP elicited a concentration-dependent inhibition of single-turnover ligation. Activity was reduced by threefold at 2 mM ADP. For the reasons noted above in discussing why high concentrations of ATP were inhibitory to steady-state ligation, the ADP inhibition cannot plausibly be attributed to sequestration of magnesium (present at 5 mM). The most likely explanation for the ADP inhibition of single-turnover ligation and ATP inhibition of steady-state ligation is that high concentrations of ATP and ADP can act as decoy acceptors for AMP transfer by ligase-adenylate, thereby diverting the ligase from nick joining and toward the synthesis of dinucleoside polyphosphates, as described by Sillero and colleagues (1, 4, 12). The Sillero lab has also shown that nucleoside triphosphates (NTPs) can act as decoy acceptors for AMP transfer. We found that single-turnover ligation by P. horikoshii ligase was reduced to 18%, 38%, and 7% of the control value when the reaction mixtures were supplemented with 2 mM CTP, GTP, and UTP, respectively (data not shown).
DISCUSSION
The present study was undertaken as an extension of our prior work reporting the characterization of an archaeal DNA ligase (M. thermautotrophicum ligase) as strictly ATP dependent (20). All archaeal ligases contain the core catalytic domain typical of ATP-dependent ligases (composed of nucleotidyltransferase and OB-fold modules) plus an N-terminal domain similar to that found in eukaryotic cellular and poxvirus DNA ligases. The N-terminal extension is absent from the minimized DNA ligases encoded by bacteriophages and Chlorella virus (15, 16, 24). Studies of the poxvirus DNA ligase implicated the N-terminal extension in the DNA-binding step of the ligase reaction (18). The recently reported crystal structure of human DNA ligase I bound to the DNA-adenylate intermediate (16) shows that (i) the N-terminal segment comprises a globular alpha-helical fold that forms part of a protein clamp around the DNA duplex and (ii) this N-terminal DNA-binding domain is conserved in archaeal DNA ligases.
All archaeal DNA ligases characterized previously have ATP-dependent adenylyltransferase and ligase activities, as expected from the structural considerations mentioned above. What is striking is the disparate, almost idiosyncratic, variations in the abilities of archaeal ligases to utilize substrates other than ATP. The reports of dual-specificity ATP/NAD+ ligases in two Thermococcus species and P. abyssi (14, 17) plus an ATP/ADP ligase in Aeropyrum pernix (6) prompted us to characterize the homologous enzyme of P. horikoshii, which we find to be ATP dependent and unable to use NAD+ or ADP for nick sealing. Thus, it appears that the properties of P. horikoshii ligase differ from those cited for P. abyssi ligase, even though the two enzymes are derived from species within the same genus and the polypeptides have identical side chains at 517 of 559 positions (92% identity). The substrate specificity of P. horikoshii ligase also differs from that of T. kodakaraensis ligase and T. fumicolans ligase, which are encoded by species within the same order as P. horikoshii and which have 80% and 78% side chain identity to P. horikoshii ligase, respectively. Taken at face value, these comparisons imply that the differences in the nucleotide specificity of archaeal ligases (e.g., between P. horikoshii ligase and P. abyssi ligase) are attributable to rather subtle changes in primary structure. Alternatively, given the recent advances in the structural biology of bacterial and eucaryal ligases and the delineation of the unique NAD+ specificity determinants in bacterial LigA proteins (which are absent from archaeal ligases), it might be prudent to delay the ratification of a proposed new clade of dual-specificity ligases pending additional biochemical studies and comparisons of crystal structures of closely related archaeal ligases bound to their nucleotide substrates.
Substrate specificity aside, the present analysis of P. horikoshii ligase highlights similarities and differences relative to well-characterized ligase enzymes. Like most other DNA ligases, P. horikoshii ligase exists predominantly as a monomeric protein in solution. It requires a divalent cation for its adenylylation and nick-sealing reactions but displays the broadest metal cofactor specificity of any known DNA ligase in the autoadenylylation reaction. P. horikoshii ligase reacts with ATP in the presence of either Mg2+, Mn2+, Ni2+, Ca2+, Cu2+, Cd2+ Co2+, or Zn2+. P. horikoshii ligase is less catholic in its reliance on only magnesium in the composite nick-sealing reaction. Although neither NAD+ nor ADP could replace ATP as the nucleotide substrate for nick joining, ADP was uniquely inhibitory to single-turnover ligation by preformed ligase adenylate. Sillero et al. (1, 4, 12) have shown that DNA and RNA ligases can donate their covalently bound adenylates to non-nucleic acid acceptors, particularly to NTPs or nucleoside diphosphates, thereby forming ApppN and AppppN dinucleotide products. The reaction mechanism is presumed to mimic the pyrophosphorolytic reversal of the step 1 ligase adenylylation reaction, which regenerates ATP (26). When present at high (millimolar) concentrations, nucleoside diphosphates and NTPs (specifically, the terminal phosphate groups) compete with the 5′ PO4 of the nicked DNA substrate for attack on the phosphorus of the ligase-adenylate intermediate. We invoke this mechanism to account for the inhibition of single-turnover ligation by ADP and the reduction in steady-state nick joining when ATP is added at a 1 mM concentration (well above the optimal range of 10 to 100 μM ATP). It is therefore sensible that NAD+ would not inhibit single-turnover ligation (as shown in Fig. 6B) because NAD+ has no unbonded terminal phosphate oxygen to serve as a nucleophile.
Acknowledgments
This research was supported by NIH grant GM63611. S.S. is an American Cancer Society Research Professor.
REFERENCES
- 1.Atencia, E. A., O. Madrid, M. A. Günther Sillero, and A. Sillero. 1999. T4 RNA ligase catalyzes the synthesis of dinucleoside polyphosphates. Eur. J. Biochem. 261:802-811. [DOI] [PubMed] [Google Scholar]
- 2.Edgell, D. R., and W. F. Doolittle. 1997. Archaea and the origins(s) of DNA replication proteins. Cell 89:995-998. [DOI] [PubMed] [Google Scholar]
- 3.Gajiwala, K., and C. Pinko. 2004. Structural rearrangement accompanying NAD+ synthesis within a bacterial DNA ligase crystal. Structure 12:1449-1459. [DOI] [PubMed] [Google Scholar]
- 4.Günther Sillero, M. A., M. Montes, A. de Diego, M. del Valle, E. A. Atencia, and A. Sillero. 2002. Thermostable Pyrococcus furiosus DNA ligase catalyzes the synthesis of (di)nucleoside polyphosphates. Extremophiles 6:45-50. [DOI] [PubMed] [Google Scholar]
- 5.Håkansson, K., A. J. Doherty, S. Shuman, and D. B. Wigley. 1997. X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell 89:545-553. [DOI] [PubMed] [Google Scholar]
- 6.Jeon, S. J., and K. Ishikawa. 2003. A novel ADP-dependent DNA ligase from Aeropyrum pernix K1. FEBS Lett. 550:69-73. [DOI] [PubMed] [Google Scholar]
- 7.Kawarabayasi, Y., M. Sawada, H. Horikawa, Y. Haikawa, Y. Hino, S. Yamamoto, M. Sekine, S. Baba, H. Kosugi, A. Hosoyama, Y. Nagai, M. Sakai, K. Ogura, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Ohfuku, T. Funahashi, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, and H. Kikuchi. 1998. Complete sequence and gene organization of the hyperthermophilic archaeon Pyrococcus horikoshii. DNA Res. 5:55-76. [DOI] [PubMed] [Google Scholar]
- 8.Kletzin, A. 1992. Molecular characterization of a DNA ligase gene of the extremely thermophilic archaeon Desulfurolobus ambivalens shows close phylogenetic relationship to eukaryotic ligases. Nucleic Acids Res. 20:5389-5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai, X., H. Shao, F. Hao, and L. Huang. 2002. Biochemical characterization of an ATP-dependent DNA ligase from the hyperthermophilic crenarchaeon Sulfolobus shibatae. Extremophiles 6:469-477. [DOI] [PubMed] [Google Scholar]
- 10.Lehman, I. R. 1974. DNA ligase: structure, mechanism, and function. Science 186:790-797. [DOI] [PubMed] [Google Scholar]
- 11.Leipe, D. D., L. Aravind, and E. V. Koonin. 1999. Did DNA replication evolve twice independently? Nucleic Acids Res. 27:3389-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madrid, O., D. Martin, E. A. Atencia, A. Sillero, and M. A. Günther Sillero. 1998. T4 DNA ligase synthesizes dinucleoside polyphosphates. FEBS Lett. 433:283-286. [DOI] [PubMed] [Google Scholar]
- 13.Makarova, K. S., and E. V. Koonin. 2003. Comparative genomics of archaea: how much have we learned in six years and what's next? Genome Biol. 4:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakatani, M., S. Ezaki, H. Atomi, and T. Imanaka. 2000. A DNA ligase from a hyperthermophilic archaeon with unique cofactor specificity. J. Bacteriol. 182:6424-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Odell, M., V. Sriskanda, S. Shuman, and D. B. Nikolov. 2000. Crystal structure of eukaryotic DNA ligase-adenylate illuminates the mechanism of nick sensing and strand joining. Mol. Cell 6:1183-1193. [DOI] [PubMed] [Google Scholar]
- 16.Pascal, J. M., P. J. O'Brien, A. E. Tomkinson, and T. Ellenberger. 2004. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature 432:473-478. [DOI] [PubMed] [Google Scholar]
- 17.Rolland, J., Y. Gueguen, C. Persillon, J. M. Masson, and J. Dietrich. 2004. Characterization of a thermophilic DNA ligase from the archaeon Thermococcus fumicolans. FEMS Microbiol. Lett. 236:267-273. [DOI] [PubMed] [Google Scholar]
- 18.Sekiguchi, J., and S. Shuman. 1997. Domain structure of vaccinia DNA ligase. Nucleic Acids Res. 25:727-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shuman, S., and C. D. Lima. 2004. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr. Opin. Struct. Biol. 14:757-764. [DOI] [PubMed] [Google Scholar]
- 20.Sriskanda, V., Z. Kelman, J. Hurwitz, and S. Shuman. 2000. Characterization of an ATP-dependent DNA ligase from the thermophilic archaeon Methanobacterium thermoautotrophicum. Nucleic Acids Res. 28:2221-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sriskanda, V., R. W. Moyer, and S. Shuman. 2001. NAD+-dependent DNA ligase encoded by a eukaryotic virus. J. Biol. Chem. 276:36100-36109. [DOI] [PubMed] [Google Scholar]
- 22.Sriskanda, V., and S. Shuman. 1998. Mutational analysis of Chlorella virus DNA ligase: catalytic roles of domain I and motif VI. Nucleic Acids Res. 26:4618-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sriskanda, V., and S. Shuman. 2002. Conserved residues in domain Ia are required for the reaction of Escherichia coli DNA ligase with NAD+. J. Biol. Chem. 277:9685-9700. [DOI] [PubMed] [Google Scholar]
- 24.Subramanya, H. S., A. J. Doherty, S. R. Ashford, and D. B. Wigley. 1996. Crystal structure of an ATP-dependent DNA ligase from bacteriophage T7. Cell 85:607-615. [DOI] [PubMed] [Google Scholar]
- 25.Waters, E., M. J. Hohn, I. Ahel, D. E. Graham, M. D. Adams, M. Barnstead, K. Y. Beeson, L. Bibbs, R. Bolanos, M. Keller, K. Kretz, X. Lin, E. Mathur, J. Ni, M. Podar, T. Richardson, G. G. Sutton, M. Simon, D. Soll, K. O. Stetter, J. M. Short, and M. Noordewier. 2003. The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc. Natl. Acad. Sci. USA 100:12984-12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiss, B., A. Thompson, and C. C. Richardson. 1968. Enzymatic breakage and joining of deoxyribonucleic acid: properties of the enzyme-adenylate intermediate in the polynucleotide ligase reaction. J. Biol. Chem. 243:4556-4563. [PubMed] [Google Scholar]
- 27.Wilkinson, A., J. Day, and R. Bowater. 2001. Bacterial DNA ligases. Mol. Microbiol. 40:1241-1248. [DOI] [PubMed] [Google Scholar]