Abstract
The first genetic, in vivo, and in vitro evidences that YrxA is the regulator of NAD de novo biosynthesis in Bacillus subtilis are hereby reported. The protein is essential to the transcription repression of the divergent operons nadBCA and nifS-yrxA in the presence of nicotinic acid and binds to their shared operator-promoter region.
In eubacteria, NAD, the primary biological cofactor for oxidation-reduction reactions, is produced by a de novo pathway or recycled from exogenous or preformed nicotinic acid (NA) (2, 4, 22, 35). In the de novo biosynthetic pathway, NAD is synthesized from aspartic acid, which is converted to quinolinic acid (QA) by the concerted action of NadB, l-aspartate oxidase, and NadA, quinolinate synthetase (3, 6, 10, 18, 19, 31, 34); QA is then converted to nicotinic acid mononucleotide (NaMN) by NadC, QA phosphoribosyltransferase (14), and subsequently adenylated and amidated to NAD by NadD, NaMN adenylyltransferase, and NadE, deamido-NAD-ammonia ligase (NAD synthetase) (13, 20, 22, 30, 35). Finally, NADP is synthesized by phosphorylation of NAD catalyzed by the kinase NadF (8).
In the salvage or recycling pathway, NA, deriving from NAD dissociation or of exogenous origin, is reconverted to NaMN by a NA phosphoribosyltransferase (PncB in Escherichia coli; previously YueK in Bacillus subtilis) and reinserted in the enzymatic chain downstream of the reaction catalyzed by NadC (22).
In a comparative study in 1968, Saxton et al. revealed that the enzymatic activities of NadB, NadA, and PncB of E. coli are subject to inhibition by NA, while the inhibited enzymes for B. subtilis are NadB, NadA, and NadC but not PncB (29).
In E. coli and Salmonella species, the regulation of NAD biosynthesis is exerted by the trifunctional protein NadR, which transcriptionally represses the nadB, nadA (de novo pathway), and pncB (salvage pathway) genes and increases the uptake of nicotinamide-ribose and its conversion to nicotinamide mononucleotide and subsequently to NAD when the NAD concentration is low (7, 11, 12, 23, 25, 38).
A protein homologous to NadR is not present in Bacillus subtilis, in which, presumably, a different mechanism of NAD biosynthesis regulation is active. In the gram-positive bacterium, the nadB, nadA, and nadC genes are organized in an operon whose operator-promoter region is overlapping the operator-promoter region of the divergent dicistronic operon nifS-yrxA (17, 32) (http://genolist.pasteur.fr/SubtiList/).
NifS is a homologue of IscS cystein desulfurases and is supposed to be involved in the in vivo maturation of Fe-S clusters, possibly necessary to assemble an active form of the QA synthetase complex formed by NadB and NadA (21, 32). YrxA was classified as a potential transcriptional regulator (1), but the protein has no homologues in gram-negative bacteria, while genes for similar proteins are present in all the sequenced genomes of bacilli and other pathogenic gram-positive bacteria (genera Listeria, Streptococcus, and Enterococcus).
In 1993, Sun and Setlow (32) showed that in the presence of NA, the transcription of the nadB and nifS genes is abolished. The pncB gene (besides nadD, nadE, and nadF) was classified as essential for Bacillus subtilis by Kobayashi et al. (16): the authors speculated that the accumulation of NA, when pncB is inactivated, might repress NAD de novo synthesis and cause the bacterium's death.
We hypothesized that YrxA, due to its similarity with other transcriptional regulators able to bind small molecules as effectors (1), is, in the presence of NA, the repressor of nadBCA and nifS-yrxA transcription, allowing the recycling of the pyridinic ring from NA present in the medium and the availability of aspartic acid for protein synthesis.
Our genetic analysis shows that the pncB gene is essential only when B. subtilis is grown in the presence of NA and that the inactivation of yrxA bypasses its requirement. We show that YrxA represses the transcription of the two operons nadBCA and nifS-yrxA in vivo and that it binds in vitro to their overlapping promoters in the site of the putative operator sequence, using NA as a cofactor.
Construction of a pncB mutant.
pncB, the gene previously named yueK, coding for NA phosphoribosyltransferase of the recycling pathway, was classified as essential in B. subtilis (16). We constructed a conditional-lethal mutant by transforming wild-type (wt) PB168 (trpC2) competent cells with the integrative plasmid pMUTIN4/pncB′, obtained by cloning into pMUTIN4 (37) the 410-bp sequence coding for the N terminus of PncB (from 3259589c to 3259999c, SubtiList coordinates [http://genolist.pasteur.fr/SubtiList/]).
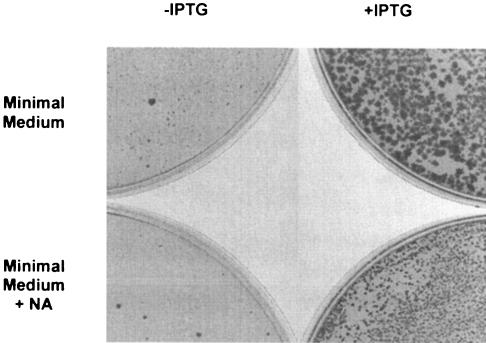
Transformants were selected on tryptose blood agar base (TBAB) medium (Difco) with erythromycin (Ery) (0.3 μg/ml) in the presence of 1 mM isopropyl-β-d-thiogalactoside (IPTG); the clones where Eryr was brought from the vector were screened on 100 μg/ml Ery. One of the clones, verified by PCR, was named PB1934 (trpC2 Ω pncB′::pMUTIN4, Eryr). When PB168 competent cells were transformed with chromosomal DNA of PB1934, Eryr colonies grew only on medium supplemented with 1 mM IPTG. This result confirmed that pncB is essential when B. subtilis is grown on rich medium (16). To investigate whether NA plays a role in causing pncB to be essential (as hypothesized in references 16 and 29), we selected Eryr transformants on minimal medium (MM) (5) with or without NA (50 μg/ml). No colonies were visible in presence of NA (Fig. 1, bottom left), while very small colonies appeared on MM plates without NA after 48 h at 37°C (Fig. 1, top left) with roughly the same frequency observed in the plates where IPTG was added (Fig. 1, top right).
FIG. 1.
Selection of Eryr (0.3 μg/ml erythromycin) colonies using DNA of PB1934 (ΩpncB′::pMUTIN4) to transform PB168 (wt) competent cells on MM (5) in the presence or absence of IPTG (1 mM) and NA (50 μg/ml). Only sporadic spontaneous Eryr mutants can be observed on the bottom left in MM with NA. Very small colonies can be observed in the absence of NA (top left) after 48 h of incubation at 37°C. When IPTG is added to the medium, the colonies observed in the presence of NA are smaller (bottom right) than the ones observed in absence of NA (top right).
Interestingly, the colonies observed in the presence of IPTG and NA (Fig. 1, bottom right) were smaller than the ones observed without NA but significantly larger than the ones grown on MM without supplements. Both 50 and 0.5 μg/ml of NA inhibited the growth of PB1934 (Table 1), but when pncB expression was induced with IPTG, the growth was comparable to that of the Eryr control strain BFS2652 (ΩynaB′::pMUTIN4 trpC2 [16]). These in vivo experiments showed that pncB is essential only when NA is present, and this gene/metabolite combination suppresses de novo NAD biosynthesis and causes the bacterium's death. NAD cannot be synthesized due to the unavailability of NaMN, the end product of the de novo (nadBCA-dependent) or recycling (pncB-dependent) pathways. In particular, NA could act as a cofactor for a transcriptional regulator that represses nadBCA transcription.
TABLE 1.
Growth of single and double mutants on MM with or without NA and 1 mM IPTGa
| Strain | Growth of strain on:
|
|||
|---|---|---|---|---|
| MM | MM + NA | MM + IPTG | MM + NA + IPTG | |
| PB1934 (ΩpncB::pMUTIN4) | +− | − | ++ | + |
| PB1935 (ΩpncB::pMUTIN4, ΔyrxA) | ++ | ++ | ++ | ++ |
| BFS2652 (ynaB::pMUTIN4) | ++ | + | ND | ND |
The same results were obtained using 50 or 0.5 μg/ml of NA. ++, colonies appear after 24 h at 37°C; +, colonies appear after 48 h at 37°C; +−, very small colonies appear after 48 h; ND, not determined.
Analysis of the YrxA primary sequence and of the genome map context of its gene.
yrxA is the second gene of the dicistronic operon nifS-yrxA, located 150 bp away from the nadBCA operon and transcribed in the opposite direction (17). This gene, coding for a putative transcriptional regulator, is also present in Bacillus anthracis (26, 27), Bacillus cereus (15), B.halodurans (36), B. licheniformis (28), and Listeria monocytogenes and Listeria innocua (9). In these organisms, where YrxA shows a high degree of similarity (identity score, >50%; similarity score, >76%), the genomic organization of the two divergent nadBCA and nifS-yrxA operons is conserved. None of the YrxA-like proteins have been functionally characterized yet; database searches (InterPro [http://www.ebi.ac.uk/interpro/] and Pfam [http://www.sanger.ac.uk/Software/Pfam/index.shtml]) revealed that the protein's primary sequence can be divided into two functional domains. The first 80 amino acids are characterized by a strong similarity with DNA-binding proteins (winged-helix DNA binding domain), while the second domain, from residue 90 to the carboxy terminus, has been classified as a potential three-histidine binding site for small effector molecules (1). The similarity data, the localization and organization of the nifS-yrxA operon, and the fact that there's no homologue in E. coli make YrxA a good candidate to be the regulator that represses nadBCA and nifS transcription in the presence of NA.
In a ΔyrxA context, the pncB gene is no longer essential.
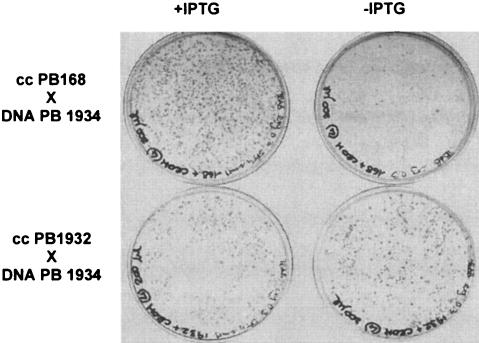
To verify the possible role of YrxA in the regulation of NAD biosynthesis, a ΔyrxA mutant was constructed by substituting the central region of the gene with the cat (chloramphenicol acetyltransferase) cassette by a PCR-based technique. The cat gene was amplified from plasmid pJM105A (24) using the oligonucleotides 5′-cat (5′-TCTTCAACTAAAGCACCCATT-3′) and 3′-cat (3′-ACAGTCGGCATTATCTCATATT-3′). A 473-bp fragment from nucleotide (nt) 368 of the coding sequence (CDS) of yrxA to nt 607 of the CDS of the downstream gene pheA was amplified using the oligonucleotides yrxA-5′cat (5′-ATGGGTGCTTTAGTTGAAGAAGGCACCCGGAAAGAAGTT-3′; underlined linker sequence homologous to the 5′ region of cat) and 3′-pheA (5′-TTAATGGTCATGATGGTCCCGCA-3′), while a 585-bp fragment from nt 707 of the nifS CDS to nt 127 of the yrxA CDS was amplified with the oligonucleotides 5′-iscS (5′-TAAATGTTCCCGGGATCGGTG-3′) and yrxA-3′cat (5′-TATGAGATAATGCCGACTGTAACCTGTCTTGAGACGTTCGCT-3′; underlined linker sequence homologous to the 3′ region of cat). The three PCR products were denatured together for three minutes at 94°C and slowly renatured by decreasing the temperature (6°C/min) to the annealing temperature of the 3′-pheA and 5′-iscS primers, used to amplify the mutagenic fragment. Five hundred nanograms of the pheA::cat::iscS mutagenic fragment were used to transform 0.5ml of B. subtilis PB168 competent cells. Cmr clones were obtained with a frequency of 1 × 102 per μg of DNA on nutrient broth (Difco) plates with 5 μg/ml chloramphenicol. One deletion-insertion ΔyrxA::cat mutant candidate, obtained by double crossover, was verified by PCR and named PB1932 (trpC2 ΔyrxA::cat, Cmr). To demonstrate that yrxA inactivation suppresses pncB essentiality and abolishes the repression role of NA, a double mutant was then created by transforming PB1932 competent cells with chromosomal DNA of PB1934. As expected, Eryr clones grew on TBAB with erythromycin (0.3 μg/ml) with or without IPTG (Fig. 2). One of the Eryr Cmr clones obtained, verified by PCR, was named PB1935 (trpC2 ΩpncB′::pMUTIN4 ΔyrxA::cat). The double mutant PB1935 also grew on MM with NA without IPTG: the addition of 0.5 or 50 μg/ml of NA did not impair the strain growth, since colonies were visible after 24 h (Table 1). PB1935 grew faster than the control strain BFS2652, suggesting that in a PncB+ strain, NA repression of de novo synthesis reduces the total NAD availability. These in vivo experiments support the hypothesis that yrxA, probably in cooperation with NA, is the repressor of NAD de novo synthesis. Constitutive expression of the nadBCA operon supports NAD synthesis even in the absence of the recycling pathway (ΩpncB′::pMUTIN4).
FIG. 2.
Selection of Eryr transformants (0.3 μg/ml erythromycin) on TBAB medium with or without IPTG (1 mM): in the top part of the figure, transformation of PB168 (wt) competent cells with chromosomal DNA of PB1934 (ΩpncB′::pMUTIN4), colonies are visible only on plates containing IPTG. When the same DNA was used to transform PB1932 (ΔyrxA) competent cells, the same number of colonies grew on TBAB plates with or without IPTG (lower part of the figure).
YrxA and NA cooperate in nadBCA transcriptional repression.
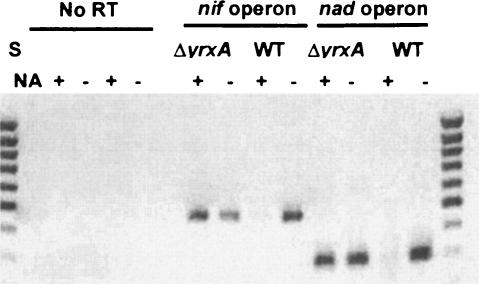
Since we hypothesized that the YrxA and NA repression action was performed at the transcriptional level, we analyzed the transcription of the nadB and nifS operons by reverse transcription (RT)-PCR under different conditions. PB168 (wt) and PB1932 (ΔyrxA) cells were grown at 37°C in PY broth (Penassay antibiotic medium 3; Difco), 0.5% glucose, to an optical density at 600 nm of 0.3; they were then washed and incubated for a further two hours at 37°C in MM with or without NA (50 μg/ml). RNA was extracted from cells by using the High Pure RNA isolation kit (Roche), and 1 μg of total RNA for each sample was retrotranscribed with the oligonucleotides NadBa2 (5′-CTCGTTCATTGCGGTCGAATG GAAATCC-3′) for the nadB transcripts and 3′-iscS (5′-TGAAGACAGGTGTTGAATGG-3′) for the nifS transcripts. cDNAs were then reamplified with primers NadBa2 and 3′NadB (5′-ATTGCAGTCATCGGTTCAGG-3′) for nadB, giving a 297-bp fragment, and with primers 3′-iscS and 5′-iscS (5′-TTATTTAGATTATGCAGCGACAACG-3′) for nifS, producing a 480-bp fragment. The nadB and nifS genes were not transcribed when the wt strain was grown in the presence of NA, while in strain PB1932 (ΔyrxA), both genes were transcribed even in the presence of NA (Fig. 3). This experiment confirms that YrxA is, in the presence of NA, the repressor of nadBCA and nifS-yrxA operons transcription. According to our results, the persistence of nad transcription regulation in the ΔnifS::erm mutant created by Sun and Setlow (32) could be explained by the absence of a polar effect on the downstream yrxA gene: the expression and the regulatory function of YrxA were evidently maintained. On the other hand, the authors did not specify if the disruption on nifS they performed introduced a strong transcription terminator: read-through from the nif promoter or from the ermC promoter itself cannot be ruled out.
FIG. 3.
RT-PCR experiments on the transcripts of the nifS/iscS and nadB genes in the PB1932 (ΔyrxA::cat) mutant and PB168 (wt) strains, grown in MM medium with or without NA (50 μg/ml). S, Mass Ruler DNA ladder (low range; Fermentas). ΔyrxA, RT-PCR on RNA extracted from PB1932 cells; WT, RT-PCR on RNA extracted from PB168 cells. A product of 297 bp indicates the presence of nadBCA operon transcripts, and a product of 480 bp indicates the presence of nifS/iscS transcripts.
YrxA binds to the overlapping promoter region of nadB and nifS.
nadB and nifS are separated by a 150-bp region including a region of imperfect dyad symmetry encompassing the −35 and −10 sequences of both genes (32) that could act as an operator for the binding of the YrxA regulatory protein repressing nadB and nifS transcription. An 87-bp DNA fragment including the putative operator and the divergent promoter region common to nadB and nifS (OP) (SubtiList coordinates 2848699 to 2848786) was amplified from PB168 chromosomal DNA with primers BSS1 (5′-CCTCCTGTTGTTTACACCTGTCT-3′) and BSS2 (5′-CTTCCATCCGTTCTCCATAAAA-3′) by incorporating [α-32P]CTP (3,000 Ci/mMol) in the PCR and used as a probe in an electrophoresis mobility shift assay (EMSA) to test if YrxA binds to the promoter region. YrxA, glutathione S-transferase (GST) tagged at the N terminus, was overexpressed into BL21(DE3) E. coli cells after the cloning into the pGEX- 6p1 expression vector (Amersham Biosciences) of the yrxA CDS, amplified from genomic wt DNA with primers 5′ByrxA- GEX1 (5′-CGGGATCCTTGACCGAAGAATTAAAGCTA-3′) and 3′yrxA-GEX (5′-CGGAATTCTTATTAATTAAAAATG CCGGCTTCT-3′), including the first of the two Met residues (Met-1 and Met-8) identified as a putative YrxA start codon (32). The YrxA-GST fusion protein was isolated by chromatography on glutathione Sepharose 4B columns (Amersham Biosciences), and after treatment with Prescission protease (Amersham Biosciences) and elution, a 90% pure YrxA protein was recovered. This 20.1-kDa YrxA protein (including five additional amino acids at the N-terminal end) was used in EMSA assays. All attempts made in expressing the untagged form of the protein failed, probably due to the instability of the protein in its native form.
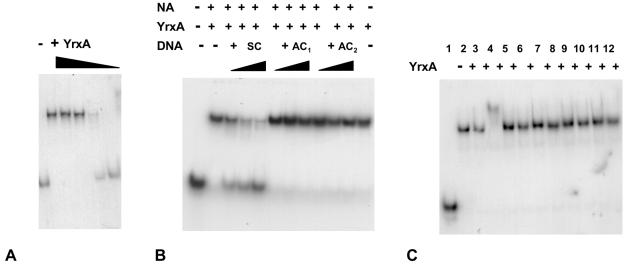
The 87-bp OP labeled fragment, purified with a PCR purification kit (QIAGEN), was incubated (37°C for 20 min) at a concentration of 1.7 nM with increasing concentrations of the YrxA protein in a 20-μl reaction volume in binding buffer (20mM Tris HCl [pH 8], 5 mM MgCl2, 100 mM KCl, 0.5 mM dithiothreitol, 0.05 mg/ml bovine serum albumin, 0.05% NP-40, 10% [vol/vol] glycerol). The reactions were loaded on a 6% polyacrylamide non denaturing gel in 0.5× Tris-Borate EDTA. Electrophoresis was performed at 70 V for 90 min.
As shown in Fig. 4A, the mobility of the 87-bp OP fragment was shifted as a function of the YrxA concentration, increasing from 3 nM to 3.4 μM. The binding was specific, since the addition to the YrxA-probe reaction of increasing concentrations (5, 10, and 20 ng) of the specific cold competitor SC (a150-bp fragment, including the 87-bp OP region, amplified with primers BSnadB 5′-CCCCTGAACCGATGACTGCA-3′ and BSnifS 5′-GGCGTTGTCGCTGCATAATC-3′), progressively eliminated the shift of labeled OP. The addition of the nonspecific cold competitors AC1 (a 318-bp fragment internal to the nadB coding region, amplified with primers NadBa2, the same used for RT-PCR, and 5′nadB 5′-CACCAATGTCTA AAAAGACGATTG-3′) and AC2, (330-bp fragment included in the nifS CDS, amplified with primers IscSex [5′-GGTTAACCA TATGATTTATTTAGATTATGCAG-3′] and Nif2 [5′-AAA GCAGCGCAATTATGAAT-3′]) had no effect on fragment mobility (Fig. 4B). Finally, to identify which molecule could act as an effector in the repression action of YrxA, NA, NA analogues, intermediates, or end products of NAD biosynthesis, were added to the reaction at a concentration of 300 μM. When 300 μM NA was added to the YrxA/DNA reaction mixture, the mobility of the labeled OP fragment had a supershift (Fig. 4C), while the addition of the other compounds didnot alter OP mobility. Both a higher concentration of YrxA (6.8 μM) alone and the addition to the reaction of NA (>150 μM) in the presence of 3.4 μM YrxA caused a “supershift” in EMSA (data not shown), while addition of NAD up to a concentration of 1.3 mM had no effect. The way in which NA favors YrxA binding to the operator-promoter region suggests a multimeric cooperative interaction, which is to be investigated further.
FIG. 4.
Electrophoresis mobility shift assays. Panel A: band shift induced by YrxA. Increasing concentrations of the protein (0.003, 0.034, 0.34, 0.77, and 3.4 μM) induce an increasing retardation of the 32P OP DNA-labeled fragment (1.7 nM) migration in the gel. Panel B: competition in EMSA between 32P-labeled 87-bp OP fragment and cold specific SC1 DNA (a 170-bp fragment spanning from just upstream to just downstream OP, including the 87-bp fragment) or nonspecific DNA (AC1, a 318-bp fragment covering a region in nadB outside OP; and AC2, a 330-bp fragment in nifS). The YrxA concentration was 50 nM, NA was 150 μM, and the increasing concentrations of competitors were, respectively, 2, 5, and 10ng in 20 μl. Panel C: band shift and “supershift” in the presence of NA and analogues, intermediates, or end products of NAD synthesis (added at a concentration of 300 μM). Lanes: 1, no cofactors, no YrxA; 2, only YrxA; 3, 150 μM NA; 4, 300 μM NA; 5, nicotinamide; 6, quinolinic acid; 7, isoniazide; 8, nicotinic acid hydrazide; 9: pyrazynamide; 10, nicotinamide mononucleotide; 11, NAD; 12, NADH. A plus sign indicates the presence of 3.4 μM YrxA.
In conclusion, we identified YrxA as the de novo NAD biosynthesis regulator in B. subtilis, and we propose to name it NadR. In the presence of NA, the protein represses transcription of the divergent operons nadBCA (for de novo biosynthesis) and nifS-yrxA (coding for the cystein desulfurase IscS, and YrxA itself) by binding to their common operator-promoter region. NA enhances the binding, as evidenced by the “supershift” shown in Fig. 4C.
This work first ascribes a biological function to the previously unknown yrxA gene and opens an interesting working hypothesis for developing new therapeutic and antimicrobial strategies for the gram-positive pathogenic bacteria (33) in which homologues of YrxA, and probably similar NAD regulation strategies, have been recognized in silico.
Acknowledgments
This work was financially supported by the University of Pavia and the Italian Ministry MIUR (Ministero dell'Istruzione, Università e Ricerca, COFIN 2002).
We thank Elisabetta Andreoli for expert technical assistance.
REFERENCES
- 1.Anantharaman, V., E. V. Koonin, and L. Aravind. 2001. Regulatory potential, phyletic distribution and evolution of ancient, intracellular, small-molecule-binding domains. J. Mol. Biol. 307:1271-1292. [DOI] [PubMed] [Google Scholar]
- 2.Begley, T. P., C. Kinsland, R. A. Mehl, A. Osterman, and P. Dorrestein. 2001. The biosynthesis of nicotinamide adenine dinucleotides in bacteria. Vitam. Horm. 61:103-119. [DOI] [PubMed] [Google Scholar]
- 3.Ceciliani, F., T. Caramori, S. Ronchi, G. Tedeschi, M. Mortarino, and A. Galizzi. 2000. Cloning, overexpression, and purification of Escherichia coli quinolinate synthetase. Protein Expr. Purif. 18:64-70. [DOI] [PubMed] [Google Scholar]
- 4.Chen, J. L., and G. J. Tritz. 1975. Isolation of a metabolite capable of differentially supporting the growth of nicotinamide adenine dinucleotide auxotrophs of Escherichia coli. J. Bacteriol. 121:212-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis, B. D., and E. S. Mingioli. 1950. Mutants of Escherichia coli requiring methionine or vitamin B12. J. Bacteriol. 60:17-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flachmann, R., N. Kunz, J. Seifert, M. Gutlich, F. J. Wientjes, A. Laufer, and H. G. Gassen. 1988. Molecular biology of pyridine nucleotide biosynthesis in Escherichia coli. Cloning and characterization of quinolinate synthesis genes nadA and nadB. Eur. J. Biochem. 175:221-228. [DOI] [PubMed] [Google Scholar]
- 7.Foster, J. W., E. A. Holley-Guthrie, and F. Warren. 1987. Regulation of NAD metabolism in Salmonella typhimurium: genetic analysis and cloning of the nadR repressor locus. Mol. Gen. Genet. 208:279-287. [DOI] [PubMed] [Google Scholar]
- 8.Garavaglia, S., A. Galizzi, and M. Rizzi. 2003. Allosteric regulation of Bacillus subtilis NAD kinase by quinolinic acid. J. Bacteriol. 185:4844-4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glaser P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P.Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F.Garcia-del Portillo, P. Garrido, L. Gaultier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F.Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 10.Griffith, G. R., J. L. Chandler, and R. K. Gholson. 1975. Studies on the de novo biosynthesis of NAD in Escherichia coli. The separation of the nadB gene product from the nadA gene product and its purification. Eur. J. Biochem. 54:239-245. [DOI] [PubMed] [Google Scholar]
- 11.Grose, J. H., U. Bergthorsson, and J. R. Roth. 2005. Regulation of NAD synthesis by the trifunctional NadR protein of Salmonella enterica. J. Bacteriol. 187:2774-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holley, E. A., M. P. Spector, and J. W. Foster. 1985. Regulation of NAD biosynthesis in Salmonella typhimurium: expression of nad-lac gene fusions and identification of a nad regulatory locus. J. Gen. Microbiol. 131: 2759-2770. [DOI] [PubMed] [Google Scholar]
- 13.Hughes, K. T., B. M. Olivera, and J. R. Roth. 1988. Structural gene for NAD synthetase in Salmonella typhimurium. J. Bacteriol. 170:2113-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes, K. T., J. R. Roth, and B. M. Olivera. 1991. A genetic characterization of the nadC gene of Salmonella typhimurium. Genetics 127:657-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. D. Ehrlich, R. Overbeek, and N. Kirpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423(6935):87-91. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi, K., S. D. Ehrlich, A. M. Albertini, G. Amati, K. K. Andersen, M.Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S.C.Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A.Danchin, M. Débarbouillé, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Masson, C. Mauël, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scanlan, W. Schumann, J. F. M. L. Seegers, J. Sekiguchi, A. Sekowska, S. J. Séror, M. Simon, P. Stragier, R. Struder, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. Van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, N. J. Cummings, R. A. Daniel, F. Denizot, K. M. Devine, A. Duesterhoft, S. D. Ehrlich, P. T. Emmerson, K. D. Entian, J. Errington, C. Fabret, E.Ferrari, D. Foulger, C. Fritz, M. Fujita, Y. Fujita, S. Fuma, A. Galizzi, N. Galleron, S.-Y. Ghim, P. Glaser, A. Goffeau, E. J. Golightly, G. Grandi, G. Guiseppi, B. J. Guy, K. Haga, J. Haiech, C. R. Harwood, A. Hénaut, H. Hilbert, S. Holsappel, S. Hosono, M.-F. Hullo, M. Itaya, L. Jones, B. Joris, D. Karamata, Y. Kasahara, M. Klaerr-Blanchard, C. Klein, Y. Kobayashi, P. Koetter, G. Koningstein, S. Krogh, M. Kumano, K. Kurita, A. Lapidus, S. Lardinois, J. Lauber, V. Lazarevic, S.-M. Lee, A. Levine, H. Liu, S. Masuda, C. Mauel, C. Medigué, N. Medina, R. P. Mellado, M. Mizuno, D. Moestl, S. Nakai, M. Noback, D. Noone, M. O'Reilly, K. Ogawa, A. Ogiwara, B. Oudega, S.-H. Park, V. Parro, T. M. Pohl, D. Portetelle, S. Porwollik, A. M. Prescott, E. Presecan, P. Pujic, B. Purnelle, G. Rapoport, M. Rey, S. Reynolds, M. Rieger, C. Rivolta, E. Rocha, B. Roche, M. Rose, Y. Sadaie, T. Sato, E. Scanlan, S. Schleich, R. Schroeter, F. Scoffone, J. Sekiguchi, A. Sekowska, S. J. Seror, P. Serror, B.-S. Shin, B. Soldo, A. Sorokin, E. Tacconi, T. Takagi, H. Takahashi, K. Takemaru, M. Takeuchi, A. Tamakoshi, T. Tanaka, P. Terpstr, A. Tognoni, V. Tosato, S. Uchiyama, M. Vandenbol, F. Vannier, A. Vassarotti, A. Viari, R. Wambutt, E. Wedler, H. Wedler, T. Weitzenegger, P. Winters, A. Wipat, H. Yamamoto, K. Yamane, K. Yasumoto, K. Yata, K. Yoshida, H.-F. Yoshikawa, E. Zumstein, H. Yoshikawa, and A. Danchin. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 18.Mattevi, A., G. Tedeschi, L. Bacchella, A. Coda, A. Negri, and S. Ronchi. 1999. Structure of L-aspartate oxidase: implications for the succinate dehydrogenase/fumarate reductase oxidoreductase family. Struct. Fold. Des. 7:745-756. [DOI] [PubMed] [Google Scholar]
- 19.Nasu, S., F. D. Wicks, and R. K. Gholson. 1982. L-Aspartate oxidase, a newly discovered enzyme in Escherichia coli, is the B protein of quinolinate synthetase. J. Biol. Chem. 257:626-632. [PubMed] [Google Scholar]
- 20.Nessi, C., A. M. Albertini, M. L. Speranza, and A. Galizzi. 1995. The outB gene of B. subtilis codes for NAD synthetase. J. Biol. Chem. 270:6181-6185. [DOI] [PubMed] [Google Scholar]
- 21.Park, J., P. C. Dorrestein, H. Zhai, C. Kinsland, F. McLafferty, and T. P. Begley. 2003. Biosynthesis of the thiazole moiety of thiamin pyrophosphate (vitamin B1). Biochemistry 42:12430-12438. [DOI] [PubMed] [Google Scholar]
- 22.Penfound, T., and J. W. Foster. 1996. Biosynthesis and recycling of NAD. p.721-730. In F. C. Neidhart, R. Curtiss, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter and H. E. Umarger (ed.), Escherichia coli and Salmonella. Cellular and molecular biology, 2nd ed. vol. 1 American Society for Microbiology, Washington, D.C. [Google Scholar]
- 23.Penfound, T., and J. W. Foster. 1999. NAD-dependent DNA-binding activity of the bifunctional NadR regulator of Salmonella typhimurium. J. Bacteriol. 181:648-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perego, M. 1993. Integrational vectors for genetic manipulation in Bacillus subtilis, p. 615-624. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington D.C.
- 25.Raffaelli, N., T. Lorenzi, P. L. Mariani, M. Emanuelli, A. Amici, S. Ruggieri, and G. Magni. 1999. The Echerichia coli NadR regulator is endowed with nicotinamide mononucleotide adenylyltransferase activity. J. Bacteriol. 181:5509-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Read, T. D., S. L. Salzberg, M. Pop, M. Shumway, L. Umayam, L. Jiang, E.Holtzapple, J. D. Busch, K. L. Smith, J. M. Schupp, D. Solomon, P. Keim, and C. M. Fraser. 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296(5575):2028-2033. [DOI] [PubMed] [Google Scholar]
- 27.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Yiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423(6935):81-86. [DOI] [PubMed] [Google Scholar]
- 28.Rey, M. W., P. Ramaiya, B. A. Nelson, S. D. Brody-Karpin, E. J. Zaretsky, M. Tang, A. Lopez de Leon, H. Xiang, V. Gusti, I. G. Clausen, P. B. Olsen, M. D. Rasmussen, J. T. Andersen, P. L. Jorgensen, T. S. Larsen, A. Sorokin, A. Bolotin, A. Lapidus, N. Galleron, S. D. Ehrlich, and R. M. Berka. 2004. Complete genome sequence of the industrial bacterium Bacillus licheniformis and comparison with closely related Bacillus species. Genome Biol. 5:R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saxton, R. E., V. Rocha, R. J. Rosser, A. J. Andreoli, M. Shimoyama, A. Kosaka, J. L. R. Chandler, and R. K. Gholson. 1968. A comparative study of the regulation of nicotinamide-adenin dinucleotide biosynthesis. Biochim. Biophys. Acta 156:77-84. [DOI] [PubMed] [Google Scholar]
- 30.Schneider, B. L., and L. J. Reitzer. 1998. Salmonella typhimurium nit is nadE: defective nitrogen utilization and ammonia-dependent NAD synthetase. J. Bacteriol. 180:4739-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seifert, J., N. Kunz, R. Flachmann, A. Laufer, K. D. Jany, and H. G. Gassen. 1990. Expression of the E. coli nadB gene and characterization of the gene product aspartate oxidase. Biol. Chem. Hoppe Seyler 371:239-248. [PubMed] [Google Scholar]
- 32.Sun, D., and P. Setlow. 1993. Cloning, nucleotide sequence, and regulation of the Bacillus subtilis nadB gene and a nifS-like gene, both of which are essential for NAD biosynthesis. J. Bacteriol. 175:1423-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutherland, S. 2003. New antibiotics for anthrax? Drug Discov. Today 8: 335-336. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki, N., J. Carlson, G. Griffith, and R. K. Gholson. 1973. Studies on the de novo biosynthesis of NAD in Eschericha coli. V. Properties of the quinolinic acid synthetase system. Biochim. Biophys. Acta 304:309-315. [DOI] [PubMed] [Google Scholar]
- 35.Switzer, R. L., H. Zalkin, and H. H. Saxild. 2002. Purine, pyrimidine, and pyridine nucleotide metabolism. p. 265-269. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 36.Takami, H., K. Nakasone, Y. Takaki, G. Maeno, R. Sasaki, N. Masui, F. Fuji, C. Hirama, Y. Nakamura, N. Ogasawara, S. Kuhara, and K. Horikoshi. 2000. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 28:4317-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 38.Zhu, N., B. M. Olivera, and J. R. Roth. 1991. Activity of the nicotinamide mononucleotide transport system is regulated in Salmonella typhimurium. J. Bacteriol. 173:1311-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]