Abstract
We have developed a novel method of genetic library construction on magnetic microbeads based on solid-phase single-molecule PCR in a fine and robust water-phase compartment formed in water-in-oil (w/o) emulsions. In this method, critically diluted DNA fragments were distributed over the emulsion as templates, where beads crosslinked with multiple primers and other PCR components were encapsulated to form multiple reaction compartments. The delivered DNA was then amplified and covalently immobilized on the beads in parallel, within individual compartments, to construct a genetic library on beads (GLOBE), which was readily applicable to a genomewide global scanning of genetic elements recognized by a defined DNA-binding protein. We constructed a GLOBE of Paracoccus denitrificans and selected gene beads that were bound to the His-tagged transcription factor PhaR by flow cytometry. As a result of flow cytometry screening with an anti-His fluorescent antibody, the PhaR target fragments were enriched 1200-fold from this library with this system. Therefore, this system is a powerful tool for analyzing the transcription network on a genomewide scale.
INTRODUCTION
Many proteins bind to specific sites in a genome to regulate gene expression and genome maintenance. Determining the binding sites of regulatory proteins on genomes is important for reconstructing transcriptional regulatory networks (1). For example, the binding of a transcription factor to its genomic targets can be assayed by chromatin immunoprecipitation (ChIP) (2). Although this assay is useful in studying the gene regulation system, it is very time consuming. Recently, novel assay combining ChIP with microarray (chip) hybridization, called the ChIP-chip method has also been developed (3). The ChIP-chip method has been applied to the analysis of transcription factors in some organisms such as Escherichia coli and humans; however, microarrays for covering the entire genome are very expensive. Moreover, such suitable microarrays are not available for many organisms.
We have developed a novel method for the high-throughput screening of DNA–protein interactions using small droplets, water-in-oil (w/o) emulsions as a reaction compartment (Figure 1). First, target DNA is diluted to one molecule per compartment on average and immobilized on microbeads conjugated with a primer by PCR in w/o emulsions. Second, the bead–DNA complex is mixed with a transcription factor containing an epitope tag, followed by adding a fluorescence-labeled anti-epitope tag antibody. Then, a fluorescent bead (the quaternary complex of bead–DNA–transcription factor–antibody) is selected by flow cytometry. We successfully enriched a DNA fragment bound to PhaR, which is a transcription factor from Paracoccus denitrificans (4,5), from the P.denitrificans genomic library with this system. Other potential applications of the system are discussed.
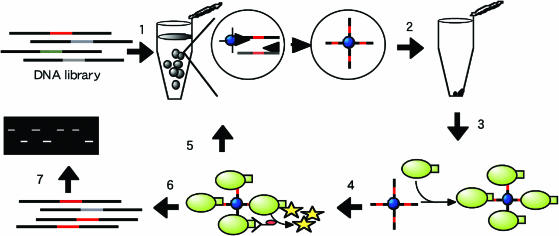
Figure 1.
Schematic representation of screening assay of DNA–protein interaction with solid-phase single-molecule PCR in w/o emulsions. 1: Target DNA mixture diluted as <1 molecule per compartment on average. The diluted DNA template solution was added to the PCR mixture including oligonucleotide-coupled beads. Subsequently, the aqueous phase is gradually added to the oil phase with stirring using a magnetic bar for 3 min at room temperature and solid-phase single-molecule PCR in w/o emulsions is carried out. 2: The emulsions are disrupted. 3: A DNA-binding protein with epitope tag is added to the beads. 4: A fluorescence-labeled anti-tag antibody is added to the complex. 5: Flow cytometry for selecting positive clones and multibead PCR with selected beads. 6: Flow cytometry for selecting positive clones and bead PCR with selected beads. 7: Gel mobility shift assay.
MATERIALS AND METHODS
Oligonucleotide primers
The sequences of primers used in this study are as follows: phaR-F-XbaI, GCTCTAGAGC GGATAACAAT TTCACACAGG AA; phaR-R-EcoRI, GGAATTCCCA AGCTTCGAAA TCCGCTTCTG CAA; F1 primer, ATCTCGATCC CGCGAAATTA ATACG; R1 primer, TCCGGATATA GTTCCTCCTT TCAG; M13M1cc, CCAGTCACGA CGTTGTA; M13 RV-N, TGTGGAATTG TGAGCGG; P.d.lib.-1, GCACTATCAC TTAACGGGGG CGTAGTGTCT TAGAGTAGGG; P.d.lib.-2, GCGGGATTAT TGAGTCTAGG CGTAGTGTCT TAGAGTAGGG; Link-pBRN, GGCGTAGTGT CTTAGAGTAG GGGGAAACAG CTATGACCAT; Link-M13M1cc, GGCGTAGTGT CTTAGAGTAG GGCCAGTCAC GACGTTGTA; P1-T10-NH2, NH2-TTTTTTTTTT CCGCGAAATT AATACGACTC AC; pBlue-Reverse-NH2, NH2-TTTTTTTTGG AAACAGCTAT GACCAT; P.d.lib.-F1-NH2, NH2-TTTTTTTTAC TATCACTTAA CGGGGG; R18, CCTTTCAGCA AAAAACCC; Guv-Rseq, TTTGTAGAGC TCATCCATGC; T7P-F, TAATACGACT CACTATAGGG; Guv-R12, CTCAAGCTTT TATTAGTGGT GG; pBlue-Reverse, TTTTTTTTGG AAACAGCTAT GACCAT; P.d.lib.-F1, ACTATCACTT AACGGGGG; P.d.1ib.-R1, GGGATTATTG AGTCTAGGC; P.d.1ib.-R2, GATTATTGAG TCTAGGCGTA; and Pp-R-N, NH2-GAACTGTTCT GTCCAACATA TAC.
Coupling oligonucleotides to beads
Magnetic bead solution (100 µl) (Dynabeads M-270 carboxylic acid, 2.8 ± 0.2 µm in diameter, Dynal Biotech, Lake Success, NY) was washed twice with 0.01 N NaOH by mixing using a rotator at room temperature. The beads were then washed three times with deionized water in the same manner and the supernatant was discarded. Twenty microliters of 5′-amino-modified oligonucleotide, P1-T10-NH2, pBlue-Reverse-NH2 or P.d.lib.-F1-NH2 solution (100 µM) and 50 µl of 2-(N-morpholino) ethane sulfonic acid (MES) buffer (0.4 M, pH 5.0) were added to the beads, and the solution was mixed using a rotator at room temperature. After 30 min, 30 µl of MES buffer including 3 mg of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride was added to the bead solution. The solution was mixed using a rotator for another 5 h at room temperature. The beads were washed four times with 200 µl of TE buffer, and finally suspended in 50 µl of TE buffer and stored at 4°C.
Solid-phase single-molecule PCR in w/o emulsions
Target DNA was diluted with TE buffer to various amounts of molecules per compartment on average. DNA concentration was determined by measuring absorbance at 260 nm. The aqueous phase consisted of 1× ExTaq buffer (Takara, Kyoto, Japan), 0.2 mM of each dNTP, 0.25 µM reverse primer, 5 U of ExTaq DNA polymerase (Takara), various amounts of template DNA (0.33, 1 or 30 molecules per compartment on average), 1 × 107 oligonucleotide-coupled beads and, when required, 2.5 nM forward primer in a total volume of 20 µl. The oil phase was freshly prepared by dissolving 1% Sun Soft No. 818SK (Taiyo Kagaku, Yokkaichi, Japan), polyglycerol esters of interesterified ricinoleic acid in 380 µl of mineral oil (Sigma).
The aqueous mixture was gradually added to the mineral oil phase in a 5 ml vial with stirring (at 750–800 r.p.m.) using a magnetic bar (10 mm × 4 mm) for 3 min at room temperature. The obtained emulsion was dispensed, 50 µl each into PCR tubes. Then solid-phase PCR was carried out in the following temperature sequence: preheating at 95°C for 10 s; 55 cycles consisting of 95°C for 15 s, 50°C for 9 min and 72°C for 30 s; and an additional extension at 72°C for 7 min. After the PCR cycles, the emulsions were precipitated by centrifugation at 17 400× g for 3 min and the oil phase was removed at the bottom of the tubes to recover the reaction beads. The emulsions were disrupted by the addition of 50 µl of B&W buffer [1 M NaCl, 5 mM Tris–HCl (pH 8.0) and 0.5 mM EDTA] (Dynal Biotech) in a 0.5 ml tube. After mixing, the beads were precipitated by centrifugation at 17 400× g for 3 min. To remove the oil phase completely, the bead suspension was washed four times with 400 µl of hexane in a 0.5 ml tube. The beads suspension was then transferred to a 1.5 ml tube, washed three times with 1 ml of TE buffer, suspended in 10 µl of TE buffer and stored at 4°C.
PCR amplification from beads
Bead–DNA conjugates were critically diluted with TE buffer, then dispensed into PCR tubes at one to three beads per tube on average. The templates were amplified separately in a 10 µl PCR mixture containing 0.25 U of ExTaq DNA polymerase (Takara) with 0.25 µM of each primer in the following temperature sequence: preheating at 94°C for 5 min; 35 cycles consisting of 94°C for 15 s, 50°C for 30 s and 72°C for 30 s; and an additional extension at 72°C for 7 min. The PCR products were extracted with phenol-chloroform, precipitated with ethanol, and dissolved in TE buffer. The DNA fragments were sequenced by the dye-terminator sequencing method with Guv-Rseq, M13M1cc, P.d.lib.-F1 and P.d.lib.-R1.
Plasmid construction
Plasmid DNA isolation, agarose gel electrophoresis and E.coli transformation were carried out as described by Sambrook et al. (6). The PhaR gene fragment was amplified by PCR from pTV119N::phaR (4) using 0.5 U of Pyrobest DNA polymerase (Takara, Kyoto, Japan) and 0.5 µM each of the primers (phaR-F-XbaI and phaR-R-EcoRI). The amplified fragment was ligated with XbaI–EcoRI-digested pET22b(+) (Novagen) to yield pETphaR-His. The fragment of KpnI–BamHI-digested pBK-PpLf, which contained upstream regulatory elements for phaP in P.denitrificans (4), was ligated with KpnI–BamHI-digested pBluescriptIISK(+) (stratagene) (pBluescript::Pp). The fragment of XhoI–SalI-digested pGFPuv (clontech) was ligated with XhoI-digested pBluescriptIISK(+) (pBluescript GFP′).
Synthesis of templates for solid-phase single-molecule PCR in w/o emulsions
Each of the GFPUV mutants (TA, 148Thr/205Ala; TS, 148Thr; TT, 148Thr/205Thr; GS, 148Gly; CS, 148Cys; and CT, 148Cys/205Thr) (R. Mizuno, unpublished data) was amplified in a 20 µl PCR mixture using 0.5 U of Pyrobest DNA polymerase under the recommended reaction conditions with 0.05 µM each of the primers (F1 and R1 primers). The PCR products were separated by electrophoresis, recovered from the agarose gel, dissolved in TE buffer, and stored at −25°C until use.
The Pp fragment, in which the PhaR target sequence is included, and the negative control fragment were amplified with pBluescript::Pp and pBluescript GFP′, respectively, in a 20 µl PCR mixture using 0.5 U of ExTaq DNA polymerase and 0.5 µM each of the primers (M13 M1cc and M13 RV-N). The PCR products were separated by electrophoresis, recovered from the polyacrylamide gel, dissolved in TE buffer, and stored at −25°C until use.
P.denitrificans genomic library construction
The sequences of the oligonucleotides for use as a linker in this study are as follows: P. d.-link-1-ATC, ATCCCCTACT CTAAGACACT ACGCC; and P.d.-link-1, GGCGTAGTGT CTTAGAGTAG GG. P. d.-link-1-ATC (300 pmol) was 5′-phosphorylated with T4 polynucleotide kinase (PNK) at 37°C for 30 min with the presence of 3 mM ATP. After mixing the reaction mixture thoroughly to inactivate the T4 PNK, 300 pmol of the P.d.-link-1 was added to the mixture. These oligonucleotides were denatured at 95°C for 3 min and annealed at room temperature.
Total genomic DNA of P.denitrificans was isolated according to Wilson (7). The total DNA (5 µg) was then digested with 24 U of Sau3AI endonuclease at 37°C for 1 h. After the inactivation of Sau3AI, the digested products were treated in a reaction mixture consisting of 45 µM dGTP, 0.025 U/µl Taq DNA polymerase (Takara), 10 mM Tris–HCl (pH 8.3), 50 mM KCl and 1.5 mM MgCl2, incubated at 74°C for 30 min to add an extra G on both 3′ ends of the Sau3AI fragments. The products were ligated with the 5′-phosphorylated linker using T4 DNA ligase (Takara). After removing unligated linker fragments, the products were amplified in a 20 µl PCR mixture using 1 U of Pyrobest DNA polymerase and 0.25 µM each of the primers (P.d.-lib-1 and P.d.-lib-2) in the following temperature sequence: preheating at 95°C for 5 min; 20 cycles consisting of 95°C for 15 s, 57°C for 10 s and 72°C for 30 s; and an additional extension at 72°C for 7 min. The PCR products with 150–1000 bp in sizes were separated by electrophoresis and recovered from the agarose gel, dissolved in TE buffer and stored at −25°C until use as templates for genetic library on beads (GLOBE) construction.
A PhaR target fragment for P.denitrificans genomic library screening was constructed as follows. pBluescript::Pp was amplified in a 20 µl PCR mixture using 0.5 U of Pyrobest DNA polymerase and 0.25 µM each of the primers (Link-pBRN and Link-M13M1cc). The PCR product was separated by electrophoresis and recovered from the agarose gel. The obtained product was amplified in a 20 µl PCR mixture using 1 U of Pyrobest DNA polymerase (Takara) and 0.25 µM each of the primers (P.d.-lib.-1 and P.d.-lib.-2) in the following temperature sequence: preheating at 95°C for 5 min; 20 cycles consisting of 95°C for 15 s, 57°C for 10 s and 72°C for 30 s; and an additional extension at 72°C for 7 min. The PCR product was separated by electrophoresis, recovered from the agarose gel, dissolved in TE buffer and stored at −25°C.
PhaR purification
E.coli BL21 (DE3) cells harboring the plasmid pETphaR-His, which contained the phaR gene of P.denitrificans with the C-terminal histidine-tag, were grown at 37°C in 250 ml of Luria–Bertani medium containing 50 µg/ml ampicillin (6). The expression of the phaR gene was induced at OD660 of 0.5 by adding 0.5 mM IPTG. After incubation at 37°C for another 4 h, the cells were harvested and stored at −25°C.
The frozen cells were thawed and suspended in 25 ml of binding buffer (20 mM sodium phosphate and 0.5 M NaCl, pH7.4) containing 0.1 mg/ml lysozyme. After incubation for 30 min on ice, the cell lysate was sonicated on ice and then centrifuged at 5800× g for 15 min at 4°C. The supernatant was subjected to HiTrap Chelating HP (Amersham Biosciences) affinity chromatography. Unbound proteins were eluted from the column by washing with the binding buffer containing 5 mM imidazole. PhaR was eluted with the elution buffer (20 mM sodium phosphate, 0.5 M NaCl and 0.5 M imidazole; pH 7.4), and dialyzed against the storage buffer (20 mM Tris–HCl, 50% glycerol and 0.1 mM DTT, pH 7.4). PhaR concentration was determined as described previously (8) using BSA as standard. Since Maehara et al. (4) suggested that the functional PhaR is composed of a homotetramer, PhaR concentration was evaluated as if PhaR forms a tetramer.
Gel mobility shift assay
Gel mobility shift assay was carried out as described previously (4) with some modifications. Of DNA fragments 0.1–0.2 pmol was added to the PhaR reaction buffer [phosphate-buffered saline (PBS): 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4•12H2O, 1.4 mM KH2PO4, pH 7.3 and 1.0 µM purified PhaR tetramer] in a total volume of 10 µl and incubated at 25°C for 30 min. The DNA–protein complex was separated from the unbound DNA fragment on a 5% native polyacrylamide gel using 0.5% Tris–borate–EDTA as the electrophoresis buffer. After electrophoresis, DNA fragments in the gel were stained with ethidium bromide and detected by UV exposure.
High-throughput screening of DNA-beads using flow cytometry
The DNA–bead complex solution (2 µl), the product of solid-phase PCR in w/o emulsions, was added to 8 µl of PBS including 1.0 µM of purified PhaR and mixed using a rotator at room temperature for 25 min. In the P.denitrificans genomic library bead screening, E.coli S30 extract was used instead of PBS. Then, the beads were collected with a magnet and washed with 1 ml of PBS. The collected beads were mixed with 100 ng of Anti-His(C-term)-FITC (Invitrogen) in 10 µl of PBS using a rotator at room temperature for 15 min under shaded conditions.
The beads were collected and suspended in 500 µl of PBS and analyzed using EPICS ELITE ESP (Beckman Coulter). The collected positive beads were suspended in TE buffer and stored at 4°C.
BIAcore affinity measurement
The in vitro affinity of the PhaR tetramer was determined using surface plasmon resonance on a BIAcore 2000 instrument (Pharmacia Biosensor). The target DNA fragment was amplified with pBluescript::Pp (this study) using 0.5 U of ExTaq DNA polymerase (Takara) under the recommended reaction conditions with 0.25 µM each of the primers (T7P-F and Pp-R-N). The amplified fragment was digested with HaeIII, and the resulting 100 bp fragment was separated by electrophoresis and recovered from the polyacrylamide gel. The fragment was immobilized on the sensor chip CM5 (Pharmacia Biosensor) by the amine coupling method according to the manufacturer's instruction. The kinetic rate constants kon and koff were calculated using the software supplied by the manufacturer. Kd was calculated from the kon and koff (Kd = koff/kon).
RESULTS AND DISCUSSION
Optimization of w/o emulsions for GLOBE construction
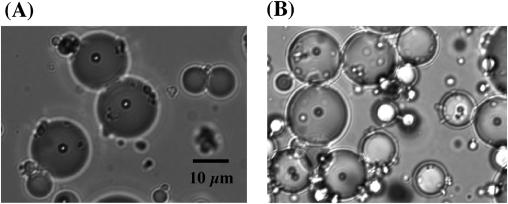
At first, we prepared standard w/o emulsions for GLOBE construction as described by Tawfik and Griffiths (9) with some modifications (the oil phase was mineral oil including 4.5% Span 80, 0.45% Tween 80 and 0.05% Triton X-100), but the emulsions were disrupted after PCR (data not shown). Then, we attempted the preparation of more stable w/o emulsions with mineral oil and another surfactant, Sun Soft No. 818SK (Taiyo Kagaku). The average diameter of the emulsion particles was 15–20 µm (2–4 pl) under the conditions described in Materials and Methods (Figure 2A). On the basis of the actual compartment size, we estimated that an emulsion comprising 20 µl of the aqueous solution and 380 µl of mineral oil contains 5 × 106 to 1.2 × 107 compartments. Since the beads encapsulated in the compartments remained after PCR (Figure 2B), the surfactant would offer a high heat resistance to the compartments in emulsions.
Figure 2.
Solid-phase PCR system encapsulated w/o emulsion. A typical encapsulated w/o emulsion before solid-phase single-molecule PCR (A) or after solid-phase single-molecule PCR (B). Beads are 2.8 ± 0.2 µm in diameter.
Confirmation of solid-phase single-molecule PCR in w/o emulsions
An equimolar mixture of DNAs for six types of GFPUV mutant (R. Mizuno, unpublished data) was diluted to give one molecule per compartment on average. Solid-phase single-molecule PCR was carried out in each compartment in the w/o emulsions with P1-T10-NH2 on beads and R18 primers. After the PCR cycles, the beads with immobilized PCR products were recovered by disrupting the emulsions. The obtained beads were extensively diluted to less than one bead per tube on average, and each DNA fragment on individual beads was separately amplified by bead PCR with the T7P-F and Guv-R12 primers. The amplified fragments were sequenced (summarized in Table 1). All types of GFPUV mutant sequence were found in 29 samples; 22 samples were sequence-amplified from a single template, and the other 7 samples were probably from a multitemplate. These multitemplate products were probable to have been obtained in solid-phase single-molecule PCR in w/o emulsions or bead PCR. Nonetheless, the results suggest that a single DNA fragment was successfully amplified and bound to a bead in the emulsions under the described picoliter-scale conditions.
Table 1.
The results of DNA sequence analysis of GFPUV mutant from solid-phase single-molecule PCR in w/o emulsions
| Mutant typea | Experiment 1 | Experiment 2 | Total |
|---|---|---|---|
| TA | 2 | 1 | 3 |
| TS | 0 | 5 | 5 |
| TT | 1 | 3 | 4 |
| GS | 6 | 1 | 7 |
| CS | 0 | 1 | 1 |
| CT | 1 | 1 | 2 |
| Multiple | 0 | 7 | 7 |
aTA, GFPUV mutant 148Thr/205Ala; TS, GFPUV mutant 148Thr; TT, GFPUV mutant 148Thr/205Thr; GS, GFPUV mutant 148Gly; CS, GFPUV mutant 148Cys; CT, GFPUV mutant 148Cys/205Thr.
Selection of PhaR binding DNA from Pp fragment and negative-control fragment mixture
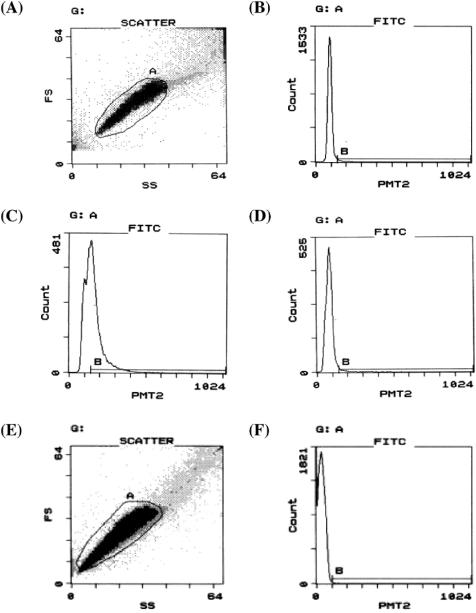
The Pp fragment, PhaR target sequence and negative-control fragment, part of the gfpuv sequence, were mixed in a molecular ratio of 1:102 or 1:103. The mixture was then diluted to 0.33 molecules per compartment on average and immobilized onto beads by solid-phase single-molecule PCR in the emulsions. The primers used in the PCR were pBlue-Reverse-NH2 (on beads), M13M1cc and free pBlue-Reverse. The obtained beads were analyzed by PhaR binding assay and flow cytometry, and positive beads (events in region B in Figure 3D) were sorted. Typical dot-plot and histogram for the beads containing Pp and negative-control fragments are shown in Figure 3A and D. The obtained beads were diluted to one bead or two beads per tube on average, and bead PCR was carried out with M13M1cc and pBlue-Reverse primers. To determine the interaction between the PCR products and PhaR, gel mobility shift assay was performed as shown in Figure 4.
Figure 3.
Flow cytometry of bead–DNA–PhaR–antibody complexes. (A) Typical dot-plot for beads containing Pp and negative-control fragments in FS (forward scatter) and SS (side scatter). Events gated in A (∼80% of total events) were subjected to the following analysis. (B) Histogram for ‘negative-control’ beads in FITC fluorescence intensity (for A-gated events). Events in region B were 1.6% of A-gated events. (C) Histogram for ‘positive-control’ beads in FITC fluorescence intensity (for A-gated events). Events in region B were 51.5% of A-gated events. (D) Typical histogram for beads containing Pp and negative-control fragments in FITC fluorescence intensity (for A-gated events). Events in region B were sorted. (E) Typical dot-plot of beads containing P.denitrificans genomic library in FS and SS. Events gated in A (∼97% of total events) were subjected to the following analysis. (F) Typical histogram for beads containing P.denitrificans genomic library in FITC fluorescence intensity (for A-gated events). Events in region B were sorted. PMT2, fluorescence channel 2 (FITC) signal intensity.
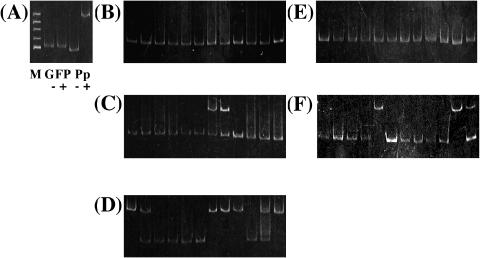
Figure 4.
Gel mobility shift assay for PCR products from two kinds of DNA mixture beads. (A) Negative and positive-control. M, ΦX174/HinfI marker; GFP, negative-control fragment; Pp, Pp fragment; −, minus-purified PhaR; and +, plus-purified PhaR. (B) Before sorting of DNA mixture beads (Pp fragment:negative-control fragment = 1:100). (C) After first sorting of DNA mixture beads (Pp fragment:negative-control fragment = 1:100). (D) After second sorting of DNA mixture beads (Pp fragment:negative-control fragment = 1:100). (E) Before sorting of DNA mixture beads (Pp fragment:negative-control fragment = 1:1000). (F) After second sorting of DNA mixture beads (Pp fragment:negative-control fragment = 1:1000).
When the Pp and negative-control fragments were mixed in a ratio of 1:102, ∼250 positive beads were pooled in the first sorting. The pooled beads were diluted to one bead per tube on average and bead PCR was carried out. The amplified fragments were found in 12 of 32 trials. Two bands were detected in some samples, which were supposed to have been formed from the multitemplate in the step of single-molecule PCR or bead PCR. Two Pp fragments were found in the reactions (Figure 4C). From the DNA on ∼50 beads pooled in the first selection, the second cycle was carried out as illustrated in Figure 1. Approximately 100 positive beads were pooled in the second sorting. The sorted beads were diluted to one bead per tube on average and bead PCR was carried out. Then the fragments amplified by the PCR were found in 12 of 80 trials. Seven Pp fragments were found in these 12 trials (Figure 4D).
When the Pp and negative-control fragment were mixed in a ratio of 1:103, ∼700 positive beads were pooled in the first sorting and used in the second cycle. Approximately 170 positive beads were pooled in the second sorting. All the sorted beads were diluted to two beads per tube on average, and bead PCR was carried out. Amplified fragments were found in 12 of 86 trials. Eventually, 3 Pp fragments were found in the 12 trials (Figure 4F). We also confirmed the sequences of the obtained clones (data not shown).
As shown in Figure 4, the Pp fragment was enriched to >50-fold (Figure 4D) and 200-fold (Figure 4F) after two cycles of sorting. The results show that our system successfully enriched the Pp fragment, the target of PhaR, from a pool of unrelated fragments.
Selection of PhaR binding DNA from P.denitrificans genomic library
We constructed a P.denitrificans genomic fragment library as illustrated in Figure 5. The average length of the fragments was estimated to be 300 bp. The PhaR target fragment and the library fragments were mixed in a molecular ratio of 1:30 000. The mixture was then diluted to 30 molecules per compartment on average and immobilized onto beads by solid-phase single-molecule PCR, because the immobilization efficiency of the library DNA on to beads was much lower than that in the previous experiment. The primers used in the PCR were P.d.lib.-F1-NH2 covalently bonded to beads, P.d.lib.-R1 and free P.d.lib.-F1. The obtained beads were mixed with 1 µM PhaR in the presence of E.coli S30 extract, which contributed to decreasing background noise, and analyzed by flow cytometry, and positive beads (events in region B in Figure 3F) were pooled. Typical dot-plot and histogram for the beads containing P.denitrificans genomic library are shown in Figure 3E and F. Approximately 150 beads were sorted from ∼112 000 A-gated events. The obtained beads were diluted to three beads per tube on average, and bead PCR was carried out with P.d.lib.-F1 and P.d.lib.-R2. To determine the interaction between the PCR products and PhaR, gel mobility shift assay was performed as shown in Figure 6.
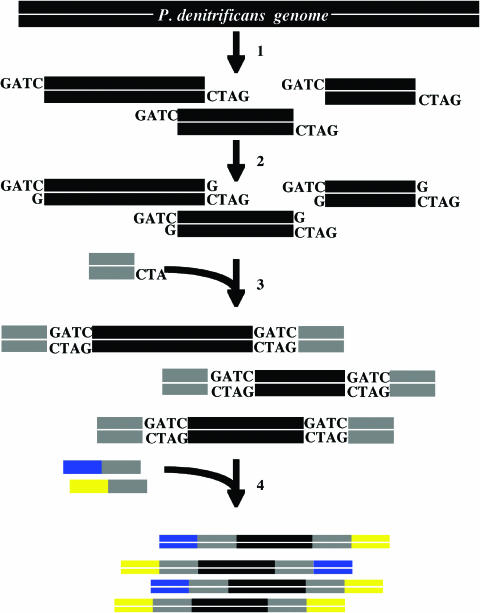
Figure 5.
P.denitrificans genomic library construction. 1: P.denitrificans total DNA is digested with Sau3AI endonuclease. 2: dGTPs are added to the P.denitrificans genomic library fragments. 3: The genomic fragments are ligated to the 5′-phosphorylated linker using DNA ligase. 4: To make the hetero-tail fragments, the linker-ligated fragments are amplified with hetero-ended primers.
Figure 6.
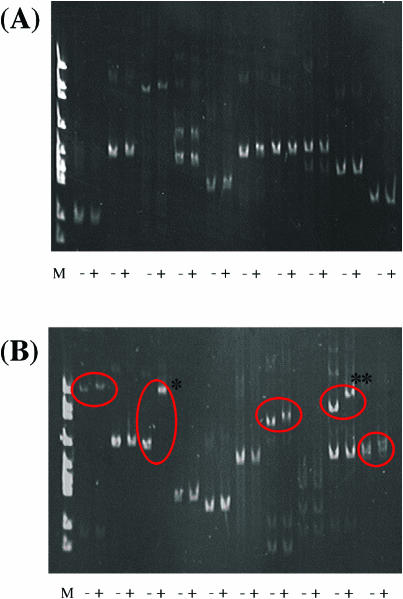
Gel mobility shift assay for PCR products from P.denitrificans genomic library mixture beads. (A) Before sorting of P.denitrificans genomic library beads (PhaR target fragment:P.denitrificans genomic library fragment = 1:30 000). (B) After sorting of P.denitrificans genomic library beads (PhaR target fragment: P.denitrificans genomic library fragment = 1:30 000). M, ΦX174/HincII marker; −, minus-purified PhaR; and +, plus-purified PhaR. Circles indicate the shift fragment by PhaR. An asterisk indicates PhaR target fragment. A double asterisk indicates a shifted fragment including a region upstream of moxZ from P.denitrificans.
Clear amplification fragments were found in 10 of 46 trials. Only 5 of 25 fragments were shifted in the presence of PhaR in the gel mobility shift assay (Figure 6B). Multiple bands were detected in some samples, probably because a multitemplate was used in the single-molecule PCR. Five gel-shifted fragments were sequenced and all of them were found to have a TGC-rich sequence, which is a putative PhaR recognition region (5). Furthermore, one spiked PhaR target fragment was found in the amplified fragments (Figure 6B, asterisk). Thus, the PhaR target fragment was enriched from 1:30 000 to 1:25. These results show that the target fragment was enriched ∼1200-fold after only one cycle of sorting from the genomic library and that our system is applicable to the screening of transcription factor recognition sequences from a genomic library.
Moreover, another shifted band (Figure 6B, double asterisk) included upstream of moxZ, which is involved in putative methanol oxidation in P.denitrificans. Interestingly, a TGC-rich region existed ∼20 bp upstream of the SD sequence of the gene in the fragment. P.denitrificans synthesizes polyhydroxyalkanoate from some alcohols including methanol (10). This result suggests that PhaR also regulates moxZ.
We also tried to enrich the target sequence of PhaR from an intact P.denitrificans genomic library, in which the PhaR target fragment was not spiked. However, we could not enrich the target sequence. On the other hand, we could not also amplify a 1.7 kb fragment containing the target sequence even from plasmid pPDPK1.7 (11) by PCR using ExTaq or Pyrobest DNA polymerase but could amplify it using LATaq (Takara), which is more suitable for the amplification of a GC-rich sequence (data not shown). Since phaC containing very high GC content region exists upstream of the PhaR target sequence (4), the failure of getting the native PhaR target sequence is supposed to be due to this high GC content region. Therefore, using such DNA polymerase suitable for amplification of high GC content region under the optimization conditions in the steps of the library construction and the solid-phase single-molecule PCR in w/o emulsions would lead successful enrichment of the target sequence.
We have developed a technology termed single-molecule DNA amplification by PCR (SM-PCR) for the construction of a protein library (12). This technique has been shown to be extremely useful in high-throughput functional screening (13–15), but the library scale is practically limited by the availability of PCR machines.
However, in vitro compartmentalization (IVC) in which genotype–phenotype linkage is achieved by compartmentalizing single genes in aqueous compartments of w/o emulsions, which enables high-throughput screening, has been reported (9,16–18).
Our novel library GLOBE can be made by direct linkage between IVC and SM-PCR on beads. GLOBE has the following advantages. First, it can be easily, stably, reproducibly and automatably obtained by solid-phase single-molecule PCR in w/o emulsions. Second, flow cytometry can be utilized for the screening of GLOBE. Since flow cytometry makes it possible to analyze and sort 100 to 10 000 samples per second, millions of samples from a DNA library can be analyzed and selected in one day using an automatic process. This screening system for GLOBE by flow cytometry is very powerful because we could enrich the target fragment of PhaR 1200-fold in only one cycle (Figure 6).
We used a transcription factor in P.denitrificans, PhaR, in this study. The Kd of the PhaR against the target sequence was measured to be ∼100 nM using BIAcore 2000 (Pharmacia Biosensor) (data not shown). Generally, this value is relatively high for a transcription factor, which suggests that our system can analyze various affinities of DNA–protein interactions.
For the same purpose, yeast one-hybrid systems (19,20) have been widely used so far. In particular, Wilson et al. (21) succeeded in identifying the DNA-binding site for NGFI-B from a rat genomic library with the system. The method is very convenient, but time consuming because it is carried out in vivo.
The ChIP-chip method has enabled the identification of transcription factor targets (2). The method can screen even binding sites for a transcription complex. However, the method can be applied only when a DNA chip of the target organism is available. Recently, a novel assay for mapping DNA–protein interactions, STAGE, has been developed (22). The method is very economical because no array is required, but it involves many complicated steps (e.g. formaldehyde cross-linking, immunoprecipitation and random amplification). On the other hand, our system can be applied to the identification of transcription factor targets in all types of organisms, though the system requires a cloned transcription factor. Moreover, our system using DNA-beads and flow cytometry for the analysis of DNA–protein interactions is very simple, rapid and effective.
Many studies related to solid-phase technology have recently been reported. Adessi et al. (23) have successfully amplified DNA on a glass surface. This solid-phase PCR is extremely useful in producing DNA chips for genome-wide screening. Yang et al. (25,26) have applied a split synthesis method to the construction of a one-bead one-compound library and screened proteins that specifically binds aptamers from large aptamer-bead-based libraries. Dressman et al. (27) have developed BEAMing (beads, emulsion, amplification and magnetics) for the detection and enumeration of genetic variants of human MID42. Very recently, Margulies et al. (28) have been succeeded in Mycoplasma genitalium genome sequencing from genomic DNA immobilized on beads by PCR in w/o emulsion.
Although we focused on DNA–protein interactions in this study, our system could be applied to the detection and analysis of other biomolecular interactions, such as protein–protein interactions, using an in vitro transcription and translation expression system. Thus, our system will be able to contribute to high-throughput transcriptome or proteome analysis.
Acknowledgments
We thank Dr Masatoshi Maki for valuable advice regarding BIAcore 2000. This work was supported in part by a Grant-in-Aid (No. 16360411) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT). Funding to pay the Open Access publication charges for this article was provided by MEXT.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pollack J.R., Iyer V.R. Characterizing the physical genome. Nature Genet. 2002;32:515–521. doi: 10.1038/ng1035. [DOI] [PubMed] [Google Scholar]
- 2.Orlando V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 2000;25:99–104. doi: 10.1016/s0968-0004(99)01535-2. [DOI] [PubMed] [Google Scholar]
- 3.Ren B., Robert F., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 4.Maehara A., Doi Y., Nishiyama T., Takagi Y., Ueda S., Nakano H., Yamane T. PhaR, a protein of unknown function conserved among short-chain-length polyhydroxyalkanoic acids producing bacteria, is a DNA-binding protein and represses. FEMS Microbiol. Lett. 2001;200:9–15. doi: 10.1111/j.1574-6968.2001.tb10685.x. [DOI] [PubMed] [Google Scholar]
- 5.Maehara A., Taguchi S., Nishiyama T., Yamane T., Doi Y. A repressor protein, PhaR, regulates polyhydroxyalkanoate (PHA) synthesis via its direct interaction with PHA. J. Bacteriol. 2002;184:3992–4002. doi: 10.1128/JB.184.14.3992-4002.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 7.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K., editors. Current protocols in molecular biology. NY: John Wiley and Sons, Inc; 1997. pp. 2.4.1–2.4.5. [Google Scholar]
- 8.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 9.Tawfik D.S., Griffiths A.D. Man-made cell-like compartments for molecular evolution. Nat. Biotechnol. 1998;16:652–656. doi: 10.1038/nbt0798-652. [DOI] [PubMed] [Google Scholar]
- 10.Yamane T., Chen X.F., Ueda S. Polyhydroxyalkanoate synthesis from alcohols during the growth of Paracoccus denitrificans. FEMS Microbiol. Lett. 1996;135:207–211. [Google Scholar]
- 11.Maehara A., Ueda S., Nakano H., Yamane T. Analyses of a polyhydroxyalkanoic acid granule-associated 16-kilodalton protein and its putative regulator in the pha locus of Paracoccus denitrificans. J. Bacteriol. 1999;181:2914–2921. doi: 10.1128/jb.181.9.2914-2921.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohuchi S., Nakano H., Yamane T. In vitro method for the generation of protein libraries using PCR amplification of a single DNA molecule and coupled transcription/translation. Nucleic Acids Res. 1998;26:4339–4346. doi: 10.1093/nar/26.19.4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rungpragayphan S., Kawarasaki Y., Imaeda T., Kohda K., Nakano H., Yamane T. High-throughput, cloning-independent protein library construction by combining single-molecule DNA amplification with in vitro expression. J. Mol. Biol. 2002;318:395–405. doi: 10.1016/S0022-2836(02)00094-3. [DOI] [PubMed] [Google Scholar]
- 14.Rungpragayphan S., Nakano H., Yamane T. PCR-linked in vitro expression: a novel system for high-throughput construction and screening of protein libraries. FEBS Lett. 2003;540:147–150. doi: 10.1016/s0014-5793(03)00251-5. [DOI] [PubMed] [Google Scholar]
- 15.Koga Y., Kato K., Nakano H., Yamane T. Inverting enantioselectivity of Burkholderia cepacia KWI-56 lipase by combinatorial mutation and high-throughput screening using single-molecule PCR and in vitro expression. J. Mol. Biol. 2003;331:585–592. doi: 10.1016/s0022-2836(03)00782-4. [DOI] [PubMed] [Google Scholar]
- 16.Doi N., Yanagawa H. STABLE: protein–DNA fusion system for screening of combinatorial protein libraries in vitro. FEBS Lett. 1999;457:227–230. doi: 10.1016/s0014-5793(99)01041-8. [DOI] [PubMed] [Google Scholar]
- 17.Sepp A., Tawfik D.S., Griffiths A.D. Microbead display by in vitro compartmentalisation: selection for binding using flow cytometry. FEBS Lett. 2002;532:455–458. doi: 10.1016/s0014-5793(02)03740-7. [DOI] [PubMed] [Google Scholar]
- 18.Cohen H.M., Tawfik D.S., Griffiths A.D. Altering the sequence specificity of Hae III methyltransferase by directed evolution using in vitro compartmentalization. Protein Eng. Des. Sel. 2004;17:3–11. doi: 10.1093/protein/gzh001. [DOI] [PubMed] [Google Scholar]
- 19.Wang M.M., Reed R.R. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature. 1993;364:121–126. doi: 10.1038/364121a0. [DOI] [PubMed] [Google Scholar]
- 20.Joachim J.L., Herskowitz I. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science. 1993;262:1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- 21.Wilson T.E., Fahrner T.J., Johnston M., Milbrandt J. Identification of DNA binding site for NGFI-B by genetic selection in yeast. Science. 1991;252:1296–1300. doi: 10.1126/science.1925541. [DOI] [PubMed] [Google Scholar]
- 22.Kim J., Bhinge A.A., Morgan X.C., Iyer V.R. Mapping DNA–protein interactions in large genomes by sequence tag analysis of genomic enrichment. Nature Methods. 2005;2:47–53. doi: 10.1038/nmeth726. [DOI] [PubMed] [Google Scholar]
- 23.Adessi C., Matton G., Ayala G., Turcatti G., Mermod J.J., Mayer P., Kawashima E. Solid phase DNA amplification: characterisation of primer attachment and amplification mechanisms. Nucleic Acids Res. 2000;28:e87. doi: 10.1093/nar/28.20.e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam K.S., Salmon S.E., Hersh E.M., Hruby V.J., Kazmierskl W.M., Knapp R. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991;354:82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 25.Yang X., Bassett S.E., Li X., Luxon B.A., Herzog N.K., Shope R.E., Aronson J., Prow T.W., Leary J.F., Kirby R., et al. Construction and selection of bead bound combinatorial oligonucleoside phosphorothioate and phosphorodithioate aptamer libraries designed for rapid PCR-based sequencing. Nucleic Acids Res. 2002;30:e132. doi: 10.1093/nar/gnf132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang X., Li X., Prow T.W., Reece L.M., Bassett S.E., Luxon B.A., Herzog N.K., Aronson J., Shope R.E., Leary J.F., et al. Immunofluorescence assay and flow-cytometry selection of bead-bound aptamers. Nucleic Acids Res. 2003;31:e54. doi: 10.1093/nar/gng054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dressman D., Yan H., Traverso G., Kinzler K.W., Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc. Natl Acad. Sci. USA. 2003;100:8817–8822. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]