Abstract
DNA microarray analysis of Clostridium acetobutylicum was used to examine the genomic-scale gene expression changes during the shift from exponential-phase growth and acidogenesis to stationary phase and solventogenesis. Self-organizing maps were used to identify novel expression patterns of functional gene classes, including aromatic and branched-chain amino acid synthesis, ribosomal proteins, cobalt and iron transporters, cobalamin biosynthesis, and lipid biosynthesis. The majority of pSOL1 megaplasmid genes (in addition to the solventogenic genes aad-ctfA-ctfB and adc) had increased expression at the onset of solventogenesis, suggesting that other megaplasmid genes may play a role in stationary-phase phenomena. Analysis of sporulation genes and comparison with published Bacillus subtilis results indicated conserved expression patterns of early sporulation genes, including spo0A, the sigF operon, and putative canonical genes of the σH and σF regulons. However, sigE expression could not be detected within 7.5 h of initial spo0A expression, consistent with the observed extended time between the appearance of clostridial forms and endospore formation. The results were compared with microarray comparisons of the wild-type strain and the nonsolventogenic, asporogenous M5 strain, which lacks the pSOL1 megaplasmid. While some results were similar, the expression of primary metabolism genes and heat shock proteins was higher in M5, suggesting a difference in metabolic regulation or a butyrate stress response in M5. The results of this microarray platform and analysis were further validated by comparing gene expression patterns to previously published Northern analyses, reporter assays, and two-dimensional protein electrophoresis data of metabolic genes (including all major solventogenesis genes), sporulation genes, heat shock proteins, and other solventogenesis-induced gene expression.
Newly developed genomic technologies allow high-throughput screening of entire transcriptomes. Several recent studies of Bacillus subtilis have utilized the DNA microarray technology for the analysis of key mutants to identify genes affected by sporulation in addition to identifying new genes in sporulation regulons (8, 18, 19, 22). A fundamental biological problem is to identify gene expression patterns for all major functional classes during the shift from exponential-phase growth to stationary phase, especially in endospore-forming organisms such as the bacilli and clostridia, and relate such changes to important physiological changes and events. The number of affected genes is potentially very large; in B. subtilis, it has been shown that inactivation of the stationary-phase gene regulator spo0A can directly and indirectly affect the expression of approximately 500 genes at least threefold during the onset of sporulation (22).
Solventogenic clostridia like Clostridium acetobutylicum produce acids during exponential-phase growth, and during stationary phase, they form granulose, take up acids to produce solvents, and sporulate. An understanding of the global events during this shift would provide invaluable information toward understanding cellular physiology at the genomic transcriptional level, thus facilitating a broader understanding of clostridial differentiation, as well as beneficial applications. Numerous papers have examined stationary-phase phenomena in clostridia, and several genes directly related to solvent formation and sporulation have been cloned. Microarrays have been successfully used to examine the temporal expression of genes during the sporulation of Bacillus anthracis, including the sporulation-specific sigma factors (45).
In this study, we performed DNA microarray analysis of C.acetobutylicum batch cultures during the exponential-growth, transitional, and early stationary phases with a newly developed, genomic-scale microarray platform (2) which provides superior sensitivity and accuracy. Samples were hybridized against a global RNA pool, and gene expression was compared to the expression from the mid-exponential phase of growth. Furthermore, we compared gene expression in the wild-type (WT) strain against that of asporogenous, nonsolventogenic strain M5, which lacks the pSOL1 megaplasmid (13).
MATERIALS AND METHODS
Strains, growth conditions, and maintenance.
C. acetobutylicum ATCC 824 (American Type Culture Collection, Manassas, VA) is the WT strain. Strain M5 lacks the pSOL1 megaplasmid (12). Strains were stored at −85°C in clostridial growth medium (CGM) (91) containing 15% glycerol and grown in an anaerobic chamber (Forma Scientific, Marietta, OH) in CGM supplemented with 80 g/liter glucose or on agar-solidified 2×YTG plates (YTG is 16 g/liter tryptone, 10 g/liter yeast extract, 4 g/liter NaCl, 5 g/liter glucose, and 15 g/liter agar, pH 5.8). WT cultures were inoculated with single colonies from plates at least 4 days old and heat shocked at 70 to 80°C for 10 min.
Analytical methods.
The methods used for monitoring cellular growth were previously described (3). Culture supernatants were analyzed for glucose, acetate, butyrate, acetoin, ethanol, acetone, and butanol on a Waters high-performance liquid chromatography system (Waters, Milford, MA) (85).
Fermentations.
A preinoculum flask was grown to an A600 of 0.2, and the culture was transferred to a BioFlo II (working volume, 3.6 liters; New Brunswick Scientific, Edison, NJ) reactor in a 10-fold dilution of CGM. The fermentation was pH controlled at a low end of 5.0 with 6 N NH4OH. Anaerobic conditions were maintained with nitrogen at flow rates of 125 ml/min (for an A600 below 1.0) and 25 ml/min (for an A600 above 1.0). Static flask cultures without pH control were carried out as previously described (2, 3).
RNA sampling, isolation, and cDNA labeling.
The methods used for RNA isolation and reverse transcription and cDNA probe labeling have been previously described (2).
RNA pool construction.
To measure differences in gene expression between time points within an experiment and to normalize signals against a universal sample, we hybridized the time course microarray samples against an oppositely labeled probe made from an RNA pool which was constructed as follows. A 500-ml bottle of CGM was inoculated with 5 ml of exponential-growth culture. RNA from all stages of growth was individually sampled, purified, and quantitated. An RNA pool was created by mixing equal quantities of purified RNA from the exponential-growth, transitional, and early stationary phases. Microarray probes were then made as previously described (2).
Microarray construction and hybridizations.
The methods used for the construction and validation of the full-genome microarrays and hybridizations were described previously (2). Briefly, microarrays were spotted with PCR-generated probes (designed to minimize cross hybridization) 150 to 500 bp in size. A total of 3,802 genes are present on the microarrays (97% genome coverage). Samples from the fermentation time course were hybridized against oppositely labeled cDNA made from the above-described RNA pool. Two hybridizations were performed at each time point. To minimize dye biases, dyes (Cy3 and Cy5) were swapped for each replicate.
Microarray analysis.
Microarray data were normalized by using a segmental nearest-neighbor logarithmic expression ratio of the mean approach (97) coded in MATLAB (MathWorks, Natick, MA). To factor out the RNA pool from microarray ratios in the time course experiment, each gene's normalized ratio was divided by the normalized ratio of the same gene at first mid-exponential time point A (Fig. 1A). The result is the ratio of expression of a gene at a given time point compared to time point A. Normalized ratios were grouped by expression similarity with either self-organizing maps (SOMs; GeneCluster, version 2.0) (80) or average-linkage hierarchical clustering (Cluster [20] or TIGR MeV, version 3.0.3 [64]). Before SOM analysis, genes with microarray signals significantly above the background at all time points were standardized by dividing each gene's ratio at each time point by the square root of the sum of the squares of the gene's ratios within an experiment. The result is that the sum of the squares of each gene's standardized ratios within an experiment is 1. Data were visualized colorimetrically with heat plots (or Eisen plots) (20) with TreeView (version 1.5) (20) or TIGR MeV (64).
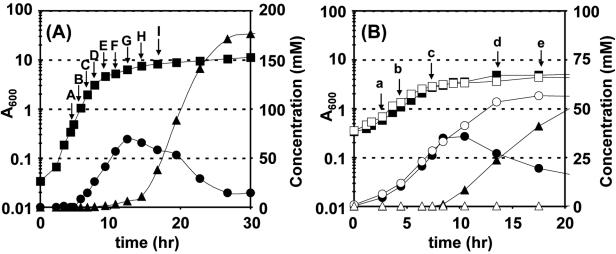
FIG. 1.
Growth curves and selected product concentrations for the WT fermentor (pH ≥5.0) culture time course (A) and the WT-M5 static flask (no pH control) comparison (B). WT results are represented by solid symbols, M5 results by open symbols. Squares, A600; circles, butyrate; triangles, butanol. Letters represent time points at which microarray analysis was performed; uppercase letters represent WT time course time points, lowercase letters represent WT-M5 time points.
RESULTS
In solventogenic clostridia, the stationary-phase program, from its earliest events (solvent formation and/or presence of clostridial forms) to forespore formation, can take 8 to 20 h (35, 47, 65). In this study, we desired to identify changes in gene expression during the transition from the exponential growth phase to the early stationary phase in order to examine early differentiation and stationary-phase phenomena in detail. RNA samples (Fig. 1A) of WT cultures were taken over a 12-h period at nine time points whereby the first three data points belong to the mid- to late-exponential growth phase (microarray time points A, B, and C) and the remaining time points (Dto I) belong to the transitional and early stationary phases. Butanol was first detected (Fig. 1A) in significant quantities at time point E. Samples were hybridized against a probe made from an RNA pool as described in Materials and Methods. To analyze the change in gene expression compared to mid-exponential-phase growth, we divided the expression ratio of each gene by the expression ratio of the gene at time point A. The resulting ratios, which are reported here, are the changes in gene expression compared to time point A.
We also analyzed samples from a time course comparison of a static-flask culture (without pH control) of the WT strain against asporogenous, nonsolventogenic strain M5 (12) (degenerate strain lacking the pSOL1 megaplasmid [13]), which we previously used to validate this microarray technology (2). RNA samples for the WT-to-M5 comparison were taken during the exponential-growth (a and b), transitional (c), and stationary (d and e) phases. The results from the WT-M5 comparison are shown alongside the WT time course expression results and are specifically discussed only if they provide additional or different evidence regarding gene expression.
Confirmation of gene expression from previous published studies.
To further (2) verify the accuracy of this microarray platform, we used results from numerous previous studies which had examined differential expression of C. acetobutylicum genes or proteins during the transition from the exponential growth phase and acidogenesis to stationary phase and solventogenesis.
Solventogenic pSOL1 megaplasmid genes aad-ctfA-ctfB (CAP0162 to CAP0164) and adc (CAP0165) and chromosomal solventogenic genes bdhA (CAC3299) and bdhB (CAC3298) (Fig. 2A) were upregulated at time point E, coinciding with the onset of solvent formation. The expression of most of these genes increases continuously throughout the stationary phase; however, differential expression of bdhA is lower than that of the other genes, peaks in early stationary phase, and then steadily decreases, although it remains at levels higher than those in the exponential phase. At time point I, expression of the sol operon (aad-ctfA-ctfB) is over 1,200-fold higher than at the early exponential-phase time point. Expression of aad-ctfA-ctfB (29, 66), adc (23, 25, 29, 66), bdhA, and bdhB (23, 66, 88) at the onset of solventogenesis has been well established. In the results presented here, at time point D, expression of adc (2.5-fold higher) and bdhB (2.3-fold higher) seems to be initiated before expression of aad-ctfA-ctfB. This is consistent with previous reports (23, 25, 29, 66) and indicates that the regulation of these genes may differ from that of the sol operon. Some of the previous studies (25, 66) were performed with transient chemostat cultures. While the present study was performed with a batch culture, the sequence of solventogenic events is apparently conserved regardless of the culture system. Expression of spo0A (CAC2071), the master regulator of solventogenesis and sporulation in solvent-forming clostridia (29, 59), is discussed later.
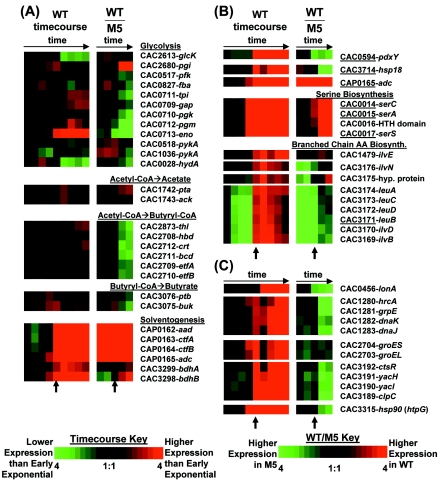
FIG. 2.
Colorimetric expression profiles, or Eisen plots (20), of genes related to primary metabolism (A), genes whose protein products were previously identified as being differentially expressed during solventogenesis by Schaffer et al. (69) (B), and heat shock proteins (C) during the WT time course and WT-M5 comparison. Genes specifically identified by Schaffer and colleagues are underlined; other related genes, such as within a putative operon or functional group, have also been included. In panel A, genes are listed in the order of the reaction pathway. In panels B and C, putative operons are listed in the order of transcription. Short arrows beneath the Eisen plots denote the onset of stationary-phase gene expression (spo0A and aad-ctfA-ctfB expression).
Schaffer and colleagues have previously identified several proteins through two-dimensional gel electrophoresis whose expression was induced during solventogenesis in a transient continuous culture (69): solventogenic acetoacetate decarboxylase (gene adc); heat shock protein Hsp18 (CAC3714); isopropylmalate dehydrogenase LeuB (CAC3171); serine biosynthesis proteins SerA (CAC0015), SerC (CAC0014), and SerS (CAC0017); and pyridoxine biosynthesis protein PdxY (CAC0594). The expression of all these genes (Fig. 2B) increases during the transition from exponential phase to stationary phase. Expression of the serine biosynthesis genes is induced during the transitional and stationary phases, with expression at time point E 130- to 320-fold higher than in the early exponential-growth phase. Expression of adc and hsp18 remains high during the stationary phase, when each is expressed at least 100-fold higher at the last microarray time point than at the first. Induction of hsp18 expression at the onset of solventogenesis in continuous cultures has also been reported (56, 66, 67). Schaffer and colleagues also report that glyceraldehyde-3-phosphate dehydrogenase (Gap, CAC0709) is in greater abundance during solventogenesis although apparently not due to nascent synthesis of protein. In the results presented here, expression of gap is modestly induced in stationary phase (see below). Expression of leuB is discussed later.
Several stress proteins have been shown to be induced prior to or at the onset of sporulation. According to the B. subtilis classification of heat shock proteins (72), the classes induced during solventogenesis in C. acetobutylicum include the class I hrcA (orfA)-grpE-dnaK-dnaJ (CAC1280-CAC1283) and groESL (CAC2704-CAC2703) transcripts (7, 56) and the previously described class III hsp18 transcript (CAC3714) (56, 66, 69). All of these transcripts were upregulated 2.5- to 9.3-fold by the onset of solventogenesis at time point E (Fig. 2C). Just before the onset of solventogenesis at time point D, some of the class I genes were first upregulated at lower levels, including hrcA (2.1-fold), groES (3.0-fold), and groEL (2.2-fold).
Other heat shock proteins were also induced during the shift to solvent production (Fig. 2C). The ctsR-yacH-yacI-clpC (CAC3192 to CAC3189) operon was induced 5- to 11-fold at time point E. We note that class III genes in B. subtilis are regulated by CtsR (15), and others have noted that the hsp18 gene in C. acetobutylicum has an exact B. subtilis-like CtsR recognition sequence directly upstream of the gene (72, 84). The class IV (regulated by neither HrcA nor CtsR) hsp90 (CAC3315, htpG) gene was induced 4.3-fold at time point D and 12-fold at time point E. The putative protease-encoding lonA (CAC0456) gene was induced 3.5- to 5.6-fold starting at time point F.
In the WT-M5 comparison, several heat shock proteins have higher expression in M5 at time point d (Fig. 2B and C), which corresponds to early stationary phase in both cultures (Fig. 1B). In our previous analysis, we reported higher expression of ctsR, hsp90, dnaJ, groES, clpC, and dnaK in M5 at time points corresponding to 10 h after the onset of sporulation in the WT strain (83). One possibility is that expression of the heat shock proteins in M5 may be a response to stress from increased butyrate accumulation (relative to the WT) at lower pH values (31) since these cultures were carried out without pH control.
Expression of primary metabolic genes.
Expression of the acetate formation genes pta (CAC1742) and ack (CAC1743) increased up to twofold during the transitional and early stationary phases (Fig. 2A). Previous Northern analysis of ack expressed in chemostat cultures indicated relatively constant expression between acidogenic and solventogenic cells (92). The expression of butyrate formation genes ptb (CAC3076) and buk (CAC3075) was slightly upregulated (1.5- to 2.3-fold) during the late exponential and early stationary phases. Northern (29) and reporter (86) analyses showed that ptb-buk transcript expression was highest in late exponential phase, corresponding to the expression pattern presented here.
Expression of the butyryl coenzyme A (butyryl-CoA) formation genes thl (CAC2873), hbd (CAC2708), crt (CAC2712), etfA (CAC2709), etfB (CAC2710), and bcd (CAC2711) generally increased in stationary phase but not more than 1.9-fold. Two previous analyses of thl expression in batch cultures have indicated similar expression patterns (29, 86), but a third study with a continuous culture indicated that thl has relatively high expression during the early exponential growth phase, decreased expression through the remaining exponential growth phase, and then higher expression corresponding to solvent production (93). It is unclear if the difference in early exponential-phase gene expression is a continuous-culture-specific effect.
The glycolytic genes for the conversion of glucose to acetyl-CoA (Fig. 2A) show variable expression patterns. Expression of the first gene in the pathway, the putative glucokinase (CAC2613), decreases 2.6-fold near the onset of stationary phase (time point F). Most of the remaining glycolytic genes showed higher expression at stationary-phase time points, including the triosephosphate isomerase gene (tpi, CAC0711; 2.0-fold higher, time point G), gap (CAC0709; 2.2-fold higher, time point H), the phosphoglycerate mutase gene (pgm, CAC0712; 2.8-fold higher, time point G), and the enolase gene (eno, CAC0713; 11-fold higher, time point H). However, expression of 3-phosphoglycerate kinase (pgk, CAC0710) changed no more than 1.5-fold (time point D). Several transcriptional start sites and transcripts have been identified in the CAC0709-to-CAC0713 locus (including gap, gap-pgk, gap-pgk-tpi, and tpi) (70), and the eno and pgm genes are apparently regulated separately. Therefore, we would not expect a common expression pattern within this genetic locus.
In acidogenesis, glycolysis-generated NADH is reduced by ferredoxin, which is, in turn, regenerated by hydrogenase. During stationary phase, NAD(P)H is required for solvent production. As NAD(P)H is shunted to solventogenesis, hydrogenase activity decreases (27, 36). This has also been shown on the transcript level, as hydA expression in acidogenic, continuous cultures is higher than in solventogenic cultures (27). In the WT time course (Fig. 2A), expression of hydA (CAC0028) decreases 2.9-fold at point F and remains low for the remaining stationary-phase time points, thus confirming prior reports.
SOM analysis of functional groups.
SOM analysis was used to look for global expression patterns across the genome. Data for genes that had signal intensities significantly greater than the background at all nine time points (3,546 genes out of a total of 3,802 on the microarray) were standardized and then organized by SOMs into 24 clusters. The expression patterns for all 24 clusters were visualized with an Eisen plot (Fig. 3A).
FIG. 3.
Eisen plots of SOM clusters of gene expression in the WT time course (A) and WT-M5 comparison (B). The clusters are grouped by average-linkage hierarchical clustering to organize similar expression patterns. Distribution of gene functional groups across the clusters is represented colorimetrically next to the Eisen plots. Functional groups: C, energy production and conversion; D, cell division and chromosome partitioning; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; G, carbohydrate transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; J, translation, ribosomal structure, and biogenesis; K, transcription; L, DNA replication, recombination, and repair; M, cell wall and membrane biogenesis; N, cell motility and secretion; O, posttranslational modification, protein turnover, and chaperones; P, inorganic-ion transport and metabolism; Q, secondary-metabolite biosynthesis, transport, and catabolism; R, general function prediction; S, function unknown; T, signal transduction mechanisms; U, hypothetical protein; W, predicted membrane protein.
In order to examine the SOM clusters for enriched functional genetic classes, we calculated the frequency at which genes in a functional category were enriched in a cluster compared to the total number of genes in the analysis within the same functional category. The functional classes used in this analysis were included in the original C. acetobutylicum annotation (51) and are based on the category-of-gene annotation (81). In cases where a gene is in two functional classes, the gene was counted twice, once for each functional category; therefore, there were more data points (n = 3,769) in the functional gene analysis than actual genes (3,546 genes) (Fig. 3A).
Clusters t12 and t18 are very similar, both containing genes whose expression level is lower in the stationary phase than that in exponential-growth phase. While the two combined clusters have 487 (12.9%) of the genes in this analysis, the clusters are enriched with 26% of all nucleotide transport and metabolism genes (category F), thus suggesting that this activity is downregulated in stationary phase. These clusters also contain 46% of the genes related to translation (category J), which suggests that, as expected, transcription of proteins for translation is higher in exponential phase.
Clusters t6 and t7 also show decreased expression at transitional- and stationary-phase time points. The two clusters contain 497 (13.2%) of the genes but are enriched with 43% of the cell motility genes (category N) and 19% of the cell wall-membrane biogenesis genes (category M). The C. acetobutylicum genome contains 108 motility-related genes, and 104 of these genes were included among the aforementioned 3,546 genes of this analysis. The majority of genes annotated as having roles in assembly of the flagella or chemotaxis are downregulated at some point during or after late exponential-phase growth. Of the 104 motility-related genes, over 40 genes were downregulated by at least threefold in transitional or stationary phase (time points F to I). In previous studies with C. acetobutylicum nonsolventogenic, asporogenous strain M5, we noted that expression of motility genes in stationary-phase cultures was higher in M5 than in the WT (83), thus suggesting that stationary-phase gene regulation associated with solventogenesis and sporulation downregulates motility gene expression.
pSOL1 gene expression.
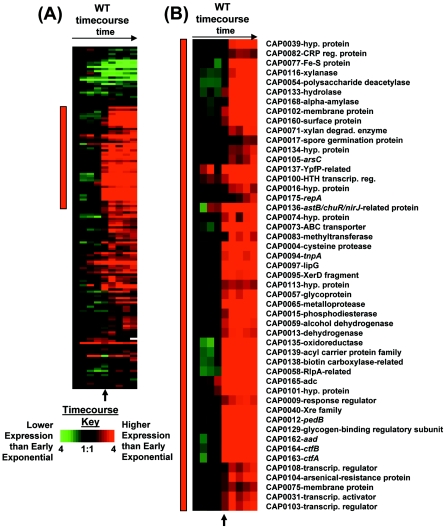
The majority of pSOL1 genes have increased expression at the beginning of stationary phase (Fig. 4A). The SOM clusters with the most pSOL1 genes are t11 (51 genes), t17 (21 genes), t16 (13 genes), t10 (11 genes), and t4 (10 genes). More than 20 genes were induced at least 10-fold at the onset of solventogenesis. Some of the strongly induced genes include (Fig. 4B) the solventogenic megaplasmid genes as previously described (Fig. 2A); CAP0058 (rare lipoprotein A-related protein), CAP0101 (hypothetical protein), CAP0102 (membrane protein), and CAP0129 (glycogen-binding regulatory subunit of S/T protein phosphatase I); and the loci CAP0135 (annotated as an oxidoreductase), CAP0136 (astB-chuR-nirJ-related protein), CAP0138 (related to biotin carboxylase N-terminal fragment), and CAP0139 (acyl carrier protein family). We previously observed that butanol stress induced expression of CAP0058 in a groESL-overexpressing strain (84) and CAP0102 in four different C. acetobutylicum strains (3).
FIG. 4.
Expression profiles of 160 pSOL1 genes (A) and a selected cluster of upregulated genes (B). The red bar in panel A indicates the location of the selected genes in panel B. The arrow indicates the onset of solventogenesis and sporulation. The colorimetric scale is the same as in Fig. 2. White squares indicate that microarray signals were too close to the background to calculate a ratio. hyp., hypothetical; reg., regulatory; degrad., degrading; transcrip. reg., transcription regulator.
Amino acid biosynthesis (functional category C).
A previous study noted that protein expression of the isopropylmalate dehydrogenase gene leuB (CAC3171) was induced at the onset of solventogenesis (69). It is part of a cluster of genes (CAC3169 to CAC3176) putatively annotated as part of the branched-chain amino acid synthesis pathway that converts pyruvate into leucine, isoleucine, and valine. These genes and the putative branched-chain amino acid transaminase (ilvE, CAC1479) show a pattern (Fig. 2B) of upregulation in stationary phase, especially at the early stationary-phase time point, where several of these genes are upregulated over 30-fold compared to exponential phase. Genes CAC3169 to CAC3176 are included in cluster t5 (highest expression at the sixth microarray time point), and CAC1479 is in cluster t17 (also highest expression at the sixth time point but sustained expression afterward). One explanation may be adaptation of membrane fluidity. Transcriptional analysis of the B. subtilis response to cold shock (30 to 90 min after a shift from 37 to 18°C) showed that several genes related to branched-chain amino acid synthesis (ilvN, ilvC, leuA, leuB, leuC, leuD, ilvD, and metC) were downregulated following the stress (38). B. subtilis is capable of converting branched-chain amino acids into branched-chain fatty acids (48), and it is well established that C. acetobutylicum adjusts membrane fluidity in response to solvent stress (5, 6, 44, 87). Thus, it seems possible that this is an adaptive response to the initial formation of solvents in early stationary-phase cultures, although literature reporting fatty acid contents of clostridia (5, 34, 44, 87) has not specifically reported the presence of branched-chain fatty acids.
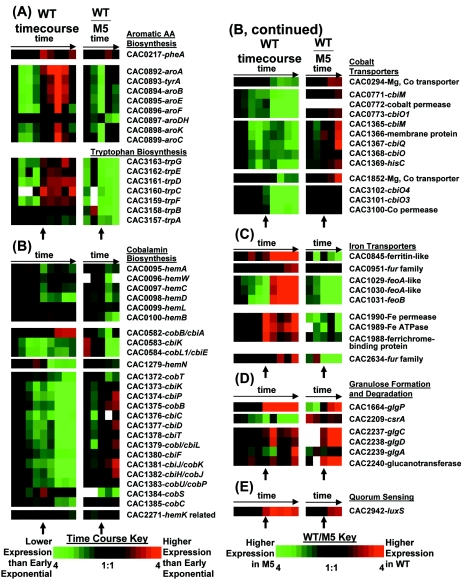
Genes required for the synthesis of the aromatic amino acids phenylalanine, tyrosine, and tryptophan are found primarily in two clusters (Fig. 5A). The aro locus, CAC0892 to CAC0899, consists of genes required for the formation of the intermediate chorismate and other precursors for phenylalanine and tyrosine synthesis; another cluster, CAC3157 to CAC3163, converts chorismate into tryptophan. Conversion of the intermediates to amino acids requires an aspartate aminotransferase (five genes are putatively annotated as such in the C. acetobutylicum genome), and phenylalanine synthesis also requires prephenate dehydratase (CAC0217). The major clusters of aromatic amino acid biosynthesis genes are upregulated during stationary phase, with maximum upregulation occurring in early stationary phase. The cluster containing CAC0892 to CAC0899 is expressed up to sevenfold more strongly in early stationary phase, and the tryptophan formation cluster containing CAC3157 to CAC3163 is expressed up to threefold more strongly. The aro locus members are grouped in clusters t2 (CAC0896 and CAC0898), t3 (CAC0897), and t4 (CAC0892 to CAC0895 and CAC899), and the tryptophan formation genes are grouped in clusters t4 (CAC3159 to CAC3163), t16 (CAC3158), and t18 (CAC3157).
FIG. 5.
Expression profiles of selected genes related to aromatic amino acid biosynthesis (A), cobalamin biosynthesis and cobalt transport (B), iron transport (C), granulose formation and degradation (D), and quorum sensing (E). Arrows indicate the onset of solventogenesis and sporulation. The colorimetric scale is the same as in Fig. 2. White squares indicate that microarray signals were too close to the background to calculate a ratio.
Translation (functional category J).
The majority of translation-related genes (54%) are grouped in downregulated clusters t12, t13, and t18 (16.7% of all genes). The C. acetobutylicum genome includes 56 genes encoding ribosomal proteins. Of the 52 genes of ribosomal proteins included in the 3,546 genes in this analysis (see above), 28 are downregulated at least threefold in stationary phase (time points G to I), and of those, 16 are downregulated at least sixfold. This indicates that reduction of cellular growth and the onset of sporulation are apparently associated with a widespread downregulation in the cell's protein synthesis machinery.
Aminoacyl-tRNA synthetases attach tRNAs with the appropriate amino acid. Transcription of the genes encoding these proteins has a mixed response to the stationary phase, with approximately half being downregulated when entering the stationary phase (data not shown). The reason for specific aminoacyl-tRNA synthetase downregulation is not clear. It is noted that the seryl-tRNA synthetase (CAC0017) is upregulated up to 180-fold more strongly in stationary phase (Fig. 2B), which is consistent with solventogenic-phase protein level reports for this protein (69).
Inorganic-ion transport and metabolism (functional category P).
SOM analysis of inorganic-ion-related genes (Fig. 3A) does not reveal any specific enrichment within a cluster. However, there are clear patterns among specific ion transporters.
Expression of genes related to inorganic-ion metabolism is dependent on the type of ion targeted by the gene. Numerous genes putatively related to cobalt metabolism (CAC0294, CAC0771 to CAC0773, CAC1365, CAC1368, CAC1369, and CAC1852; Fig. 5B) had decreased expression in later stages of growth. Cobalt is required for cobalamin (vitamin B12) activity, and the C. acetobutylicum genome contains the genes for its (anaerobic) biosynthesis (53, 63). A large number of these genes are also downregulated in later stages of growth, which may suggest coregulation between these two classes of genes. Cobalamin is a necessary cofactor for various reactions involving rearrangements including glycerol dehydratases for glycerol metabolism and transmethylation for the formation of methionine from homocysteine (28, 32).
Other inorganic-ion transport-related systems are upregulated in stationary phase, including several genes related to iron uptake (Fig. 5C). Two genes (CAC0951 and CAC2634) related to the iron uptake regulator Fur are upregulated during the transition to stationary phase. A putative operon encoding two FeoA-like proteins (CAC1029 and CAC1030) and an FeoB-like GTPase (CAC1031) was upregulated at least 18-fold in stationary phase. The ferrous uptake system feo, which was initially characterized in Escherichia coli (39), is believed to be responsible for anaerobic iron uptake. Clostridium perfringens (75) and Clostridium thermocellum (Department of Energy Joint Genome Institute, http://img.jgi.doe.gov) contain orthologs for both feoA and feoB, and Clostridium tetanii (9) contains an feoB ortholog. Other iron uptake genes upregulated in stationary phase include a putative iron transport operon (CAC1988 to CAC1990), a gene encoding a product similar to the iron storage protein ferritin (CAC0845), and a ferrichrome transport permease (CAC0788). Iron limitation has been shown to inhibit sporulation in B. subtilis (1). Specifically, the aconitase protein, a citric acid cycle gene required in B. subtilis for sporulation (14), requires iron (24), so it is possible that upregulation of iron uptake genes coincides with this metabolic demand. Iron limitation has also been shown to decrease acetone formation (37) as the acetoacetate decarboxylase enzyme requires iron (90).
Lipid transport and metabolism (functional category I).
A large cluster of genes (data not shown) was identified that contains genes annotated as being related to fatty acid synthesis. The annotations include two 3-oxoacyl-(acyl carrier protein) synthases (CAC2008 [pksF] and CAC2011 [fabH]), 3-hydroxyacyl-CoA dehydrogenase (CAC2009 [mmgB]), two enoyl-CoA hydratases (CAC2012 and CAC2016 [both named fadB]), an acyl carrier protein (CAC2017), and malonyl CoA-acyl carrier protein transacylase (CAC2019). The upregulated lipid metabolism genes described above are part of a large locus of 47 genes (CAC1980 to CAC2026), nearly all of which show strong upregulation in stationary phase. Twenty-nine of these genes are in cluster t17, and 12 are in cluster t23, both indicating higher expression in stationary phase (Fig. 3A). All but 3 are on the same DNA strand, and 37 of the genes are upregulated at least fivefold in stationary phase. We have previously shown that fatty acid synthesis genes on a different locus (CAC3568 to CAC3579) were upregulated in spo0A-overexpressing strain 824(pMSPOA) (3). Since 824(pMSPOA) undergoes sporulation earlier than the plasmid control strain (29), this gene expression pattern was attributed to sporulation. Here, genes CAC3568 to CAC3573 had 1.5- to 2.2-fold increased expression at time point F (data not shown), near the onset of sporulation. In B. subtilis, fatty acid synthesis is required for sporulation (71) and various genes related to fatty acid metabolism have been shown to be directly regulated by σE (10, 19) and Spo0A (22, 49). Thus, it may be inferred that fatty acid metabolism might also be regulated by a stationary-phase regulator like Spo0A in C. acetobutylicum.
Expression analysis of functional groups in WT versus M5 cultures.
The results presented here confirm the expression of some known stationary-phase events in C. acetobutylicum. Another way to understand stationary-phase phenomena is to compare the WT strain with M5, which is unable, due to loss of the megaplasmid, to sporulate or make solvents, both of which are stationary-phase events. We performed SOM analysis of the WT-M5 data set based on the analysis of the experiment shown in Fig. 1B. A small subset (the pSOL1 genes) of these data was used to validate the microarrays (2), but otherwise, these data have not been biologically analyzed. Microarray analysis of M5 cultures with partial-genome arrays (ca. 25% coverage) has been previously published (83); note that in this present analysis the ratios are reported oppositely (WT/M5 compared to M5/WT previously). Genes that contained a signal above the background in at least one channel for all five time points (3,064 genes out of 3,802 present on the microarray) were standardized and organized with SOMs, and functional groups across SOM clusters (n = 3,251 functional categorizations for 3,064 genes) were analyzed as described above for the WT time course microarray analysis (Fig. 3B).
Of the 127 pSOL1 genes included in the set of 3,064 genes, 109 are included in the three similar clusters c14, c18, and c19 (as indicated by the close proximity on the hierarchical tree of Fig. 3B), all indicating lower expression in M5 at four or five time points. Several solventogenic genes are included in clusters c14 and c19, including the megaplasmid genes aad, ctfA, ctfB, and adc and the chromosomal butanol dehydrogenase gene bdhA. Previous transcriptional analysis of M5 showed significantly decreased expression of these solventogenic genes (83). Also included in these clusters is the putative sigD sigma factor (CAC2143; regulator of flagella, motility, and autolysin expression in B. subtilis).
Nearly 50% of the genes annotated as motility related (category N) are included in cluster c17 (down in M5 during the exponential phase) or cluster c18 (down in M5 at all five time points). We previously noted that that lower expression of motility-related genes in M5 occurs during exponential phase (83). The biological interpretation of this result may be that, during the exponential-growth phase, WT cultures are more motile, but during the stationary phase, WT expression of motility genes decreases (as discussed above), thus leading to decreased expression relative to M5. Another nonmotile and nonsolventogenic C. acetobutylicum mutant, DP4-X, has been described as having an altered flagellin and lacking three solventogenic enzymes which probably correspond to the aad, ctfA and ctfB, and adc genes. These results seem to describe a degenerate mutant like M5 (55).
Clusters c5, c10, c11, and c15 contain genes with higher expression in M5, especially during the stationary phase (Fig. 3B). These clusters contain 26% of the genes in the SOM analysis but are enriched with 34% of the genes related to carbohydrate transport and metabolism, including the glycolytic genes pfk, gap, pgk, tpi, pgm, and eno and all five genes used in the conversion of acetoacetyl-CoA to butyryl-CoA (hbd, etfA, etfB, bcd, and crt) (Fig. 2A), a pattern we have previously reported (higher expression in M5) (82). Also included in clusters c5, c10, c11, and c15 are 41% of the genes related to energy production (category C) and 45% of the genes related to coenzyme transport and metabolism (category H). Finally, 46% of the category O genes (posttranslational modification, protein turnover, and chaperone genes) belong also to clusters c5, c10, c11, and c15. Included are many chaperone transcripts, such as the operon hrcA-grpE-dnaK-dnaJ, clpC, hsp90, hsp18, protease-encoding clpA (CAC1824), and two lonA-annotated genes (CAC2637 and CAC0456). All these genes have the same general expression pattern, with significantly higher expression in M5 at the two stationary-phase time points. The chaperone operon groESL (CAC2703 and CAC2704) is in cluster c0 (higher in M5), but expression in M5 does not exceed WT expression by more than 1.9-fold.
Forty percent of the amino acid synthesis and metabolism genes (category E) are in clusters c1, c5, and c6 (higher expression in M5) and cluster c9 (downregulated in M5). in accordance with the KEGG Pathway database (www.genome.jp/kegg/pathway.html), significant numbers of genes in clusters c1, c5, and c6 were identified as being part of branched-chain amino acid synthesis (10 genes), histidine metabolism (8 genes), and aromatic amino acid biosynthesis (11 genes). While expression during exponential phase of several branched-chain amino acid synthesis genes is higher in M5 than in the WT, the expression levels are similar during stationary phase (Fig. 2B). Since it was previously shown that expression of these genes was upregulated in the WT in the stationary phase, the diminished difference in expression of the genes in stationary phase is likely due to upregulation in the WT cultures. Thus, we conclude that in the WT, expression of branched-chain amino acid synthesis genes is correlated with stationary-phase gene expression.
In stationary phase, clusters c3, c8, and c9 exhibit higher expression in the WT than in the M5 strain. While these clusters contain 23% of the genes analyzed, they contain 46% of the translation and ribosomal structure and biogenesis genes (category J). Thirty-three of these genes are specifically annotated as ribosomal proteins, and eight encode aminoacyl-tRNA synthetases, including seryl-tRNA synthetase (CAC0017). This seems to contradict what was reported during the WT time course (Fig. 3A). One possible explanation is that both the WT and M5 strains downregulate the expression of ribosomal proteins in stationary phase, but downregulation is more profound in M5. All four of the previously mentioned serine biosynthesis genes were assigned to cluster c9, confirming that expression of this operon is associated with stationary phase (Fig. 2B) (69).
Clusters c3, c8, and c9 also contain many sporulation-related genes, including the sporulation-specific transcriptional factor spo0A (CAC2071), the putative spoIIA operon (CAC2306 to CAC2308) that includes sigF (CAC2306), spoIIGA (CAC1694), and sigE (CAC1695), sigG (CAC1696), and nine predicted stage V sporulation-related genes. abrB (CAC0310) (74), the transitional stage gene regulator that is antagonistic to spo0A, has roughly the opposite expression of spo0A, being located in cluster c10 (higher expression in M5 in stationary phase). Higher expression of sporulation genes in the WT is expected due to the M5 asporogenous phenotype.
Expression of sporulation genes.
The time course of sporulation events in B. subtilis is more thoroughly studied than in any clostridia. Research on the early sporulation events in clostridia has been recently reviewed (17). Much of this research to date has examined the expression of Spo0A, which is the master regulator of stationary-phase gene expression in both the bacilli and the clostridia (29, 30, 59). The C. acetobutylicum sigma factors σA, σE, σG, and σK have been cloned (68, 94) and shown to be expressed in a temporal fashion similar to that in B. subtilis (65). Since sporulation is a complex process involving hundreds of genes, we wanted to examine if orthologous sporulation genes in C. acetobutylicum were expressed temporally like the corresponding genes in B. subtilis. We take Spo0A activation (phosphorylation) as the beginning of the differentiation (sporulation) program. In B. subtilis, the end of exponential-phase growth coincides with several early sporulation events, including transcription of the histidine kinase kinA (57), expression of spo0A (57), and repression of abrB (a key target of phosphorylated Spo0A) (33). Here, we use the induction of the now well-established phosphorylated Spo0A targets (29, 59) solventogenic genes aad-ctfA-ctfB and adc as corresponding to the onset (T0) of sporulation, and this corresponds to time point E of the WT time course analysis (t = 9.25 h) and time point c (t = 8.4 h) of the WT-M5 analysis. These time points approximately correspond to the first appearance of clostridial cigar-shaped forms (data not shown). B. subtilis literature on sporulation and gene expression frequently refers to T0 as the onset of sporulation, and numbers larger than zero indicate later time points (for example, T1 denotes 1 h after the onset of sporulation). Selecting this time point on a growth curve can be inexact, as morphological changes are not readily apparent.
We have previously published a transcriptional analysis (83) of the spo0A knockout strain SKO1 (29). Figure 6 displays the expression patterns of selected sporulation genes for the WT-versus-SKO1 comparison in addition to the present WT time course and the WT-versus-M5 comparison. The WT-SKO1 results were normalized as described in the original analysis (83); also, the expression ratios were inverted to WT/SKO1.
FIG. 6.
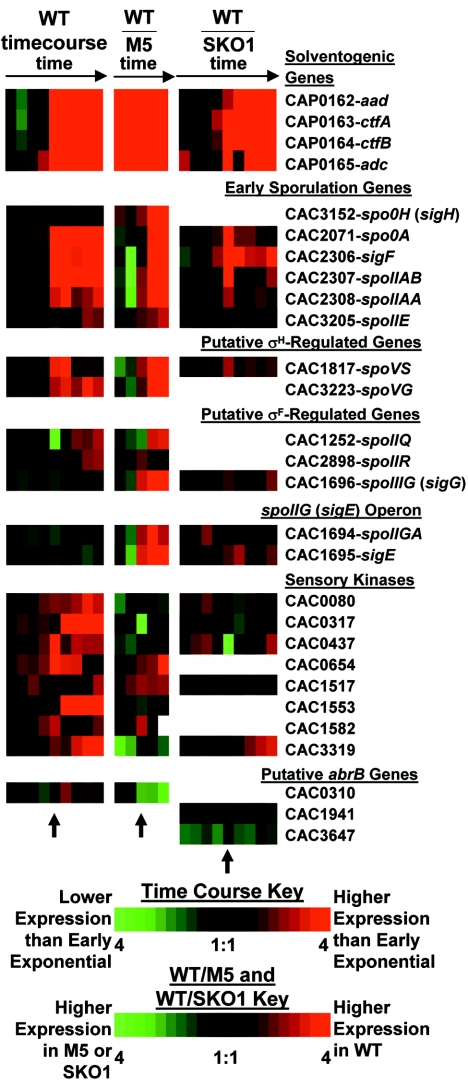
Expression profiles of solventogenic and sporulation-related genes in the WT time course, the WT-M5 comparison, and the previously published WT-SKO1 comparison (83). Note that the expression ratios of the WT-SKO1 comparison are inverted with respect to the previously published values (83). The colorimetric scale is the same as in Fig. 2. White squares indicate that either microarray signals were too close to the background to calculate a ratio (all three experiments) or the gene was not present on the partial-genome microarray (83) (WT-SKO1 comparison only).
The first sigma factor associated with the sporulation cascade is σH (encoded by the spo0H/sigH gene), which directs the expression of some of the earliest expressed sporulation genes, including spo0A (57) and the sigF operon (96). Expression of the corresponding sigH gene in B. subtilis is driven by the primary sigma factor σA, and peak expression of spo0H occurs at 40 min before the onset of sporulation, after which expression decreases (89). It has been recently suggested that in C. acetobutylicum, spo0H expression is constitutive (17). Expression of the putative C. acetobutylicum spo0H/sigH gene (CAC3152) is slightly higher at time points E (onset of sporulation) and G (3 h after the onset of sporulation) (Fig. 6). Expression of CAC3152 is higher in WT cells than in M5 cells and steadily increases throughout the stationary phase. Thus, the expression pattern of the putative sigH gene does not exactly match the orthologous B. subtilis gene. In order to examine if σH is functional in C. acetobutylicum, we next tried to analyze the expression of the orthologous C. acetobutylicum σH regulon. Twenty-one operons in B. subtilis are known to be directly regulated by σH (summarized in reference 8). Of the known σH-regulated genes, a limited number (dnaG-sigA, ftsAZ, spoIIAA-spoIIAB-sigF, spo0A, spoVG, and spoVS) have orthologs in C. acetobutylicum. Since the dnaG-sigA and ftsAZ (26) operons in B. subtilis are also known to be expressed by σA promoters and it is already known that spo0A is expressed at the onset of sporulation, we used as a canonical set of σH regulon genes in C. acetobutylicum the spoVG (CAC3223) and spoVS (CAC1817) genes. In B. subtilis, spoVS expression is induced around T0 to T1 (60) and expression of spoVG is induced near the onset of sporulation (89, 100). Hidden Markov models trained with B. subtilis σH-binding motifs as previously described (53) identified potential σH-binding motifs upstream of these putative C. acetobutylicum spoVG and spoVS genes (data not shown). In the C. acetobutylicum time course (Fig. 6), spoVS shows induced expression at time points D (1.5 h before the onset of sporulation), E (the onset of sporulation), and F (1.5 h after the onset of sporulation). spoVG has induced and sustained expression starting at the onset of sporulation. Both genes are more strongly expressed in stationary-phase WT cells than in M5 cells. In the WT-SKO1 comparison, spoVS expression coincides with the initial expression of spo0A and aad-ctfA-ctfB. These results strongly suggest that the putative spoVG and spoVS genes have a role in stationary-phase activities and, owing to their early expression and similarities to B. subtilis data, may also be regulated by σH. Thus, sigH seems to be expressed and active just before T0.
Forespore-specific sigma factor σF is encoded in the spoIIAA-spoIIAB-sigF operon, is known to be regulated by both σH and Spo0A (95), and is typically expressed within an hour of the onset of sporulation in B. subtilis (21). In the time course C. acetobutylicum microarray data, the genes spoIIAA (CAC2308), spoIIAB (CAC2307), and sigF (CAC2306) are highly expressed starting at the onset of sporulation (time point E), as expected. In the M5 experiment, the transcripts are expressed more strongly in WT cells at the stationary-phase time points. In the WT-SKO1 comparison, expression of spoIIAA-spoIIAB-sigF coincides with the onset of spo0A and aad-ctfA-ctfB expression. Two C. acetobutylicum genes homologous to B. subtilis σF-regulated genes were upregulated 3 to 5 h after sigF expression in the WT time course experiment: spoIIQ (CAC1252; 2.6-fold higher expression at time point I) and spoIIR (CAC2898; 2.4-fold higher expression at time point I). In B. subtilis, expression of spoIIR and spoIIQ has been shown to begin 90 to 120 min after the onset of sporulation (40, 46). The expression of spoIIQ and spoIIR indicates σF functionality. A third σF-regulated gene, spoIIIG/sigG (encoding the σG sigma factor), is discussed later.
spoIIE is a serine phosphatase necessary for sporulation which dephosphorylates SpoIIAA so that it can release SpoIIAB from σF (16). Expression is driven by a σA promoter but requiring phosphorylated Spo0A (99). In B. subtilis, expression of spoIIE starts approximately 1 h after the onset of sporulation, with the highest expression approximately 1 h later; spoIIE has a temporal expression pattern similar to that of the spoIIA (sigF) and spoIIG (sigE) operons (99). Expression of spoIIE in C. acetobutylicum (CAC3205) has been shown to coincide with solvent formation, and antisense downregulation of the gene has been shown not to affect solvent formation (73). In the experiments presented here, spoIIE expression starts 3 to 5 h following sigF expression and the onset of sporulation and peaks approximately 5 h after the onset of sporulation (2.2-fold higher expression than the initial time point).
In B. subtilis, expression of the mother cell-specific sigma factor σE begins approximately 2 h after the onset of sporulation (41) and significant expression of spoIIIG (sigG) begins 150 to 210 min after the onset of sporulation (79). In the experiments presented here, expression of the sigE operon (spoIIGA-sigE; CAC1694 and CAC1695) and the sigG transcript (CAC1696) does not exceed 1.3-fold higher expression than the first microarray time point. If we examine several homologous genes known to be expressed from a σE-binding site in B. subtilis (i.e., spoIIP, spoIID, spoIIM, spoIIID, the spoIIIA operon, spoIVFB, spoVD, spoVE, and spoVR), we find that there is no clear pattern of expression in either the WT-M5 comparison or WT time course microarrays (data not shown). These results seem to indicate that σE and σG are not active in the sporulating clostridial cell up to 7.5 h after the onset of sporulation (and solventogenesis). In WT cells, Northern analysis of sigE and sigG had indicated expression approximately 10 h after the onset of sporulation (29). Expression data comparing the WT and M5 strains indicate that spoIIGA, sigE, and sigG are all expressed more strongly in WT cells than in M5 cells (2.6- to 18-fold more strongly at time point e, 9 h after the onset of solventogenesis), indicating that the genes are expressed in stationary-phase WT cultures.
Phase-contrast microscopy and differential interference contrast microscopy were performed to compare gene expression with cellular morphology. Swollen clostridial cells started to appear at 6.75 h (microarray time point C), with a greater abundance at 12.4 h (point I), and forespores and endospores began to appear at 26.7 h. The long delay in forespore appearance coincides with the apparent inability to detect sigE expression (Fig. 6). Consistent with this observation, microscopy of C. acetobutylicum P262 (now Clostridium sp. strain NCP262) indicated a 15- to 20-h delay between initial clostridial-form formation and forespore formation (35, 47). Collectively, these data suggest that forespore formation in C. acetobutylicum requires several more hours than in B. subtilis.
It is well established that clostridia do not contain an orthologous phosphorelay system or any identifiable sensory kinases similar to those (KinA, KinB, KinC, KinD, and KinE) employed by B. subtilis to phosphorylate Spo0A (51, 77, 78). We have previously suggested that the expression pattern of the C. acetobutylicum gene CAC3319 (annotated as encoding a signal transduction histidine kinase) might indicate a role similar to that of the aforementioned B. subtilis sensory kinases (83, 84). With the present time course microarray data, we can identify other candidate proteins that may act as sensory kinases for sporulation. For example, in B. subtilis, kinA is expressed maximally within 40 min of the onset of sporulation (4, 57) and expression of kinC starts at the onset of sporulation and peaks 1 h later (43). Therefore, it would be expected that expression of the putative kinA gene should be highest near the onset of sporulation and solventogenesis, corresponding to time point E or a slightly earlier time point. Of the 41 genes annotated in the C. acetobutylicum genome as being sensory histidine kinases, 8 are temporally upregulated near the onset of sporulation and solventogenesis (Fig. 6). In the case of M5, it is unclear whether loss of the megaplasmid would affect the expression of the putative sensory kinases. Of the eight potential sensory kinases, three (CAC0654, CAC1517, and CAC1582) are expressed more strongly in WT cells than in M5 cells during the transitional and stationary growth phases.
C. acetobutylicum contains three homologs (abrB0310, abrB1941, and abrB3647) for the transitional-state regulator abrB (51), and recent experiments suggest that abrB0310 might be the true transitional-state regulator (74). Because the three sequences are very similar (3), we designed the full-genome microarray to contain a probe for abrB0310 with the expectation that it could hybridize to all three abrB genes. The previous partial-genome microarrays contained probes for abrB1941 and abrB3647. In the WT-M5 and WT-SKO1 comparisons (Fig. 6), the expected result was observed with higher expression in both SKO1 (abrB3647) and M5 (abrB0310), which suggests that these asporogenic strains are unable to downregulate abrB expression. However, abrB0310 expression in the WT time course unexpectedly was highest at time point F, which is after the onset of sporulation. Since maximum expression of abrB in B. subtilis occurs 2 h before the onset of sporulation (33), it is unclear if the microarray probe is picking up signals from all three genes.
Putative granulose formation genes?
Starting in early stationary phase, solvent-forming clostridia produce granulose (61, 62), a glycogen-like polymer used widely by bacteria for energy storage. Synthesis of the polymer via the ADP-glucose pathway requires ADP-glucose pyrophosphorylase (EC 2.7.7.27) and granulose synthase (EC 2.4.1.21). Operons containing these genes have been shown to be expressed by the sporulation-specific σH promoter in Bacillus stearothermophilus (42) and B. subtilis (8). The C. acetobutylicum genome contains a genetic locus (CAC2237, ADP-glucose pyrophosphorylase, glgC; CAC2238, ADP-glucose pyrophosphorylase, glgD; CAC2239, granulose synthase, glgA) bearing a sequential resemblance to these proteins. Operon prediction of the C. acetobutylicum genome (53) suggests a single transcript containing genes CAC2237 to CAC2240, where the annotation of CAC2240 suggests that it is a glucanotransferase; also, no σH promoter was located near the beginning of the transcript. Microscopy of the WT cultures indicated the first appearance of cigar shapes at 6.75 h (time point C) and a greater abundance at 12.4 h (time point I). In stationary phase, WT genes CAC2237, CAC2238, and CAC2240 are expressed up to 6- to 20-fold more strongly than in M5, confirming the correlation of granulose formation with solvent formation (61). At point F of the time course (1.5 h after the onset of sporulation), genes CAC2237, CAC2238, and CAC2240 are expressed two- to threefold more strongly than in mid-exponential-phase growth, generally with higher expression in all stationary-phase samples compared to exponential-phase growth, further suggesting a correlation with stationary-phase gene expression. Unexpectedly, CAC2239 generally had an expression pattern that was the opposite of that of the other genes, except for time point I, where its expression is 2.5-fold stronger than exponential-phase gene expression. Also in the C. acetobutylicum genome is a granulose phosphorylase (CAC1664, glgP) whose putative function is to depolymerize granulose and a putative carbon storage regulator (CAC2209, csrA), which in E. coli represses the expression of both glycogen biosynthesis (glgA) and degradation (glgP) genes (98). Expression of the putative csrA gene is downregulated at least threefold at time points E, H, and I, corresponding to the times during which granulose formation is expected. The putative glgP gene is upregulated starting at time point E and has 20-fold-induced expression at time point F. The reason for the contradictory expression of glgP and glgA is not apparent.
luxS expression.
Functional autoinducer systems have been demonstrated in Clostridium difficile (11) and C. perfringens (52). In C. perfringens, the luxS gene has been demonstrated to be involved in toxin production. Autoluminesence assays of C. perfringens indicate that maximum autoinducer (AI-2) activity occurs during mid-exponential-phase growth (52). The protein thought to be responsible for autoinducer production is S-ribosylhomocysteinase, which is encoded by luxS. Sequence alignment of the protein encoded by the putative C. acetobutylicum gene luxS (CAC2942, 474 bp) and the B. subtilis protein (BSU30670) with BLASTP indicates that, over 158 amino acids, there exist 37% identity (50 amino acids) and 53% positive (72 amino acids) matches. During the time course (Fig. 1), expression of CAC2942 is roughly proportional to cell density (Fig. 5E) and is highest during the transitional and early stationary phases, where, at time points F to I, expression is 3 to 3.5 higher than in the early exponential phase. This result suggests that expression of luxS is dependent on the growth phase and may be associated with stationary-phase phenomena.
DISCUSSION
With the present genomic microarrays, we were able to confirm almost all previously reported transcriptional kinetics but, significantly, provide a genomic insight into novel expression patterns of functional groups, operons, and individual genes as related to differentiation and stationary-phase phenomena. These include solvent formation and primary metabolism, sporulation, heat shock proteins, amino acid biosynthesis, translation-related proteins, motility, most pSOL1 genes, cobalamin biosynthesis, cobalt and iron uptake, lipid biosynthesis, granulose formation, and quorum sensing. We envision this data set as being a resource for anyone interested in differences in exponential-phase growth and stationary-phase gene expression for the categories listed previously and others not discussed in this paper, including glycosyltransferases, DNA replication and repair, cell division and partitioning, etc. Information about these classes and specific genes can be readily obtained from the supplemental material and is also available at our website (www.papoutsakisresearch.northwestern.edu).
Comparison of metabolic gene expression in the WT time course and the WT-M5 experiment indicates that M5 cultures have increased expression of glycolysis, butyryl-CoA formation, and butyrate formation genes in stationary-phase cultures (Fig. 2A). This seems to indicate a difference in the regulation of primary metabolism due to the loss of the megaplasmid. The continued production of butyrate in M5 seems to coincide with increased expression of heat shock proteins (Fig. 2B and C), which can be shown, by separate experiments, to induce a stress response.
Expression of some B. subtilis branched-chain amino acid synthesis genes is downregulated by the stationary-phase gene repressor CodY (50, 76), which presumably can also repress the expression of sporulation genes during exponential-phase growth (50, 58). Western blot assays of B. subtilis CodY indicated no change in protein expression during sporulation (58). The putative C. acetobutylicum codY gene (CAC1785) in the time course experiment has steadily decreasing expression at points B to H (mid-exponential growth phase through early stationary phase) compared to time point A (data not shown).
In B. subtilis, engulfment is completed 100 min after sporulation initiation, spoIIIG expression (σG, a prespore-specific event) starts at 120 min, and stage III is reached 150 min after sporulation onset (54). We were unable to confidently detect expression of sigE (CAC1695) or sigG (spoIIIG) in time course cultures within 7.5 h of spo0A and aad-ctfA-ctfB expression. Previous results have provided supporting evidence that spore formation is protracted in C. acetobutylicum. Continuous cultures of C. acetobutylicum have shown a 7- to 11-h lag from initial solvent production to significant sigE expression and forespore formation (65). As previously mentioned, other solventogenic clostridia have shown a 19-h delay between initial clostridial formation and forespore formation in a defined medium (47). The doubling time of C. acetobutylicum fermentation in this experiment was 1.0 h. While B. subtilis cells growing on rich medium would have a shorter doubling time, the magnitude of the difference in the forespore formation time between the organisms could not be explained by differences in growth rates alone.
. .
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation grant BES-0331402, Department of Energy grant DE-FG36-03GO13160, and an NIH/NIGMS Biotechnology Training Grant (T32-GM08449) fellowship to K.V.A.
We thank Carles Paredes for discussion of results and hidden Markov model calculations and Iwona Spath for the microscopy studies.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alen, C., and A. L. Sonenshein. 1999. Bacillus subtilis aconitase is an RNA-binding protein. Proc. Natl. Acad. Sci. USA 96:10412-10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alsaker, K. V., C. J. Paredes, and E. T. Papoutsakis. Design, optimization and validation of genomic DNA microarrays for examining the Clostridium acetobutylicum transcriptome. Biotechnol. Bioprocess Eng., in press.
- 3.Alsaker, K. V., T. R. Spitzer, and E. T. Papoutsakis. 2004. Transcriptional analysis of spo0A overexpression in Clostridium acetobutylicum and its effect on the cell's response to butanol stress. J. Bacteriol. 186:1959-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antoniewski, C., B. Savelli, and P. Stragier. 1990. The spoIIJ gene, which regulates early developmental steps in Bacillus subtilis, belongs to a class of environmentally responsive genes. J. Bacteriol. 172:86-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baer, S. H., H. P. Blaschek, and T. L. Smith. 1987. Effect of butanol challenge and temperature on lipid composition and membrane fluidity of butanol-tolerant Clostridium acetobutylicum. Appl. Environ. Microbiol. 53:2854-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baer, S. H., D. L. Bryant, and H. P. Blaschek. 1989. Electron spin resonance analysis of the effect of butanol on the membrane fluidity of intact cells of Clostridium acetobutylicum. Appl. Environ. Microbiol. 55:2729-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bahl, H., H. Muller, S. Behrens, H. Joseph, and F. Narberhaus. 1995. Expression of heat shock genes in Clostridium acetobutylicum. FEMS Microbiol. Rev. 17:341-348. [DOI] [PubMed] [Google Scholar]
- 8.Britton, R. A., P. Eichenberger, J. E. Gonzalez-Pastor, P. Fawcett, R. Monson, R. Losick, and A. D. Grossman. 2002. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 184:4881-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brüggemann, H., and G. Gottschalk. 2004. Insights in metabolism and toxin production from the complete genome sequence of Clostridium tetani. Anaerobe 10:53-68. [DOI] [PubMed] [Google Scholar]
- 10.Bryan, E. M., B. W. Beall, and C. P. Moran. 1996. A σE-dependent operon subject to catabolite repression during sporulation in Bacillus subtilis. J. Bacteriol. 178:4778-4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carter, G. P., D. Purdy, P. Williams, and N. P. Minton. 2005. Quorum sensing in Clostridium difficile: analysis of a luxS-type signalling system. J. Med. Microbiol. 54:119-127. [DOI] [PubMed] [Google Scholar]
- 12.Clark, S. W., G. N. Bennett, and F. B. Rudolph. 1989. Isolation and characterization of mutants of Clostridium acetobutylicum ATCC 824 deficient in acetoacetyl-coenzyme A:acetate/butyrate:coenzyme A-transferase (EC 2.8.3.9) and in other solvent pathway enzymes. Appl. Environ. Microbiol. 55:970-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornillot, E., R. V. Nair, E. T. Papoutsakis, and P. Soucaille. 1997. The genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 reside on a large plasmid whose loss leads to degeneration of the strain. J. Bacteriol. 179:5442-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Craig, J. E., M. J. Ford, D. C. Blaydon, and A. L. Sonenshein. 1997. A null mutation in the Bacillus subtilis aconitase gene causes a block in Spo0A-phosphate-dependent gene expression. J. Bacteriol. 179:7351-7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derre, I., G. Rapoport, and T. Msadek. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol. Microbiol. 31:117-131. [DOI] [PubMed] [Google Scholar]
- 16.Duncan, L., S. Alper, F. Arigoni, R. Losick, and P. Stragier. 1995. Activation of cell-specific transcription by a serine phosphatase at the site of asymmetric division. Science 270:641-644. [DOI] [PubMed] [Google Scholar]
- 17.Dürre, P., and C. Hollergschwandner. 2004. Initiation of endospore formation in Clostridium acetobutylicum. Anaerobe 10:69-74. [DOI] [PubMed] [Google Scholar]
- 18.Eichenberger, P., M. Fujita, S. T. Jensen, E. M. Conlon, D. Z. Rudner, S. T. Wang, C. Ferguson, K. Haga, T. Sato, J. S. Liu, and R. Losick. 2004. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2:1664-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eichenberger, P., S. T. Jensen, E. M. Conlon, C. van Ooij, J. Silvaggi, J. E. Gonzalez-Pastor, M. Fujita, S. Ben-Yehuda, P. Stragier, J. S. Liu, and R. Losick. 2003. The σE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 327:945-972. [DOI] [PubMed] [Google Scholar]
- 20.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Errington, J., and J. Mandelstam. 1986. Use of a lacZ gene fusion to determine the dependence pattern of sporulation operon spoIIA in spo mutants of Bacillus subtilis. J. Gen. Microbiol. 132:2967-2976. [DOI] [PubMed] [Google Scholar]
- 22.Fawcett, P., P. Eichenberger, R. Losick, and P. Youngman. 2000. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 97:8063-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feustel, L., S. Nakotte, and P. Dürre. 2004. Characterization and development of two reporter gene systems for Clostridium acetobutylicum. Appl. Environ. Microbiol. 70:798-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fortnagel, P., and E. Freese. 1968. Inhibition of aconitase by chelation of transition metals causing inhibition of sporulation in Bacillus subtilis. J. Biol. Chem. 243:5289-5295. [PubMed] [Google Scholar]
- 25.Gerischer, U., and P. Dürre. 1992. mRNA analysis of the adc gene region of Clostridium acetobutylicum during the shift to solventogenesis. J. Bacteriol. 174:426-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzy-Tréboul, G., C. Karmazyn-Campelli, and P. Stragier. 1992. Developmental regulation of transcription of the Bacillus subtilis ftsAZ operon. J. Mol. Biol. 224:967-979. [DOI] [PubMed] [Google Scholar]
- 27.Gorwa, M. F., C. Croux, and P. Soucaille. 1996. Molecular characterization and transcriptional analysis of the putative hydrogenase gene of Clostridium acetobutylicum ATCC 824. J. Bacteriol. 178:2668-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halpern, J. 1985. Mechanisms of coenzyme-B12-dependent rearrangements. Science 227:869-875. [DOI] [PubMed] [Google Scholar]
- 29.Harris, L. M., N. E. Welker, and E. T. Papoutsakis. 2002. Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824. J. Bacteriol. 184:3586-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang, I. H., M. Waters, R. R. Grau, and M. R. Sarker. 2004. Disruption of the gene (spoOA) encoding sporulation transcription factor blocks endospore formation and enterotoxin production in enterotoxigenic Clostridium perfringens type A. FEMS Microbiol. Lett. 233:233-240. [DOI] [PubMed] [Google Scholar]
- 31.Hüsemann, M. H. W., and E. T. Papoutsakis. 1988. Solventogenesis in Clostridium acetobutylicum fermentations related to carboxylic-acid and proton concentrations. Biotechnol. Bioeng. 32:843-852. [DOI] [PubMed] [Google Scholar]
- 32.Jeter, R., J. C. Escalante-Semerena, D. Roof, B. Olivera, and J. Roth. 1987. Synthesis and use of vitamin B12, p. 551-556. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 33.Jiang, M., W. Shao, M. Perego, and J. A. Hoch. 2000. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 38:535-542. [DOI] [PubMed] [Google Scholar]
- 34.Johnston, N. C., and H. Goldfine. 1983. Lipid composition in the classification of the butyric acid-producing clostridia. J. Gen. Microbiol. 129: 1075-1081. [DOI] [PubMed] [Google Scholar]
- 35.Jones, D. T., A. van der Weshuizen, S. Long, E. R. Allcock, S. J. Reid, and D. R. Woods. 1982. Solvent production and morphological changes in Clostridium acetobutylicum. Appl. Environ. Microbiol. 43:1434-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones, D. T., and D. R. Woods. 1986. Acetone-butanol fermentation revisited. Microbiol. Rev. 50:484-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Junelles, A. M., R. Janatiidrissi, H. Petitdemange, and R. Gay. 1988. Iron effect on acetone butanol fermentation. Curr. Microbiol. 17:299-303. [Google Scholar]
- 38.Kaan, T., G. Homuth, U. Mader, J. Bandow, and T. Schweder. 2002. Genome-wide transcriptional profiling of the Bacillus subtilis cold-shock response. Microbiology 148:3441-3455. [DOI] [PubMed] [Google Scholar]
- 39.Kammler, M., C. Schon, and K. Hantke. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 175:6212-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karow, M. L., P. Glaser, and P. J. Piggot. 1995. Identification of a gene, spoIIR, that links the activation of σE to the transcriptional activity of σF during sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 92: 2012-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kenney, T. J., and C. P. Moran. 1987. Organization and regulation of an operon that encodes a sporulation-essential sigma factor in Bacillus subtilis. J. Bacteriol. 169:3329-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kiel, J. A. K. W., J. M. Boels, G. Beldman, and G. Venema. 1991. Molecular cloning and mucleotide-sequence of the glycogen branching enzyme gene (glgB) from Bacillus stearothermophilus and expression in Escherichia coli and Bacillus subtilis. Mol. Gen. Genet. 230:136-144. [DOI] [PubMed] [Google Scholar]
- 43.LeDeaux, J. R., and A. D. Grossman. 1995. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J. Bacteriol. 177:166-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lepage, C., F. Fayolle, M. Hermann, and J. P. Vandecasteele. 1987. Changes in membrane-lipid composition of Clostridium acetobutylicum during acetone butanol fermentation—effects of solvents, growth temperature and pH. J. Gen. Microbiol. 133:103-110. [Google Scholar]
- 45.Liu, H. B., N. H. Bergman, B. Thomason, S. Shallom, A. Hazen, J. Crossno, D. A. Rasko, J. Ravel, T. D. Read, S. N. Peterson, J. Yates, and P. C. Hanna. 2004. Formation and composition of the Bacillus anthracis endospore. J. Bacteriol. 186:164-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Londoño-Vallejo, J. A., C. Fréhel, and P. Stragier. 1997. spollQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol. Microbiol. 24:29-39. [DOI] [PubMed] [Google Scholar]
- 47.Long, S., D. T. Jones, and D. R. Woods. 1983. Sporulation of Clostridium acetobutylicum P262 in a defined medium. Appl. Environ. Microbiol. 45:1389-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mansilla, M. C., L. E. Cybulski, D. Albanesi, and D. de Mendoza. 2004. Control of membrane lipid fluidity by molecular thermosensors. J. Bacteriol. 186:6681-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Molle, V., M. Fujita, S. T. Jensen, P. Eichenberger, J. E. Gonzalez-Pastor, J. S. Liu, and R. Losick. 2003. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 50:1683-1701. [DOI] [PubMed] [Google Scholar]
- 50.Molle, V., Y. Nakaura, R. P. Shivers, H. Yamaguchi, R. Losick, Y. Fujita, and A. L. Sonenshein. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 185:1911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nölling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, J. Hitti, Y. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohtani, K., H. Hayashi, and T. Shimizu. 2002. The luxS gene is involved in cell-cell signalling for toxin production in Clostridium perfringens. Mol. Microbiol. 44:171-179. [DOI] [PubMed] [Google Scholar]
- 53.Paredes, C. J., I. Rigoutsos, and E. T. Papoutsakis. 2004. Transcriptional organization of the Clostridium acetobutylicum genome. Nucleic Acids Res. 32:1973-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Partridge, S. R., and J. Errington. 1993. The importance of morphological events and intercellular interactions in the regulation of prespore-specific gene-expression during sporulation in Bacillus subtilis. Mol. Microbiol. 8:945-955. [DOI] [PubMed] [Google Scholar]
- 55.Petersen, D. J., and G. N. Bennett. 1991. Enzymatic characterization of a nonmotile, nonsolventogenic Clostridium acetobutylicum ATCC 824 mutant. Curr. Microbiol. 23:253-258. [Google Scholar]
- 56.Pich, A., F. Narberhaus, and H. Bahl. 1990. Induction of heat shock proteins during the initiation of solvent formation in Clostridium acetobutylicum. Appl. Microbiol. Biotechnol. 33:697-704. [Google Scholar]
- 57.Predich, M., G. Nair, and I. Smith. 1992. Bacillus subtilis early sporulation genes kinA, spo0F, and spo0A are transcribed by the RNA polymerase containing σH. J. Bacteriol. 174:2771-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ratnayake-Lecamwasam, M., P. Serror, K. W. Wong, and A. L. Sonenshein. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15:1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ravagnani, A., K. C. Jennert, E. Steiner, R. Grunberg, J. R. Jefferies, S. R. Wilkinson, D. I. Young, E. C. Tidswell, D. P. Brown, P. Youngman, J. G. Morris, and M. Young. 2000. Spo0A directly controls the switch from acid to solvent production in solvent-forming clostridia. Mol. Microbiol. 37:1172-1185. [DOI] [PubMed] [Google Scholar]
- 60.Resnekov, O., A. Driks, and R. Losick. 1995. Identification and characterization of sporulation gene spoVS from Bacillus subtilis. J. Bacteriol. 177:5628-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reysenbach, A. L., N. Ravenscroft, S. Long, D. T. Jones, and D. R. Woods. 1986. Characterization, biosynthesis, and regulation of granulose in Clostridium acetobutylicum. Appl. Environ. Microbiol. 52:185-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robson, R. L., R. M. Robson, and J. G. Morris. 1974. Biosynthesis of granulose by Clostridium pasteurianum. Biochem. J. 144:503-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodionov, D. A., A. G. Vitreschak, A. A. Mironov, and M. S. Gelfand. 2003. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem. 278:41148-41159. [DOI] [PubMed] [Google Scholar]
- 64.Saeed, A. I., V. Sharov, J. White, J. Li, W. Liang, N. Bhagabati, J. Braisted, M. Klapa, T. Currier, M. Thiagarajan, A. Sturn, M. Snuffin, A. Rezantsev, D. Popov, A. Ryltsov, E. Kostukovich, I. Borisovsky, Z. Liu, A. Vinsavich, V. Trush, and J. Quackenbush. 2003. TM4: A free, open-source system for microarray data management and analysis. BioTechniques 34:374-378. [DOI] [PubMed] [Google Scholar]
- 65.Santangelo, J. D., A. Kuhn, A. Treuner-Lange, and P. Dürre. 1998. Sporulation and time course expression of sigma-factor homologous genes in Clostridium acetobutylicum. FEMS Microbiol. Lett. 161:157-164. [DOI] [PubMed] [Google Scholar]
- 66.Sauer, U., and P. Dürre. 1995. Differential induction of genes related to solvent formation during the shift from acidogenesis to solventogenesis in continuous culture of Clostridium acetobutylicum. FEMS Microbiol. Lett. 125:115-120. [Google Scholar]
- 67.Sauer, U., and P. Dürre. 1993. Sequence and molecular characterization of a DNA region encoding a small heat shock protein of Clostridium acetobutylicum. J. Bacteriol. 175:3394-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sauer, U., A. Treuner, M. Buchholz, J. D. Santangelo, and P. Dürre. 1994. Sporulation and primary sigma factor homologous genes in Clostridium acetobutylicum. J. Bacteriol. 176:6572-6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schaffer, S., N. Isci, B. Zickner, and P. Dürre. 2002. Changes in protein synthesis and identification of proteins specifically induced during solventogenesis in Clostridium acetobutylicum. Electrophoresis 23:110-121. [DOI] [PubMed] [Google Scholar]
- 70.Schreiber, W., and P. Dürre. 2000. Differential expression of genes within the gap operon of Clostridium acetobutylicum. Anaerobe 6:291-297. [Google Scholar]
- 71.Schujman, G. E., R. Grau, H. C. Gramajo, L. Ornella, and D. de Mendoza. 1998. De novo fatty acid synthesis is required for establishment of cell type-specific gene transcription during sporulation in Bacillus subtilis. Mol. Microbiol. 29:1215-1224. [DOI] [PubMed] [Google Scholar]
- 72.Schumann, W., M. Hecker, and T. Msadek. 2002. Regulation and function of heat-inducible genes in Bacillus subtilis, p. 359-368. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 73.Scotcher, M. C., and G. N. Bennett. 2005. SpoIIE regulates sporulation but does not directly affect solventogenesis in Clostridium acetobutylicum ATCC 824. J. Bacteriol. 187:1930-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Scotcher, M. C., F. B. Rudolph, and G. N. Bennett. 2005. Expression of abrB310 and sinR, and effects of decreased abrB310 expression on the transition from acidogenesis to solventogenesis, in Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 71:1987-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shivers, R. P., and A. L. Sonenshein. 2004. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol. Microbiol. 53:599-611. [DOI] [PubMed] [Google Scholar]
- 77.Stephenson, K., and J. A. Hoch. 2002. Evolution of signalling in the sporulation phosphorelay. Mol. Microbiol. 46:297-304. [DOI] [PubMed] [Google Scholar]
- 78.Stragier, P. 2002. A gene odyssey: exploring the genomes of endospore-forming bacteria, p. 519-525. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 79.Sun, D. X., R. M. Cabrera-Martinez, and P. Setlow. 1991. Control of transcription of the Bacillus subtilis spoIIIG gene, which codes for the forespore-specific transcription factor σG. J. Bacteriol. 173:2977-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tamayo, P., D. Slonim, J. Mesirov, Q. Zhu, S. Kitareewan, E. Dmitrovsky, E. S. Lander, and T. R. Golub. 1999. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc. Natl. Acad. Sci. USA 96:2907-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tatusov, R. L., M. Y. Galperin, D. A. Natale, and E. V. Koonin. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tomas, C. A. 2003. DNA-array based transcriptional analysis of solvent tolerance and degeneration in Clostridium acetobutylicum: groESL overexpression, butanol challenges and a novel DNA-array normalization and gene identification method. PhD thesis. Northwestern University, Evanston, Ill.
- 83.Tomas, C. A., K. V. Alsaker, H. P. J. Bonarius, W. T. Hendriksen, H. Yang, J. A. Beamish, C. J. Paredes, and E. T. Papoutsakis. 2003. DNA array-based transcriptional analysis of asporogenous, nonsolventogenic Clostridium acetobutylicum strains SKO1 and M5. J. Bacteriol. 185:4539-4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tomas, C. A., J. A. Beamish, and E. T. Papoutsakis. 2004. Transcriptional analysis of butanol stress and tolerance in Clostridium acetobutylicum. J. Bacteriol. 186:2006-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tomas, C. A., N. E. Welker, and E. T. Papoutsakis. 2003. Overexpression of groESL in Clostridium acetobutylicum results in increased solvent production and tolerance, prolonged metabolism, and large changes in the cell's transcriptional program. Appl. Environ. Microbiol. 69:4951-4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tummala, S. B., N. E. Welker, and E. T. Papoutsakis. 1999. Development and characterization of a gene expression reporter system for Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 65:3793-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vollherbst-Schneck, K., J. A. Sands, and B. S. Montenecourt. 1984. Effect of butanol on lipid composition and fluidity of Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 47:193-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Walter, K. A., G. N. Bennett, and E. T. Papoutsakis. 1992. Molecular characterization of two Clostridium acetobutylicum ATCC 824 butanol dehydrogenase isozyme genes. J. Bacteriol. 174:7149-7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Weir, J., M. Predich, E. Dubnau, G. Nair, and I. Smith. 1991. Regulation of spo0H, a gene coding for the Bacillus subtilis σH factor. J. Bacteriol. 173:521-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Westheimer, F. H. 1963. Mechanism of enzymic decarboxylation of acetoacetic acid. Proc. Chem. Soc. Lond. 1963:253-261. [Google Scholar]
- 91.Wiesenborn, D. P., F. B. Rudolph, and E. T. Papoutsakis. 1988. Thiolase from Clostridium acetobutylicum ATCC 824 and its role in the synthesis of acids and solvents. Appl. Environ. Microbiol. 54:2717-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Winzer, K., K. Lorenz, and P. Dürre. 1997. Acetate kinase from Clostridium acetobutylicum: a highly specific enzyme that is actively transcribed during acidogenesis and solventogenesis. Microbiology 143:3279-3286. [DOI] [PubMed] [Google Scholar]
- 93.Winzer, K., K. Lorenz, B. Zickner, and P. Dürre. 2000. Differential regulation of two thiolase genes from Clostridium acetobutylicum DSM 792. J. Mol. Microbiol. Biotech. 2:531-541. [PubMed] [Google Scholar]
- 94.Wong, J., C. Sass, and G. N. Bennett. 1995. Sequence and arrangement of genes encoding sigma factors in Clostridium acetobutylicum. Gene 153: 89-92. [DOI] [PubMed] [Google Scholar]
- 95.Wu, J. J., M. G. Howard, and P. J. Piggot. 1989. Regulation of transcription of the Bacillus subtilis spoIIA locus. J. Bacteriol. 171:692-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu, J. J., P. J. Piggot, K. M. Tatti, and C. P. Moran. 1991. Transcription of the Bacillus subtilis spoIIA locus. Gene 101:113-116. [DOI] [PubMed] [Google Scholar]
- 97.Yang, H., H. Haddad, C. Tomas, K. Alsaker, and E. T. Papoutsakis. 2003. A segmental nearest neighbor normalization and gene identification method gives superior results for DNA-array analysis. Proc. Natl. Acad. Sci. USA 100:1122-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang, H. H., M. Y. Liu, and T. Romeo. 1996. Coordinate genetic regulation of glycogen catabolism and biosynthesis in Escherichia coli via the CsrA gene product. J. Bacteriol. 178:1012-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.York, K., T. J. Kenney, S. Satola, C. P. Moran, H. Poth, and P. Youngman. 1992. Spo0A controls the σA-dependent activation of Bacillus subtilis sporulation-specific transcription unit spoIIE. J. Bacteriol. 174: 2648-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zuber, P., and R. Losick. 1987. Role of AbrB in Spo0A-dependent and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J. Bacteriol. 169:2223-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.