Abstract
In the present study, we investigate the functions of the hupGHIJ operon in the synthesis of an active [NiFe] hydrogenase in the legume endosymbiont Rhizobium leguminosarum bv. viciae. These genes are clustered with 14 other genes including the hydrogenase structural genes hupSL. A set of isogenic mutants with in-frame deletions (ΔhupG, ΔhupH, ΔhupI, and ΔhupJ) was generated and tested for hydrogenase activity in cultures grown at different oxygen concentrations (0.2 to 2.0%) and in symbiosis with peas. In free-living cultures, deletions in these genes severely reduced hydrogenase activity. The ΔhupH mutant was totally devoid of hydrogenase activity at any of the O2 concentration tested, whereas the requirement of hupGIJ for hydrogenase activity varied with the O2 concentration, being more crucial at higher pO2. Pea bacteroids from the mutant strains affected in hupH, hupI, and hupJ exhibited reduced (20 to 50%) rates of hydrogenase activity compared to the wild type, whereas rates were not affected in the ΔhupG mutant. Immunoblot experiments with HupL- and HupS-specific antisera showed that free-living cultures from ΔhupH, ΔhupI, and ΔhupJ mutants synthesized a fully processed mature HupL protein and accumulated an unprocessed form of HupS (pre-HupS). Both the mature HupL and the pre-HupS forms were located in the cytoplasmic fraction of cultures from the ΔhupH mutant. Affinity chromatography experiments revealed that cytoplasmic pre-HupS binds to the HupH protein before the pre-HupS-HupL complex is formed. From these results we propose that hupGHIJ gene products are involved in the maturation of the HupS hydrogenase subunit.
The protein core of [NiFe] hydrogenases is composed of two different subunits. The large subunit contains the catalytic site consisting of a heterobinuclear NiFe metallocenter, and the small subunit from most [NiFe] hydrogenases holds three Fe-S clusters (43). Multiple genes are required for the synthesis of [NiFe] hydrogenases, and most of them are conserved among different bacteria (40, 41). The role of proteins encoded by these genes in the synthesis process is only partially known. Analysis of Escherichia coli hydrogenase 3 revealed that Hyp proteins are involved in the biosynthesis of the Ni-Fe cofactor, a process which ends with the processing of the large subunit by an endopeptidase which removes a C-terminal tail from the protein (3). In contrast, no auxiliary proteins have been identified that are required for synthesis of a functional small subunit of the [NiFe] hydrogenases, although most of them contain three iron-sulfur clusters (two 4Fe-4S and one 3Fe-4S) that conduct electrons from the H2-activating center in the large subunit to the physiological electron acceptor on the surface of the enzyme (11). Different proteins are known that facilitate the assembly of Fe-S clusters into other Fe-S proteins (29, 44).
In symbiosis with peas, Rhizobium leguminosarum bv. viciae strain UPM791 induces an H2 uptake [NiFe] hydrogenase whose genetic determinants are grouped in a cluster (hupSLCDEFGHIJKhypABFCDEX) required for the Hup+ phenotype (33). The hydrogenase structural genes hupSL and most of the accessory genes show high sequence similarity with the corresponding genes from other bacteria (12, 40). Unlike the situation in Bradyrhizobium japonicum (21), R. leguminosarum hupSL gene expression is observed only in pea bacteroids, and it is controlled by the nitrogenase regulatory protein NifA (4). In contrast, hyp genes are induced in microaerobic as well as in symbiotic conditions by the transcriptional activator FnrN (13, 15). The entire R. leguminosarum hydrogenase gene cluster has been engineered for expression in free-living microaerobic cells by replacing the NifA-dependent hupSL promoter by the FnrN-dependent fixN promoter (6) in order to facilitate the analysis of gene functionality.
The R. leguminosarum hydrogenase gene cluster contains a subcluster of genes, hupGHIJ (30), whose specific role in hydrogenase synthesis is still unknown. This subcluster functions as an operon under the control of a promoter (P3) located upstream of hupG (23). Genes homologous to hupGHIJ are also present in other aerobic bacteria containing H2 uptake [NiFe] hydrogenases such as B. japonicum, Azotobacter vinelandii, Rhodobacter capsulatus, and Ralstonia eutropha among others (40). In addition, HupG and HupH show homology to proteins from E. coli (HyaE and HyaF of hydrogenase 1), and a homologue to the gene encoding HupJ (HybE) is present in the gene cluster coding for hydrogenase 2 in the same bacterium. Genes homologous to hupGHIJ have not been reported in anaerobic bacteria such as Desulfovibrio spp. (31). The hupI gene encodes a rubredoxin-type protein (7, 30), whereas no similarities, outside of equivalent hydrogenase-related proteins, have been reported for HupG, HupH, and HupJ proteins.
In this work we show that hupGHIJ gene products are required for hydrogenase activity in R. leguminosarum microaerobic cells but are not involved in the synthesis of a mature large subunit (HupL) of the hydrogenase. Based on these results, and on the evidence provided by the identification of a HupS-HupH complex in microaerobically induced hydrogenase-active cultures, a role for hupGHIJ gene products in the maturation of the hydrogenase small subunit in R. leguminosarum is proposed.
MATERIALS AND METHODS
Chemicals.
All enzymes were purchased from Roche (Roche Applied Sciences, Mannheim, Germany) and were used according to manufacturer's indications. Media constituents were from Oxoid Ltd. (Basingstoke, United Kingdom). All other chemicals were of reagent or electrophoresis grade.
Bacterial strains, plasmids, media, and growth conditions.
Strains and plasmids used in this study are listed in Table 1. R. leguminosarum strains were routinely grown at 28°C in YMB (42). E. coli DH5a was used for standard cloning procedures (14). E. coli S17.1 (36) was used for conjugative plasmid transfer between E. coli and R. leguminosarum. For cell extract preparations, cultures were grown on MM medium (39). Antibiotic concentrations used were as follows (μg · ml−1): ampicillin, 100; kanamycin, 50; tetracycline, 5 (for R. leguminosarum) or 10 (for E. coli). A stoppered-tube technique, adapted to 200-ml flasks with 45-ml cultures, was routinely used for hydrogenase induction assays with free-living microaerobic cells. To this end, cultures were previously grown aerobically in YMB medium to an optical density at 600 nm (OD600) of 0.2. The flasks were then tightly capped, evacuated, and flushed three times with a mixture of 0.8% O2 in N2 and finally incubated for 16 h at 28°C. To study the effect of O2 concentration on hydrogenase activity the stoppered-tube system was adapted to continuous flushing with different O2-N2 mixtures. To induce hydrogenase in larger cultures of cells, a fermentor (Microferm; New Brunswick) was used. Initially, R. leguminosarum cultures were aerobically grown to an OD600 of 0.35, and then hydrogenase was induced by a continuous flow of 0.2% O2 in N2 until an OD600 of ca. 2 was reached.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant genotype or phenotype | Reference or source |
|---|---|---|
| R. leguminosarum | ||
| UPM791 | 128C53 wild type; Strr Nod+ Fix+ Hup+ | 32 |
| UPM1155 | UPM791 (Δhup/hyp cluster) Hup− | This lab |
| SM61 | UPM791 tatBC mutant | 25 |
| Escherichia coli | ||
| DH5α | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1Δ(lacZYA-argF)U169 φ80dlacZΔM15 | 14 |
| S17.1 | thi pro hsdR−hsdM+recA RP4 2-Tc::Mu-Km::Tn7 (Spr Smr) | 36 |
| Plasmids | ||
| pALPF1 | pAL618 with P1 promoter replaced by fixN promoter | 6 |
| pALPF15 | pALPF1 ΔhupS | This work |
| pALPF2 | pALPF1 ΔhupL | This work |
| pALPF4 | pALPF1 ΔhupD | This work |
| pALPF6 | pALPF1 ΔhupG | This work |
| pALPF7 | pALPF1 ΔhupH | This work |
| pALPF8 | pALPF1 ΔhupI | This work |
| pALPF9 | pALPF1 ΔhupJ | This work |
| pKD3 | Template plasmid harboring FLP-mediated excision sequence flanking Cmr gene | 9 |
| pKD13 | Template plasmid harboring FLP-mediated excision sequence flanking Kmr gene | 9 |
| pPM70 | pKD3 derivative plasmid containing Strep-tag II sequence for N terminus end fusion | This work |
| pALPF34 | pALPF1 derivative plasmid carrying hupHstrep gene | This work |
| pBBR1MCS-2 | Broad-host-range plasmid; Kmr mob+ | 17 |
| pPM1350 | pBBR1MCS2 derivative plasmid containing a DNA fragment harboring PfixN promoter from R. leguminosarum | This work |
| pPM125 | pBBR1MCS2 derivative plasmid containing the hupG, hupHstrep, and hupI genes in an EcoRI DNA fragment | This work |
| pPM164 | pPM1350 derivative plasmid containing an NdeI-XbaI fragment harboring hupI under the control of PfixN | This work |
| pPM165 | pPM1350 derivative plasmid containing an NdeI-XhoI fragment harboring hupK under the control of PfixN | This work |
| pPM166 | pPM1350 derivative plasmid containing an NdeI-XhoI fragment harboring hupGstrep, hupH, and hupI under the control of PfixN | This work |
| PCR2.1-TOPO | PCR cloning vector | Invitrogen |
Plant tests and enzyme assays.
Pea (Pisum sativum L. cv. Frisson) plants were used as host for R. leguminosarum bv. viciae. Conditions for plant inoculation, growth in nitrogen-free nutrient solution under bacteriologically controlled conditions, and bacteroid preparation were previously described (20). The nitrogen-free plant nutrient solution was supplemented with 20 mM NiCl2 from day 10 after seedling inoculation. Hydrogenase activity was measured in bacteroid suspensions and in free-living cells by an amperometric method with oxygen as the terminal electron acceptor (32). Protein contents of cell extracts were determined by the bicinchoninic acid method (37) with bovine serum albumin as the standard. For whole cells, the same method was followed after alkaline digestion in 1 N NaOH at 90°C for 10 min.
Recombinant DNA techniques.
DNA manipulations including purification, restriction, ligation, agarose gel electrophoresis, PCR amplification, and transformation into E. coli cells were carried out by standard methods (34). Oligonucleotides used for PCR and sequencing reactions were obtained from Sigma-Genosys (Haverhill, United Kingdom).
Generation of in-frame deletion mutants in R. leguminosarum hup genes.
In-frame deletions of hup genes were generated in plasmid pALPF1 by the one-step procedure of Datsenko and Wanner (9) based on the phage λ Red recombinase. Primers used for deletions are presented in Table 2. These primers are homologous to DNA regions adjacent to the genes to be deleted and to template plasmids (pKD3 and pKD13) containing an antibiotic resistance gene that is flanked by recombinase target sites. In order to avoid polarity effects, specific precautions were taken at the primer design step to ensure that the deletions did not affect the ribosome binding sites of the overlapping genes. The desired deletions were confirmed by PCR amplification of the corresponding plasmid pALPF1 region containing the target genes followed by electrophoretic mobility assays.
TABLE 2.
Primers used in this work
| Primer | Sequence (5′-3′) | Use |
|---|---|---|
| HUPS5 | ACTGCCGAGACTTTTTATGACGTCATTCGCCGCCAGGGGATTCCGGGGATCCGTCGACC | hupS deletion |
| HUPS3 | GAATCGTCATGATGGTGTTTCCCGCCCAGAATGTTAAGGTGTAGGCTGGAGCTGCTTC | |
| HUPL5 | AAGCGCTTGACCACCAAGCGCGAAAAAGCTGACGCTTAAGTGTAGGCTGGAGCTGCTTC | hupL deletion |
| HUPL3 | TTATCGGACCTGGACCCTGGCCATTTCTTGCCCATCCGGCATATGAATATCCTCCTTAGT | |
| HUPD5 | GATCTCGGGTGAACGGCTCTTCAAGGACAGGGAGGATTAGGTGTAGGCTGGAGCTGCTTC | hupD deletion |
| HUPD3 | GCAGTTCAACGCGGCCGGTTCGGCACGGCGCTCGTAACGCATATGAATATCCTCCTTAGT | |
| HUPG5 | TCGCAATGTTTTGCATCGTGATTTGTGGAGGAGACAATGATTCCGGGGATCCGTCGACC | hupG deletion |
| HUPG3 | TTCATCGCGCGATCTCCTTGCCGGCATGGGTGATCTCGAGTGTAGGCTGGAGCTGCTTC | |
| HUPH5 | CGAGATCACCCATGCCGGCAAGGAGATCGCGCGATGAGTGTAGGCTGGAGCTGCTTC | hupH deletion |
| HUPH3 | CGCTCATTTGAAATAGGCCTCCCAGATGTCCCGCAGTCGATATGAATATCCTCCTTAGT | |
| HUPI5 | CGACTGCGGGACATCTGGGAGGCCTATTTCAAATGAGCGATTCCGGGGATCCGTCGACC | hupI deletion |
| HUPI3 | CCATCGCCAAGCCTCATGAACTTCGACTGCAGGGCATCGGTGTAGGCTGGAGCTGCTTC | |
| HUPJ5 | CTGCAGCTCGAGATGCGCTACCGGGAGATTTATGCGACCATTCCGGGGATCCGTCGACC | hupJ deletion |
| HUPJ3 | CCGAGAAGGAATGTCATTCCGATGCCTCCTCGCGCCCGCGTGTAGGCTGGAGCTGCTTC | |
| HUPH5 | CGAGATCACCCATGCCGGCAAGGAGATCGCGCGATGAGTGTAGGCTGGAGCTGCTTC | pALPF34 |
| TAGH | GGCGCCTTCGCCCTCGGGGGCAACCCAAAAACCAGCCTTCTTCTCGAACTGCGGGTGGC | |
| TAGI | CTCATATGTGGAGC CACCCGCAGTTCGAGAAGAGCGCCTTCGAGAATTT | pPM164 |
| MAN2 | GCGGTCGCATAAATCT | |
| KNDE | GCCACAGTCGTCATATGACATTCCTTCTCGGGGC | pPM165 |
| MANK3 | TGCCGCGCCGTCTCACAG | |
| TAGG | CTCATATGTGGAGCCACCCGCAGTTCGAGAAGCCATCTGCCCTGGTCCG | pPM166 |
| MAN2 | GCGGTCGCATAAATCT | |
| MANF5 | GCGATGCTCGGCTTGCTG | pPM125 |
| MAN2 | GCGGTCGCATAAATCT | |
| STREP1 | GTGTAGGCTGGAGCTGCTTC | pPM70 |
| STREP2 | CTTTTCGAACTGCGGGTGGCTCCAGTTCATATGAATATCCTCC |
Generation of Strep-tag II-HupH fusion.
The Strep-tag II peptide, once fused to a protein, allows one-step protein purification because the tag interacts specifically with an immobilized variant of streptavidin called Strep-Tactin (IBA, Göttingen, Germany). To generate the Strep-tag II-HupH fusion protein, a modification of the Datsenko and Wanner deletion system (9) was used. The modification consisted in insertion of the sequence coding for the Strep-tag II peptide (WSHPQFEK) in the 5′ end of the antibiotic resistance gene of the pKD3 plasmid using primers STREP1 and STREP2, thus generating the new template plasmid pPM70. Subsequently, pPM70 was used as the template plasmid for in-frame fusing of the Strep-tag II sequence to the 5′ end of hupH from pALPF1 plasmid using primers HUPH5 and TAGH (Table 2). The resulting pALPF1 derivative plasmid (pALPF34) harbors a hydrogenase gene cluster encoding a Strep-tag II-HupH fusion protein.
Generation of plasmids expressing HupG, HupH, HupI, and HupK proteins in free-living R. leguminosarum cells.
In order to express hup genes in microaerobically grown cultures of R. leguminosarum, the PfixN promoter from pALPF1 was cloned in pBBR1MCS-2 vector plasmid (17) using PF1 and PF2 primers (6). PfixN is expressed in microaerobic conditions under the control of the FnrN protein. The resulting plasmid (pPM1350) was later used to clone hupI, hupK, and hupGHI genes isolated by PCR amplification from the pALPF1 plasmid using specific primers (Table 2). The resulting pPM1350 derivative plasmids were designated pPM164, pPM165, and pPM166, respectively.
Cell fractionation for protein localization.
Cells (1 g) from R. leguminosarum hydrogenase-induced cultures were suspended in 4 ml of buffer W (100 mM Tris-HCl, pH 8, 150 mM NaCl) containing a protease inhibitor mixture (Complete-mini; Roche Molecular Biochemicals, Mannheim, Germany). Cells were disrupted by three passages through a French pressure cell (SLM Aminco, Silver Spring, MD) at 100 MPa, and then the cell lysate was cleared for 20 min at 13,000 × g. The resulting supernatant was centrifuged at 135,000 × g for 1 h at 4°C in a TL-100 ultracentrifuge (Beckman Inc., Palo Alto, California.). The pellet, containing the cell membranes, was resuspended in the same volume of buffer W and precipitated again by an additional centrifugation at 135,000 × g for 1 h at 4°C.
Western immunoblot analysis.
Protein portions (30 μg) from the soluble and the membrane fractions were resolved by either sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or native PAGE in 10% polyacrylamide and subsequently transferred to polyvinylidene difluoride membranes. HupL, HupS, and HypB proteins were identified immunologically as previously described (5) using antisera raised against B. japonicum HupL and R. leguminosarum HupS and HypB, respectively.
Purification of Strep-tag II-HupH fusion protein.
Cell extracts from R. leguminosarum UPM1155 derivative strains containing plasmid pPM125 (Table 1) were obtained by French pressure cell disruption as above and subsequent centrifugation (12,000 × g for 30 min). The soluble fraction was applied to a 1-ml Strep-Tactin Superflow column (IBA, Göttingen, Germany) and developed by gravity flow. After the column was washed five times with 1 ml of buffer W to remove unbound proteins, the tagged protein was eluted six times with 0.5 ml of buffer W supplemented with 2.5 mM d-desthiobiotin. Eluted fractions were resolved by SDS-PAGE (12% polyacrylamide), and Strep-tagII-HupH, -HupL, and -HupS were identified by immunoblotting with Strep-Tactin conjugated to alkaline phosphatase (1:2,500; IBA, Göttingen, Germany) and antisera against HupL and HupS, respectively.
RESULTS
Contribution of hupGHIJ genes to free-living and symbiotic hydrogenase activity.
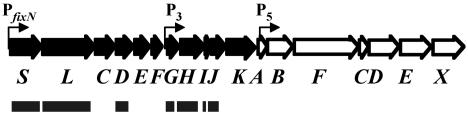
Since hup genes from R. leguminosarum bv. viciae are not expressed in free-living cells, in-frame deletion mutations in each of the hupG, hupH, hupI, and hupJ genes were generated in plasmid pALPF1 (Fig. 1). This plasmid contains the whole hup cluster from strain UPM791 under the control of the promoter of the fixN gene from the same strain, thus allowing microaerobic expression of hydrogenase activity in free-living cells and in bacteroids of Hup− strains of R. leguminosarum bv. viciae and other rhizobial species (6). The resulting pALPF1 mutant plasmid derivatives were transferred by conjugation into the Hup− R. leguminosarum UPM1155 strain. This strain is a UPM791 derivative strain with the entire hup/hyp gene cluster deleted that was constructed in our laboratory (our unpublished results). The hydrogenase activity of the corresponding transconjugant strains was tested in vegetative cells grown in microaerobic conditions (0.8% oxygen) as specified in Materials and Methods. pALPF1 mutant plasmid derivatives containing in-frame deletions in hupS or hupL hydrogenase structural genes, and in hupD, encoding HupL endopeptidase, were also constructed and used as controls (Fig. 1).
FIG. 1.
R. leguminosarum UPM791 hydrogenase gene cluster cloned in plasmid pALPF1. hup and hyp genes are shown by full and empty horizontal arrows, respectively, and designated by capital letters. The locations of characterized promoters in the hydrogenase gene cluster are shown by bent horizontal arrows. DNA fragments deleted in pALPF1 derivative plasmids containing single-gene deletions are shown by horizontal bars.
Hydrogenase activities associated with mutant plasmids affected in each of the hupG, hupH, hupI, and hupJ genes were severely reduced in vegetative cells as regards the hydrogenase activity associated with wild-type plasmid pALPF1 (Table 3). Particularly drastic for hydrogenase activity in free-living conditions was the mutation in the hupH gene, since strain UPM1155(pALPF7) (ΔhupH) exhibited no hydrogenase activity whereas mutation of the hupG gene caused only a 50% reduction of hydrogenase activity under the same conditions.
TABLE 3.
Relative free-living and symbiotic hydrogenase activities associated with pALPF1 derivative plasmids containing deletions in hup genes
| Plasmid | Hydrogenase activitya
|
|
|---|---|---|
| Free-living microaerobic cells | Pea bacteroids | |
| pALPF15 (ΔhupS) | <1 | <1 |
| pALPF2 (ΔhupL) | <1 | <1 |
| pALPF4 (ΔhupD) | <1 | <1 |
| pALPF6 (ΔhupG) | 45 | 100 |
| pALPF7 (ΔhupH) | <1 | 50 |
| pALPF8 (ΔhupI) | 10 | 30 |
| pALPF9 (ΔhupJ) | 5 | 40 |
Hydrogenase activities associated with pALPF1 derivative plasmids were measured in UPM1155 transconjugant strains and expressed as percentages of hydrogenase activity associated with the wild-type pALPF1 plasmid. The absolute values (100%) of hydrogenase activity for the UPM1155 (pALPF1) strain were (nmol of H2 · h−1 · mg protein−1) 6,520 ± 730 in microaerobic cultures and 4,490 ± 100 in pea bacteroids. Free-living cultures were bubbled with 0.8% O2. Values are averages of three replicates.
The UPM1155(pALPF1) derivative strains were also used to inoculate pea plants, and the hydrogenase activity was similarly measured in bacteroids. Remarkably, levels of hydrogenase activity exhibited by pea bacteroids from strains UPM1155(pALPF6) (ΔhupG), UPM1155(pALPF7) (ΔhupH), UPM1155(pALPF8) (ΔhupI), and UPM1155(pALPF9) (ΔhupJ) were much higher than those observed in free-living cells. In particular, wild-type levels of hydrogenase activity were detected in bacteroids of the strain containing the hupG deletion, and only a 50% reduction in activity was associated with the hupH deletion mutant. Strains carrying deletions in hupI and hupJ showed activity levels between 30 and 40% of those in the wild-type strain (Table 3).
Effect of oxygen concentration on the contribution of hupGHIJ genes to hydrogenase activity.
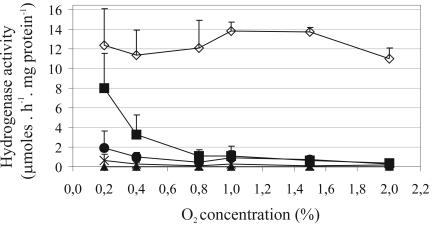
Since hupGHIJ genes appeared less relevant for hydrogenase activity in symbiosis than in free-living conditions and since the concentration of free oxygen inside the nodule is extremely low and precisely regulated (19), we decided to investigate the requirement of hupGHIJ genes for hydrogenase activity at different O2 concentrations in free-living conditions. The assay was performed with bacterial cultures continuously bubbled with a gas mixture containing O2 concentrations ranging from 0.2% to 2%. We first investigated the effect of O2 on hydrogenase activity in the wild-type strain UPM1155(pALPF1). Maximum levels of hydrogenase activity were observed in 1.0 to 1.5% O2 cultures, whereas small variations were observed at the remaining O2 concentrations assayed (Fig. 2).
FIG. 2.
Effect of hupGHIJ genes on the O2 tolerance of hydrogenase induction in free-living cultures of R. leguminosarum. Hydrogenase activity of UPM1155 transconjugant strains containing pALPF1 or pALPF1 derivative plasmids with in-frame deletions in hupG, hupH, hupI, or hupJ genes were determined in cell cultures bubbled with gas mixtures containing different O2 concentrations. The values correspond to the average of three replicate cultures, and error bars represent standard deviations. Symbols: ⋄, UPM1155(pALPF1); ▪, UPM1155(pALPF6) (ΔhupG); ▴, UPM1155(pALPF7) (ΔhupH); •, UPM1155(pALPF8) (ΔhupI); ×, UPM1155(pALPF9) (ΔhupJ).
The same analysis was performed with strains containing the hup-deleted mutant plasmids (Fig. 2). The hydrogenase activity of cultures from ΔhupG, ΔhupI, and ΔhupJ mutant strains increased as the O2 concentration decreased. This gradual increase was particularly evident in cultured cells from the ΔhupG strain and less so for the strains containing the hupI and hupJ deletions. No hydrogenase activity was detected in cells from the ΔhupH strain at any of the O2 concentrations assayed. These results indicate that HupH is essential for hydrogenase activity in free-living R. leguminosarum cells and that the requirement of hupGIJ genes for the hydrogenase activity of free-living cultures increases at higher free-O2 concentrations in the medium.
Effect of hupGHIJ genes on the maturation of hydrogenase structural proteins.
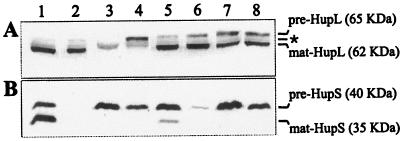
The potential role of hupGHIJ genes in the maturation of hydrogenase subunits was investigated immunochemically for HupL (Fig. 3A) and HupS (Fig. 3B) in total cell extracts from 0.8% O2 microaerobic cultures of strains containing pALPF1 or pALPF1 derivative plasmids with deletions in each of the hupGHIJ genes. The unprocessed and processed forms of both HupS and HupL subunits were identified in free-living cells of the wild-type strain (Fig. 3A and B, lane 1). The processed form of HupL (mature HupL) was present in free-living cells from the hupG, hupH, hupI, and hupJ mutants (Fig. 3A, lanes 5 to 8), including the strain containing the hupH mutation, which exhibited no hydrogenase activity (Fig. 3A, lane 6). These results suggest that the hupGHIJ genes are not required for any biosynthetic step previous to full processing of HupL.
FIG. 3.
Effect of hupGHIJ genes in the processing of hydrogenase subunits. Shown is the immunodetection of hydrogenase subunits (HupL and HupS) in cell extracts from microaerobically induced cultures of R. leguminosarum UPM1155 transconjugant strains containing pALPF1 or pALPF1 derivative plasmids with in-frame deletions in hup genes. Each lane was loaded with crude cell extracts containing 30 μg of total proteins from cell cultures bubbled with 0.8% O2. Antibodies generated against HupL (A) and HupS (B) were used. Lines on the right side indicate the positions and molecular sizes of the unprocessed (pre) and the mature (mat) forms of the structural hydrogenase proteins and the presence of an unspecific anti-HupL reactive band (*). Lanes: 1, UPM1155(pALPF1); 2, UPM1155(pALPF15) (ΔhupS); 3, UPM1155(pALPF2) (ΔhupL); 4, UPM791(pALPF4) (ΔhupD); 5, UPM791(pALPF6) (ΔhupG); 6, UPM791(pALPF7) (ΔhupH); 7, UPM791(pALPF8) (ΔhupI); 8, UPM791(pALPF9) (ΔhupJ).
In contrast, the band corresponding to the processed form of HupS (mature HupS) was not detected in strains containing mutations in hupH, hupI, or hupJ (Fig. 3B, lanes 6 to 8), which exhibited low or no hydrogenase activity. In these mutant strains most of the HupS protein was in the unprocessed form (pre-HupS). A weak band corresponding to the processed form was observed associated with the hupG deletion, which showed the highest hydrogenase activity (50%) among mutants (Fig. 3B, lane 5). The processed form of HupL (Fig. 3A, lane 2) and the unprocessed form of HupS (Fig. 3B, lane 3) were observed in cell extracts from hupS and hupL mutants, respectively. As expected, the unprocessed forms of both subunits were detected in the hupD mutant (Fig. 3A and B, lane 4). These results support the idea that the hupGHIJ genes are required either for maturation of pre-HupS or for the formation or stability of the pre-HupS-HupL complex previous to its translocation to the membrane or for both processes. This was further investigated with the hupH mutant, the only one that did not have residual hydrogenase activity.
HupH is required for translocation of hydrogenase structural proteins to the membrane.
The subcellular localization of structural subunits was investigated in the hupH mutant with UPM1155(pALPF1) and SM61(pALPF1) as controls. SM61 is a tatBC mutant affected in hydrogenase translocation to the cytoplasmic membrane (25). In the mutant strain containing the hupH deletion, HupL was located exclusively in the soluble fraction (Fig. 4A, lane 4). As expected, the large subunit in the wild type was located in the membrane fraction (Fig. 4A, lane 1), whereas in the tatBC mutant HupL was detected only in the soluble fraction (Fig. 4A, lane 6). The presence of HupL in the soluble fraction of the wild-type strain (Fig. 4A, lane 2) may be due to residual levels of HupL that has not been translocated.
FIG. 4.
Subcellular location of HupS and HupL in R. leguminosarum containing a deletion in the hupH gene. Hydrogenase subunits were immunologically detected using antisera generated against HupL (A) and HupS (B). SDS-PAGE gels were loaded with cell membrane (lanes 1, 3, and 5) or soluble (lanes 2, 4, and 6) fractions from microaerobically grown free-living culture cells. Lanes 1 and 2, UPM1155(pALPF1); lanes 3 and 4, UPM1155(pALPF7) (ΔhupH); lanes 5 and 6, SM61(pALPF1). pre and mat are as defined for Fig. 3.
On the other hand, the analysis of the subcellular localization of HupS subunit was consistent with the results observed for HupL. As expected, the small subunit was associated with the membrane cell fraction in the wild-type strain and with the soluble cell fraction in the tatBC mutant strain (Fig. 4B, lanes 1 and 6, respectively). In the strain containing the hupH deletion, HupS was not detected at all in the membrane fraction and was hardly detected in the soluble fraction (Fig. 4B, lanes 3 and 4). The weak bands corresponding to HupS in the ΔhupH and tatBC mutant strains could be due to the high lability of the HupS protein caused by the action of nonspecific cytoplasmic proteases (see below). These results are consistent with those obtained with the large subunit and indicate that HupH is required for translocation of hydrogenase to the membrane.
Formation of HupS-HupL complex requires HupH.
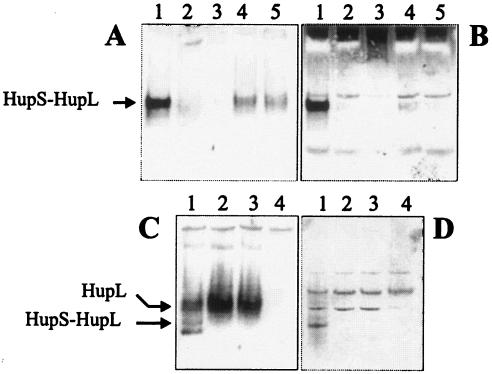
The effect of HupH on HupS-HupL complex formation was first investigated by immunological analyses of native polyacrylamide gels loaded with membrane fractions of the ΔhupH mutant, using UPM1155(pALPF1) and UPM1155(pALPF15) (ΔhupS) as controls. Strains carrying hupG and hupI deletions were also included. HupS-HupL complexes were detected by using both HupL (Fig. 5A) and HupS (Fig. 5B) antisera.
FIG. 5.
Effect of HupH protein on the assembly of the HupS-HupL complex in free-living cultures of R. leguminosarum. The analysis was performed on native gels loaded with membrane (A and B) or soluble (C and D) fractions from cultures microaerobically induced for hydrogenase activity. The heterodimer complex was immunologically identified with HupL (A and C) or HupS (B and D) antisera. Lanes for panels A and B: 1, UPM1155(pALPF1); 2, UPM1155(pALPF7) (ΔhupH); 3, UPM1155(pALPF2) (ΔhupL); 4, UPM1155(pALPF6) (ΔhupG); 5, UPM1155(pALPF8) (ΔhupI); lanes for panels C and D: 1, UPM1155(pALPF1); 2, UPM1155(pALPF7) (ΔhupH); 3, UPM1155(pALPF15) (ΔhupS); 4, UPM1155(pALPF2) (ΔhupL).
A strongly labeled band was detected by both antisera in the membrane fraction from the wild-type strain (Fig. 5A and B, lane 1). This band, likely corresponding to the HupS-HupL complex, was not detected in membrane fractions from the mutant strain containing the hupH deletion (Fig. 5A and B, lane 2) or in the control ΔhupL strain (Fig. 5A and B, lane 3). This result suggests that the HupH protein is required for heterodimeric complex formation. This observation might also apply to HupG and HupI since only weak bands corresponding to the potential HupS-HupL complex were detected in mutant strains containing the hupG or hupI deletions (Fig. 5A and B, lanes 4 and 5). The presence of these faint bands likely accounts for the low hydrogenase activity induced by ΔhupG and ΔhupI mutant strains (Table 3). The absence of the HupS-HupL complex in the membrane fraction of the hupH mutant is consistent with the observed subcellular localization of hydrogenase structural proteins (Fig. 4A and B).
The possibility that a HupS-HupL complex is formed in the soluble fraction but not transported to the periplasm was also investigated in native gels by using HupL and HupS antisera. The results obtained indicate the presence of a band identified with both antisera in the soluble cell fraction from the wild-type strain (Fig. 5C and D, lanes 1) but not from the ΔhupS and ΔhupL strains used as controls (Fig. 5C and D, lanes 3 and 4). This band can be attributed to a HupS-HupL complex and was not detected in the soluble cell fraction from the mutant strain containing the hupH deletion (Fig. 5C and D, lane 2). In contrast to the HupL protein, clearly identified in these native gels (Fig. 5C, lane 2), a band corresponding to the HupS protein was not detected in the hupH mutant (Fig. 5D, lane 2). Taken together, these results with membrane and soluble cell fractions suggest that HupH is required for pre-HupS-HupL complex formation.
The level of HupS decreases in the absence of a functional HupH protein.
The fact that HupS frequently appeared as a weak band in extracts from mutant strains containing the hupH deletion (Fig. 3B, lane 6) compared, for instance, with the band appearing in cell extracts from the ΔhupL mutant (Fig. 3B, lane 3) prompted us to investigate the effect of HupH on HupS protein accumulation.
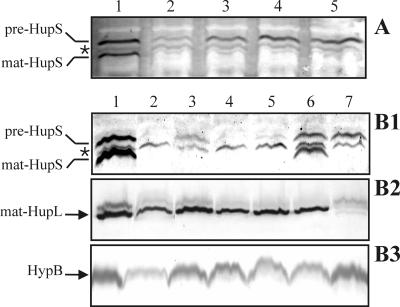
First, immunoblot analyses of HupS were performed on extracts from ΔhupG, ΔhupH, ΔhupI, and ΔhupJ mutants using extracts from the ΔhupD mutant as a control (Fig. 6A). Cell extracts from the strain containing the ΔhupH mutation clearly exhibited a band corresponding to the unprocessed form of HupS (Fig. 6A, lane 2), but much weaker than the corresponding band in cell extracts from strains containing the ΔhupG, ΔhupI, ΔhupJ, or ΔhupD mutations (Fig. 6A, lanes 1, 3, 4, and 5).
FIG. 6.
Effect of HupH on the accumulation of HupS. Cell extracts from microaerobically grown (0.8% oxygen) cultures of R. leguminosarum UPM1155 transconjugant strains were subjected to Western immunoblotting analysis after PAGE separation. Samples of cell extracts containing 30 μg of total proteins were applied to each lane. HupS (A and B1), HupL (B2), and HypB (B3) proteins were detected by using the corresponding antisera. Arrows on the left side of panels indicate the position of the unprocessed (pre) and the mature (mat) forms of the structural hydrogenase proteins; the presence of an unspecific band resulting from cross-reactivity with HupS-antiserum is indicated by an asterisk. (A) Analysis of mutants. Lanes: 1, UPM1155(pALPF6) (ΔhupG); 2, UPM1155(pALPF7) (ΔhupH); 3, UPM1155(pALPF8) (ΔhupI); 4, UPM1155(pALPF9) (ΔhupJ); 5, UPM1155(pALPF4) (ΔhupD). (B1 to B3) Complementation analysis of mutants. Lanes: 1, UPM1155(pALPF1); 2, UPM1155(pALPF15) (ΔhupS); 3, UPM1155(pALPF7) (ΔhupH); 4, UPM1155(pALPF7/pPM164) (ΔhupH/hupG); 5, UPM1155(pALPF7/pPM165) (ΔhupH/hupK); 6, UPM1155(pALPF7/pPM166) (ΔhupH/hupGHI); 7, UPM1155(pALPF4) (ΔhupD).
Second, wild-type HupS levels could be restored by introduction of the hupH gene (Fig. 6B1). The decrease of HupS levels in the hupH mutant background was corrected by complementation with plasmid pPM166 containing hupGHI genes (Fig. 6B1, lane 6), but not by plasmid pPM164 or pPM165, containing the hupI and hupK genes, respectively (Fig. 6B1, lanes 4 and 5). The effect on HupS accumulation associated with plasmid pPM166 could not be due to the hupG gene since the unprocessed form of HupS was stable in the ΔhupG strain (Fig. 3, lane 5). Similarly, HupH-dependent accumulation of immature HupS was not related to formation of the HupS-HupL complex, since pre-HupS levels were also high in a ΔhupD background (Fig. 6B1, lane 7) where no HupL processing took place. Other nonspecific reasons for the low intensity of the pre-HupS band in the ΔhupH background were ruled out by assaying HupL and HypB levels in the same extracts (Fig. 6B2 and B3, respectively). Since there are no reasons to postulate a change on the level of hupS translation, our interpretation of the above results is that the presence of HupH is required for HupS stability.
HupH forms a complex with HupS.
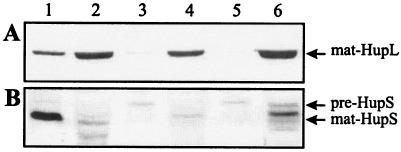
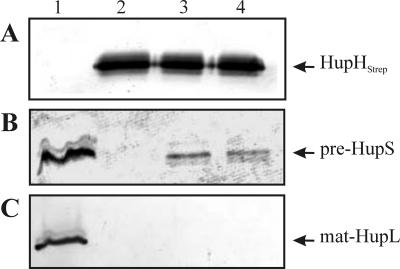
Since HupH is essential for hydrogenase activity in free-living conditions but seems not to be needed for HupL maturation, we decided to analyze its involvement in HupS maturation. To investigate the potential formation of a HupS-HupH complex, we used an affinity chromatography-based approach. To this end, a Strep-tag II sequence was fused to the N terminus of the HupH protein. We expected that the resulting construct, designated HupHstrep and encoded by plasmid pALPF34, would allow us a one-step affinity purification of proteins interacting with HupH. Since foreign protein overexpression in E. coli frequently has limitations, we decided to work with the original host, R. leguminosarum. First, we checked for functionality of the HupHstrep modified protein. The pALPF34 plasmid induced in free-living cells of R. leguminosarum UPM1155 the same levels of hydrogenase activity associated with the wild-type plasmid, pALPF1, and the HupHstrep protein encoded by pALPF34 could be detected using a Strep-Tactin-alkaline phosphatase conjugate (data not shown). Next, the EcoRI fragment containing the hupGHstrepI genes and the upstream P3 promoter was isolated from pALPF34 and cloned in a pBBR1MCS-2 broad-host-range plasmid. The resulting plasmid, pPM125, complemented the ΔhupH mutation for hydrogenase activity (data not shown). This indicates both that hupGHstrepI genes are transcribed in microaerobic cultures, likely from a functional P3 promoter (23), and that the tagged HupH protein complements the hupH mutation. Finally, the resulting pPM125 plasmid was introduced into UPM1155 transconjugant strains carrying plasmids with the ΔhupS mutation (pALPF15), as a control, and ΔhupL (pALPF2) or ΔhupD (pALPF4) mutations to favor the accumulation of a potential HupS-HupHstrep complex. Cell extracts were loaded into a Strep-Tactin column, and, after standard washes, proteins bound to the column were eluted with desthiobiotin (2.5 mM) and separated in SDS gels. HupHStrep, HupS, and HupL proteins were identified by immunoblotting with the corresponding antisera. The eluted fractions from all strains harboring the pPM125 plasmid contained large amounts of the tagged HupHstrep protein (Fig. 7A, lanes 2, 3, and 4).
FIG. 7.
Analysis of the formation of a HupS-HupHStrep complex. Proteins eluted from a Strep-Tactin column loaded with cell extracts from different strains were separated in a 15% SDS-PAGE gel (lanes 2, 3, and 4). Lane 1 contains crude cell extracts from microaerobically grown cells. HupHStrep (A), HupS (B), and HupL (C) proteins were identified using a streptavidin-alkaline phosphatase conjugate and antisera against HupS and HupL, respectively. Plasmid pPM125 encodes the HupH-Strep-tag fusion. Lanes: 1, UPM1155(pALPF1); 2, UPM1155(pALPF15/pPM125) (ΔhupS/hupGHI); 3, UPM1155(pALPF2/pPM125) (ΔhupL/hupGHI); 4, UPM1155(pALPF4/pPM125) (ΔhupD/hupGHI).
A band of an apparent molecular mass of 35 kDa and reactive with the anti-HupS antiserum was present in the eluate from cell extracts from ΔhupL and ΔhupD strains (Fig. 7B, lanes 3 and 4, respectively), but not from the control ΔhupS strain (Fig. 7B, lane 2). This result is consistent with the presence of a cytoplasmic unprocessed form of HupS (pre-HupS) protein that has been retained by the HupHstrep fusion protein. The amount of the HupS protein in the purified HupS-HupHstrep complex may represent only a portion of the total pre-HupS pool, since the wild-type copy of hupH is still present in the strain and its product may compete with the tagged HupH for available pre-HupS. Copurification of HupL with HupHstrep was not observed in any case (Fig. 7C, lanes 2, 3, and 4). These results indicate the formation of a complex between pre-HupS and HupHstrep proteins, prior to HupS-HupL complex assembly.
DISCUSSION
The results reported in this work suggest a role for the hupGHIJ gene products in the maturation of HupS, the hydrogenase small iron-sulfur subunit, and show the formation of a HupS-HupH complex in R. leguminosarum. These results are specially relevant since, apart from the structural component and the Tat system (25), no other proteins with a role in synthesis of a functional iron-sulfur subunit of [NiFe] hydrogenase have been identified.
Immunoblot analyses of in-frame deletion mutants revealed that the R. leguminosarum HupGHIJ proteins are not required for synthesis of a processed large hydrogenase subunit (HupL) but that instead they are involved in the maturation of the hydrogenase small subunit of this bacterium. Consistent with these results is the fact that an E. coli mutant lacking the HyaE protein, a homologue of R. leguminosarum HupG, was unable to process HyaA, the small subunit of Hyd1 hydrogenase (26). Also, a mutant with a deletion in hybE, a hupJ-homologous gene in the E. coli hydrogenase 2 gene cluster, was able to C-terminally process the large subunit HybC (16). Similarly, HoxO and HoxQ, the Ralstonia eutropha homologues of HupG and HupH, respectively, have also been shown to be required for hydrogenase activity in this aerobic bacterium (2) and to interact with HoxK, the hydrogenase small subunit (T. Schubert, M. Bernhard, O. Lenz, and B. Friedrich, Abstr. 7th Int. Hydrogenase Conf., abstr. P3-5, 2004).
The requirement of the HupGHIJ proteins for hydrogenase synthesis seems to be related to the O2 concentration in the medium during hydrogenase induction and appears crucial when hydrogenase is induced under high O2. These proteins might be fully or partially replaced by housekeeping Fe-S cluster biosynthetic proteins at low free-O2 concentrations in the media or in symbiosis. In this regard, it is known that some Fe-S clusters are labile under oxidative conditions and require repair or resynthesis by specific proteins, as it is the case for Erwinia chrysanthemi SufC (27) and A. vinelandii IscA (18). Circumstantial evidence supporting a connection between the oxygen level in the hydrogenase-inducing environment and the role of HupGHIJ proteins in hydrogenase synthesis is the absence of these proteins in anaerobic, [NiFe] hydrogenase-containing bacteria such as Desulfovibrio species (31).
Regarding the potential participation of HupGHIJ proteins in reduction chemistry leading to the assembly or maintenance of the Fe-S clusters into pre-HupS, it should be noted that HupI shows homology to rubredoxins (7, 30). Although the precise physiological function of rubredoxins remains elusive, especially in aerobic bacteria, they have been repeatedly associated with electron transfer reactions to diverse substrates with a wide range of reduction potentials (35, 38). Also, HupG and homologous proteins contain a structural domain related to thioredoxins and thiol-disulfide isomerases, proteins that participate in redox reactions, via the reversible oxidation of an active center disulfide bond (COG0526) (22). There is also some evidence indicating that HupH, HupI, and HupJ have related roles. First, HyaF2, homologous to HupH in the hydrogenase I gene cluster of Salmonella enterica, is likely a HupH-I fusion protein that contains the functional domain characteristic of rubredoxins at the C terminus (24). Second hupJ from some bacteria encodes a combined protein with the N terminus homologous to HupI and the remainder of the protein homologous to HupJ (1, 8).
The involvement of HupH on the maturation of a pre-HupS subunit able to form a periplasm-translocatable complex with the HupL subunit is evident from the experiments with the ΔhupH mutant. Besides the requirement of HupH for the translocation of the HupS-HupL complex to the membrane, we have also found evidence indicating that HupH binds the pre-HupS subunit. This binding may be required in the process of Fe-S cluster incorporation to the pre-HupS protein, with the likely participation of HupG, HupI, and HupJ proteins. In addition, the binding of HupH to HupS may be needed either to mediate the interaction between the pre-HupS and HupL modules or to prevent the formation of a defective complex before the completion of both the HupL and pre-HupS moieties and the subsequent wasteful export of incompletely folded or immature enzyme. Such a role has been proposed for E. coli HyaE and HybE chaperone-like proteins based on their interactions with the Tat signal peptide-bearing hydrogenase precursors HyaA of hydrogenase 1 and HybO of hydrogenase 2, respectively (10, 16). This role would be also similar to that of the DmsD chaperone, which binds the Tat signal peptides of the dimethyl sulfoxide and trimethylamine N-oxide reductases (28).
In conclusion, in this work we demonstrate that the R. leguminosarum HupGHIJ proteins are involved in the maturation of the hydrogenase small subunit. We propose that they play a common role related to the incorporation or maintenance of the iron-sulfur clusters in the pre-HupS form and that the relevance of this role is dependent on the oxygen levels of the hydrogenase induction conditions. It is also clear from this work that HupH forms a complex with pre-HupS. Unraveling the precise role of HupGHIJ proteins in HupS maturation will require additional experimentation that will shed light on the biosynthetic process of this complex but fascinating metalloenzyme.
Acknowledgments
We are grateful to R. J. Maier for the gift of the anti-HupL antiserum.
This work has been supported by funds from Programa de Grupos Estratégicos (III PRICYT) of the Comunidad Autónoma de Madrid and Spain's Ministry of Science and Education (projects AGL2001-2295 to T.R.-A., BIO2004-00251 to J.M.P., and BIO2004-05385 to J.I.).
REFERENCES
- 1.Baginsky, C., J. M. Palacios, J. Imperial, T. Ruiz-Argúeso, and B. Brito. 2004. Molecular and functional characterization of the Azorhizobium caulinodans ORS571 hydrogenase gene cluster. FEMS Microbiol. Lett. 237:399-405. [DOI] [PubMed] [Google Scholar]
- 2.Bernhard, M., E. Schwartz, J. Riertdorf, and B. Friedrich. 1996. The Alcaligenes eutrophus membrane-bound hydrogenase gene locus encodes functions involved in maturation and electron transport coupling. J. Bacteriol. 176:4522-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blokesch, M., A. Paschos, E. Theodoratou, A. Bauer, M. Hube, S. Huth, and A. Bock. 2002. Metal insertion into NiFe-hydrogenases. Biochem. Soc. Trans. 30:674-680. [DOI] [PubMed] [Google Scholar]
- 4.Brito, B., M. Martínez, D. Fernández, L. Rey, E. Cabrera, J. M. Palacios, J. Imperial, and T. Ruiz-Argüeso. 1997. Hydrogenase genes from Rhizobium leguminosarum bv. viciae are controlled by the nitrogen fixation regulatory protein NifA. Proc. Natl. Acad. Sci. USA 94:6019-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brito, B., J. M. Palacios, E. Hidalgo, J. Imperial, and T. Ruiz-Argüeso. 1994. Nickel availability to pea (Pisum sativum L.) plants limits hydrogenase activity of Rhizobium leguminosarum bv. viciae bacteroids by affecting the processing of the hydrogenase structural subunits. J. Bacteriol. 176:5297-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brito, B., J. M. Palacios, J. Imperial, and T. Ruiz-Argueso. 2002. Engineering the Rhizobium leguminosarum bv. viciae hydrogenase system for expression in free-living microaerobic cells and increased symbiotic hydrogenase activity. Appl. Environ. Microbiol. 68:2461-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, J. C., and L. E. Mortenson. 1992. Two open reading frames (ORFs) identified near the hydrogenase structural genes in Azotobacter vinelandii, the first ORF may encode for a polypeptide similar to rubredoxins. Biochim. Biophys. Acta 1131:122-124. [DOI] [PubMed] [Google Scholar]
- 8.Colbeau, A., P. Richaud, B. Toussaint, J. F. Caballero, C. Elster, C. Delphin, R. Smith, J. Chabert, and P. M. Vignais. 1993. Organization of the genes necessary for hydrogenase expression in Rhodobacter capsulatus. Sequence analysis and identification of two hyp regulatory mutants. Mol. Microbiol. 8:15-29. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubini, A., and F. Sargent. 2003. Assembly of Tat-dependent [NiFe] hydrogenases: identification of precursor-binding accessory proteins. FEBS Lett. 549:141-146. [DOI] [PubMed] [Google Scholar]
- 11.Fontecilla-Camps, J. C., M. Frey, E. Garcin, C. Hatchikian, Y. Montet, C. Piras, X. Vernede, and A. Volbeda. 1997. Hydrogenase: a hydrogen-metabolizing enzyme. What do the crystal structures tell us about its mode of action? Biochimie 79:661-666. [DOI] [PubMed] [Google Scholar]
- 12.Friedrich, B., and E. Schwartz. 1993. Molecular biology of hydrogen utilization in aerobic chemolithotrophs. Annu. Rev. Microbiol. 47:351-383. [DOI] [PubMed] [Google Scholar]
- 13.Gutiérrez, D., Y. Hernando, J. M. Palacios, J. Imperial, and T. Ruiz-Argüeso. 1997. FnrN controls symbiotic nitrogen fixation and hydrogenase activities in Rhizobium leguminosarum bv. viciae UPM791. J. Bacteriol. 179:5264-5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 15.Hernando, Y., J. M. Palacios, J. Imperial, and T. Ruiz-Argüeso. 1995. The hypBFCDE operon from Rhizobium leguminosarum bv. viciae is expressed from an Fnr-type promoter that escapes mutagenesis of the fnrN gene. J. Bacteriol. 177:5661-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jack, R. L., G. Buchanan, A. Dubini, K. Hatzixanthis, T. Palmer, and F. Sargent. 2004. Coordinating assembly and export of complex bacterial proteins. EMBO J. 23:3962-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 18.Krebs, C., J. N. Agar, A. D. Smith, J. Frazzon, D. R. Dean, B. H. Huynh, and M. K. Johnson. 2001. IscA, an alternate scaffold for Fe-S cluster biosynthesis. Biochemistry 40:14069-14080. [DOI] [PubMed] [Google Scholar]
- 19.Kuzma, M. M., S. Hunt, and D. B. Layzell. 1993. Role of oxygen in the limitation and inhibition of nitrogenase activity and respiration rate in individual soybean nodules. Plant Physiol. 101:161-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leyva, A., J. M. Palacios, T. Mozo, and T. Ruiz-Argüeso. 1987. Cloning and characterization of hydrogen uptake genes from Rhizobium leguminosarum. J. Bacteriol. 169:4929-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maier, R. J., F. J. Hanus, and H. J. Evans. 1979. Regulation of hydrogenase in Rhizobium japonicum. J. Bacteriol. 137:825-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33(Database issue):D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martínez, M., B. Brito, J. Imperial, and T. Ruiz-Argüeso. 2004. Characterization of a new internal promoter (P3) for Rhizobium leguminosarum hydrogenase accessory genes hupGHIJ. Microbiology 150:665-675. [DOI] [PubMed] [Google Scholar]
- 24.McClelland, M., K. E. Sanderson, S. W. Clifton, P. Latreille, S. Porwollik, A. Sabo, R. Meyer, T. Bieri, P. Ozersky, M. McLellan, C. R. Harkins, C. Wang, C. Nguyen, A. Berghoff, G. Elliott, S. Kohlberg, C. Strong, F. Du, J. Carter, C. Kremizki, D. Layman, S. Leonard, H. Sun, L. Fulton, W. Nash, T. Miner, P. Minx, K. Delehaunty, C. Fronick, V. Magrini, M. Nhan, W. Warren, L. Florea, J. Spieth, and R. K. Wilson. 2004. Comparison of genome degradation in paratyphi A and typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat. Genet. 36:1268-1274. [DOI] [PubMed] [Google Scholar]
- 25.Meloni, S., L. Rey, S. Sidler, J. Imperial, T. Ruiz-Argüeso, and J. M. Palacios. 2003. The twin-arginine translocation (Tat) system is essential for Rhizobium-legume symbiosis. Mol. Microbiol. 48:1195-1207. [DOI] [PubMed] [Google Scholar]
- 26.Menon, N. K., J. Robbins, J. C. Wendt, K. T. Shanmugan, and A. E. Przybyla. 1991. Mutational analysis and characterization of the Escherichia coli hya operon, which encodes [NiFe] hydrogenase 1. J. Bacteriol. 173:4851-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nachin, L., L. Loiseau, D. Expert, and F. Barras. 2003. SufC: an unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. EMBO J. 22:427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oresnik, I. J., C. L. Ladner, and R. J. Turner. 2001. Identification of a twin-arginine leader-binding protein. Mol. Microbiol. 40:323-331. [DOI] [PubMed] [Google Scholar]
- 29.Patzer, S. I., and K. Hantke. 1999. SufS is a NifS-like protein, and SufD is necessary for stability of the [2Fe-2S] FhuF protein in Escherichia coli. J. Bacteriol. 181:3307-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rey, L., E. Hidalgo, J. Palacios, and T. Ruiz-Argüeso. 1992. Nucleotide sequence and organization of an H2-uptake gene cluster from Rhizobium leguminosarum bv viciae containing a rubredoxin-like gene and four additional open reading frames. J. Mol. Biol. 228:998-1002. [DOI] [PubMed] [Google Scholar]
- 31.Rousset, M., V. Magro, N. Forget, B. Guigliarelli, J. P. Belaich, and E. C. Hatchikian. 1998. Heterologous expression of the Desulfovibrio gigas [NiFe] hydrogenase in Desulfovibrio fructosovorans MR400. J. Bacteriol. 180:4982-4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruiz-Argüeso, T., F. J. Hanus, and H. J. Evans. 1978. Hydrogen production and uptake by pea nodules as affected by strains of Rhizobium leguminosarum. Arch. Microbiol. 116:113-118. [Google Scholar]
- 33.Ruiz-Argüeso, T., J. Imperial, and J. M. Palacios. 2000. Uptake hydrogenases in root nodule bacteria, p. 489-507. In E. W. Triplett (ed.), Prokaryotic nitrogen fixation: a model system for analysis of a biological process. Horizon Scientific Press, Wymondham, United Kingdom.
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 35.Shen, G., J. Zhao, S. K. Reimer, M. L. Antonkine, Q. Cai, S. M. Weiland, J. H. Golbeck, and D. A. Bryant. 2002. Assembly of photosystem I. I. Inactivation of the rubA gene encoding a membrane-associated rubredoxin in the cyanobacterium Synechococcus sp. PCC 7002 causes a loss of photosystem I activity. J. Biol. Chem. 277:20343-20354. [DOI] [PubMed] [Google Scholar]
- 36.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 37.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 38.Solomon, E. I., T. C. Brunold, M. I. Davis, J. N. Kemsley, S. K. Lee, N. Lehnert, F. Neese, A. J. Skulan, Y. S. Yang, and J. Zhou. 2000. Geometric and electronic structure/function correlations in non-heme iron enzymes. Chem. Rev. 100:235-350. [DOI] [PubMed] [Google Scholar]
- 39.Thorne, S. H., and H. D. Williams. 1997. Adaptation to nutrient starvation in Rhizobium leguminosarum bv. phaseoli: analysis of survival, stress resistance, and changes in macromolecular synthesis during entry to and exit from stationary phase. J. Bacteriol. 179:6894-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vignais, P. M., B. Billoud, and J. Meyer. 2001. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 25:455-501. [DOI] [PubMed] [Google Scholar]
- 41.Vignais, P. M., and A. Colbeau. 2004. Molecular biology of microbial hydrogenases. Curr. Issues Mol. Biol. 6:159-188. [PubMed] [Google Scholar]
- 42.Vincent, J. M. 1970. A manual for the practical study of root-nodule bacteria. Blackwell Scientific Publications, Ltd., Oxford, United Kingdom.
- 43.Volbeda, A., M. H. Charon, C. Piras, E. C. Hatchikian, M. Frey, and J. C. Fontecilla-Camps. 1995. Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas. Nature 373:580-587. [DOI] [PubMed] [Google Scholar]
- 44.Zheng, L., V. L. Cash, D. H. Flint, and D. R. Dean. 1998. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J. Biol. Chem. 273:13264-13272. [DOI] [PubMed] [Google Scholar]