Obtaining and maintaining proper levels of iron are major challenges for most organisms (reviews may be found in references 2, 9, and 10). The redox features that make iron such a versatile and valuable cofactor can also lead to formation of extremely toxic and highly reactive oxygen species, particularly the hydroxyl radical, which can damage any cellular component. Hence, bacteria and other organisms need powerful and sophisticated mechanisms to acquire iron but also to keep its reactivity in check. Most cells produce iron storage proteins, such as ferritin, where the reactivity of stored iron is lessened.
Iron is plentiful yet scarce: it forms a major part of the Earth's crust but has very limited solubility. Under oxygenated, nonacidic aqueous conditions, ferric iron [Fe(III)] prefers to form barely soluble iron hydroxides, well known as rust. To solubilize iron from these complexes and acquire levels adequate for growth, bacteria and other free-living microorganisms frequently secrete siderophores. These catechol, hydroxamate, or carboxylate compounds bind ferric iron with high affinity and maintain it in a soluble state whence it can be brought into the cell by high-affinity active transport systems. The genes for siderophore biosynthesis and transport are usually under transcriptional control in response to the cellular pool of iron. Repression at high iron levels is probably as important for cell health as is the derepression at limiting iron levels to maintain an intracellular iron pool that satisfies the metabolically crucial roles iron plays, while decreasing the risk of toxicity. Iron is a cofactor or structural component of myriad enzymes participating in most of the important steps of metabolism. As the metal in heme and in Fe-S complexes in many proteins, iron is crucial for electron transport, the tricarboxylic acid cycle, photosynthesis, nitrogen fixation, DNA synthesis, and so on. Conversely, both free iron and heme can participate in redox reactions that generate hydroxyl radicals and other damage.
Fur protein, an iron-dependent repressor.
The Fur protein plays a key role in the transcriptional response to iron in Escherichia coli and other gram-negative bacteria. Identified by their importance for iron-dependent repression of siderophore synthesis and transport genes, Fur homologues are present in many bacteria. Other iron-dependent repressors are in the DtxR family, first identified as being involved in repression of the diphtheria toxin gene in Corynebacterium diphtheriae. The Fur and DtxR families are unrelated in sequence, but the structures of their DNA-binding domains are quite similar (18). Their C-terminal dimerization domains differ and force a different geometry on the DNA interaction domains. Some Fur family proteins respond to other signals besides iron. Bacillus subtilis cells contain three homologues, iron-responsive Fur, zinc-responsive Zur, and peroxide, or oxidative stress-responsive PerR (references cited in reference 4). Fur proteins bind to DNA when they are loaded with a divalent cation, mainly Fe(II). Fur proteins often have multiple sites for cations, a regulatory site which in the case of Pseudomonas aeruginosa Fur can bind Zn or Fe and a structural site which binds Zn quite tightly (18). The DNA target recognized by iron-loaded Fur has an unusual trimeric repeat structure, with the consensus sequence GATNAT-GATNAT-CAANATC (2, 3). No functional sites are shorter than this, although some sites possess more repeats. Current evidence indicates that two Fur dimers bind to this recognition sequence, in such a way that one monomer in each dimer binds to opposite faces of the middle repeat.
Fur-regulated promoters have been identified and studied with the aid of lac fusions, by the Fur titration assay (20), and recently by global transcriptional profiling using DNA microarrays. The fur mutants in many bacteria exhibit derepressed expression of the genes for siderophore production and transport. They also display numerous unexpected phenotypes, and Fur is essential in P. aeruginosa, Neisseria, and some other bacteria. These phenotypes include the inability to grow on the respiratory substrate succinate and impaired survival after oxidative and acid stresses. Perhaps most surprising is the effect of the fur mutation on cellular iron levels. Although a fur mutant shows derepressed siderophore production and iron uptake, the cellular level of iron is only about 30% that of the isogenic fur+ strain. This lower pool size is associated with corresponding decreases in the levels of the major iron-storing ferritin protein FtnA and of numerous iron-containing metabolic proteins (15). Microarray analyses reveal that transcription of a large number of genes is affected by iron supply, and many but not all of these responses are dependent on Fur function. In E. coli, B. subtilis, P. aeruginosa, and Neisseria meningitidis, there was substantial repression by iron of 53, 37, 118, and 80 genes, respectively; conversely, induction by iron was reported for 48, ∼100, 87, and 153 genes, respectively (4, 8, 15, 17). As expected, these transcript changes operate so that iron limitation results in increased synthesis of iron acquisition systems and decreased synthesis of iron storage proteins and many iron-containing metabolic proteins. Iron excess results in Fur-dependent and Fur-independent decreases in iron transport, increases in iron storage in the ferritin-like FtnA and similar storage proteins, and increased production of iron-containing enzymes (Fig. 1).
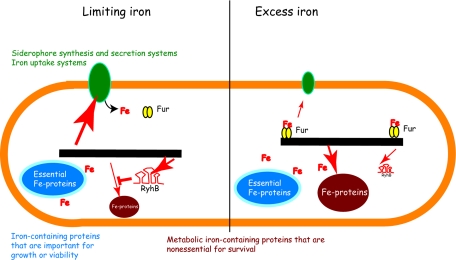
FIG. 1.
Schematic representation of the contribution of the Fur repressor and RyhB RNA to expression of genes involved in iron uptake and iron storage. When iron is limiting, the unloaded Fur protein is inactive as a repressor. This results in derepressed transcription of genes involved in siderophore synthesis and high-affinity iron uptake. Also derepressed is the gene for the small RNA RyhB. This RNA binds to and accelerates the turnover of mRNAs that encode iron-containing proteins, such as the iron-containing superoxide dismutase SodB. When iron is plentiful, the ferrous-iron-bound Fur protein binds to DNA targets to repress transcription of the transport genes and of RyhB. This allows elevated expression of iron-containing proteins and several iron storage proteins. When RyhB was expressed from the arabinose promoter in cells with sufficient iron, it caused decreased transcript levels for some genes by increased turnover of the RNA, as in the limiting-iron case. In addition, a number of other genes showed decreased expression as a result of Fur-dependent repression, which was increased owing to the decreased production of iron-sequestering proteins. Thus, RyhB affected gene expression in separate Fur-independent and Fur-dependent mechanisms.
The mechanism of Fur action as a repressor at the siderophore and iron transport promoters is well defined. Fur-binding sites in the iron-repressible promoters overlap the promoter sites for RNA polymerase. How does Fur bring about iron-dependent gene induction? Many examples of transcriptional regulatory proteins acting as activator and repressor are available. Is that also the case for Fur?
Small RNAs join the scene.
Descriptions of bacterial gene regulation have focused too long on the primacy of specific regulatory proteins which bind to DNA targets near a promoter and thereby affect the binding or clearance of RNA polymerase. The many examples of transcriptional attenuation and other forms of posttranscriptional control have shown how RNA folding or interactions play an effective role in gene control. Alternative RNA structures that influence transcription termination or ribosome binding can be favored by the binding of regulatory factors, ranging from ribosomes to RNA-binding proteins, such as CsrA or TRAP. Several recent dramatic additions to the role of RNA in bacterial gene control are riboswitches and small RNAs. Riboswitches are regions in the leaders of mRNAs which bind specific small-molecule ligands and change the conformation of the RNA to modulate RNA stability or ribosome binding. Small, noncoding regulatory RNAs (sRNAs) control expression of target genes typically by base pairing to a region of the target RNA transcript (7). The consequences of mRNA-sRNA binding differ in various systems (21). The most common effects are blockage of the ribosome-binding site on the mRNA and targeting of the RNA-RNA duplex for degradation by structure-dependent RNases, such as RNase E or RNase III (1, 11). Another possible outcome is stabilization of an mRNA against 3′-exonucleolytic degradation. A characteristic feature of many sRNAs is their dependence on the bacterial Hfq protein. Hfq (named for “host factor for phage Qβ replication”) is an RNA-binding chaperone that promotes the pairing of partially complementary RNA molecules (6, 16, 19, 22). The actions of some well-studied sRNAs, including DsrA, MicF, MicC, and OxyS, have been reviewed (7, 13).
The sRNA called RyhB is intimately involved in iron regulation in E. coli (5, 11, 12). It was discovered in genomic scans for potential sRNAs encoded in intergenic regions (23). The 90-nt RyhB RNA is highly conserved in enteric genomes and folds into three stem-loop structures (6). Two clues to the role of RyhB were provided by the presence of a typical Fur site overlapping the −10 promoter element of the ryhB gene and the finding that overexpression of RyhB resulted in an inability to grow on succinate, reminiscent of the Fur phenotype (12). The succinate-negative growth response of a fur mutant is overcome by deletion of ryhB, indicating that the requirement for Fur protein for growth on succinate is associated with the production of RyhB. Transcription of the ryhB gene is repressed by the Fe-Fur complex. Expression of RyhB is antagonistic with expression of the sdhCDAB operon, encoding succinate dehydrogenase. Strong complementarity exists between RyhB and the ribosome-binding site on the sdh mRNA corresponding to the ribosome-binding site for sdhD. As expected, RyhB requires Hfq for its function. The presence of RyhB accelerates the rate of turnover of some of its target mRNAs, and conversely, RyhB turns over much more rapidly in the presence of its target RNAs than when their transcription is blocked by rifampin. The turnover is associated with the action of RNase E (11). Thus, the induction by the Fe-Fur complex of some iron-containing proteins can now be ascribed to the repression of synthesis of RyhB, which would otherwise cause the accelerated turnover of the mRNA for those proteins. A similar process has been described for the action of two sRNAs, called PrrF1 and PrrF2, in the control of iron-inducible genes in P. aeruginosa (24).
RyhB regulates iron-related genes.
To dissociate the action of RyhB from the level of iron, thus avoiding the growth limitation resulting from iron starvation or toxicity, the ryhB gene was placed under the control of the arabinose-inducible araBAD promoter (11). Here, too, the level of RyhB quickly declined when the arabinose promoter was shut off. Decay was slowed in the presence of the transcription inhibitor rifampin, suggesting that RyhB degradation was promoted by the presence of its binding targets.
The arabinose-inducible RyhB expression system provided a valuable tool for identifying the targets of RyhB action and testing whether Fur and iron supplementation are directly or indirectly involved. An article by Eric Massé, Carin Vanderpool, and Susan Gottesman from the Université de Sherbrooke and the National Institutes of Health in this issue describes the changes in the global transcriptional profiles in response to arabinose-dependent induction of RyhB in fur+ and fur mutant strains (14). The strains used were deleted for the chromosomal copy of ryhB, so that all RyhB production came from the plasmid-borne arabinose-regulated gene. As expected, many gene transcripts were decreased when RyhB was expressed. Most of the previously recognized targets of RyhB action were confirmed and many new ones were identified, comprising a total of 56 genes in at least 18 operons that are depressed by RyhB. These RyhB-depressed genes were shut off upon RyhB expression whether Fur was present or not and are considered direct targets for RyhB action. These transcripts were subject to rapid turnover within 3 min as a result of RyhB binding. The changes in their transcript levels as detected in microarrays were often fairly modest, and only half of the operons changed more than fourfold. The greatest change was seen for the iron-containing superoxide dismutase SodB.
Surprisingly, another group of genes was depressed by RyhB only when Fur was present. This group of indirect RyhB targets comprise 29 genes in 10 operons, many of which are involved in iron uptake. Several were previously known to be regulated by Fur. A simple explanation for the operation of this class of genes postulates that when RyhB production blocks the synthesis of its iron-containing direct target proteins, the internal pool of iron can increase. The elevated iron pool allows increased occupancy of Fur, and thus a higher degree of repression of all Fur-regulated genes. Consistent with this view, the decrease in the level of an operon in this group declined only 7.5 min after RyhB induction, later than the decline in a direct RyhB target gene. Thus, the major function of RyhB action is to help direct scarce supplies of iron under limiting conditions away from nonessential metabolic processes and towards some essential iron-containing proteins, such as ribonucleotide reductase (Fig. 1).
An interesting corollary of this proposal helps explain the existence in E. coli of two systems for insertion of iron-sulfur complexes into proteins. The Isc pathway is a direct target of RyhB, whereas the Suf pathway is an indirect, Fur-dependent target. Because synthesis of the Isc pathway is shut off by RyhB, it must not be required to function under conditions of low internal iron. It operates during normal growth for insertion of iron-sulfur clusters into nonessential genes. Conversely, synthesis of the Suf complex is repressed only by the Fe-Fur complex, meaning that it is active in cells with limiting iron, which in turn suggests that it can satisfy the needs for synthesis of essential iron-containing proteins.
A small third group of 15 genes in 10 operons showed modestly increased expression (around threefold or less) in response to RyhB production. Most of these genes have no obvious relationship to iron metabolism or content, and their variations may have no biological significance. The important member of this group is ftnA, encoding the cytoplasmic iron storage protein, ferritin. Induction of this gene under conditions of iron excess is dependent on Fur. It is likely that the increase in ftnA expression upon induction of RyhB is the result of the iron-sparing activity invoked in explanation of the indirect RyhB effect. The increased internal iron pool would increase the loading of Fur. The paper by Massé et al. (14) does not suggest a mechanism whereby iron-loaded Fur could increase gene expression. Recent findings from the lab of Simon Andrews suggest that Fur binding to an upstream region in the ftnA promoter relieves the inhibitory effect of nucleoid-binding proteins.
Although this mechanism might explain iron induction of this gene, it is clear that iron induction of most other genes is a product of the control of the small RNA RyhB. Operation of this RNA explains the low iron levels in fur mutants, because loss of Fur repression leads to accumulation of sufficient levels of RyhB to depress expression of many proteins that contain or store iron. It is further likely that our appreciation of the participation of small RNAs in gene regulation is only beginning. Thus, an sRNA plays a crucial role complementary to that of Fur protein to properly coordinate the uptake, storage, and distribution of iron in E. coli.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Afonyushkin, T., B. Vecerek, I. Moll, U. Blasi, and V. R. Kaberdin. 2005. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res. 33:1678-1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 3.Baichoo, N., and J. D. Helmann. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 184:5826-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baichoo, N., T. Wang, R. Ye, and J. D. Helmann. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 45:1613-1629. [DOI] [PubMed] [Google Scholar]
- 5.Davis, B. M., M. Quinones, J. Pratt, Y. Ding, and M. K. Waldor. 2005. Characterization of the small untranslated RNA RyhB and its regulon in Vibrio cholerae. J. Bacteriol. 187:4005-4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geissmann, T. A., and D. Touati. 2004. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 23:396-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottesman, S. 2004. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 58:303-328. [DOI] [PubMed] [Google Scholar]
- 8.Grifantini, R., S. Sebastian, M. Frigimelica, E. Bartolini, A. Muzzi, R. Rappuoli, G. Grandi, and C. A. Genco. 2003. Identification of iron-activated and -repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. Proc. Natl. Acad. Sci. USA 100:9542-9547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 10.Massé, E., and M. Arguin. 2005. Ironing out the problem: new mechanisms of iron homeostasis. Trends Biochem. Sci. 30:462-468. [DOI] [PubMed] [Google Scholar]
- 11.Massé, E., F. E. Escorcia, and S. Gottesman. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17:2374-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Massé, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Massé, E., N. Majdalani, and S. Gottesman. 2003. Regulatory roles for small RNAs in bacteria. Curr. Opin. Microbiol. 6:120-124. [DOI] [PubMed] [Google Scholar]
- 14.Massé, E., C. K. Vanderpool, and S. Gottesman. 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 187:6962-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McHugh, J. P., F. Rodriguez-Quinones, H. Abdul-Tehrani, D. A. Svistunenko, R. K. Poole, C. E. Cooper, and S. C. Andrews. 2003. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 278:29478-29486. [DOI] [PubMed] [Google Scholar]
- 16.Moll, I., T. Afonyushkin, O. Vytyytska, V. R. Kaberdin, and U. Blasi. 2003. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA 9:1308-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ochsner, U. A., P. J. Wilderman, A. I. Vasil, and M. L. Vasil. 2002. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 45:1277-1287. [DOI] [PubMed] [Google Scholar]
- 18.Pohl, E., J. C. Haller, A. Mijovilovich, W. Meyer-Klaucke, E. Garman, and M. L. Vasil. 2003. Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol. Microbiol. 47:903-915. [DOI] [PubMed] [Google Scholar]
- 19.Schumacher, M. A., R. F. Pearson, T. Møller, P. Valentin-Hansen, and R. G. Brennan. 2002. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 21:3546-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stojiljkovic, I., A. J. Baumler, and K. Hantke. 1994. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. J. Mol. Biol. 236:531-545. [DOI] [PubMed] [Google Scholar]
- 21.Storz, G., J. A. Opdyke, and A. Zhang. 2004. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 7:140-144. [DOI] [PubMed] [Google Scholar]
- 22.Vecerek, B., I. Moll, T. Afonyushkin, V. R. Kaberdin, and U. Blasi. 2003. Interaction of the RNA chaperone Hfq with mRNAs: direct and indirect roles of Hfq in iron metabolism of Escherichia coli. Mol. Microbiol. 50:897-909. [DOI] [PubMed] [Google Scholar]
- 23.Wassarman, K. M., F. Repoila, C. Rosenow, G. Storz, and S. Gottesman. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 15:1637-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilderman, P. J., N. A. Sowa, D. J. FitzGerald, P. C. FitzGerald, S. Gottesman, U. A. Ochsner, and M. L. Vasil. 2004. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. USA 101:9792-9797. [DOI] [PMC free article] [PubMed] [Google Scholar]