Abstract
The cap-dependent endonuclease of the influenza viral RNA polymerase, which produces the capped RNA primers that initiate viral mRNA synthesis, is comprised of two active sites, one for cap binding and one for endonuclease cleavage.We identify the amino acid sequences that constitute these two active sites and demonstrate that they are located on different polymerase subunits. Binding of the 5′ terminal sequence of virion RNA (vRNA) to the polymerase activates a tryptophan-rich, cap-binding sequence on the PB2 subunit. At least one of the tryptophans functions in cap binding, indicating that this active site is probably similar to that of other known cap-binding proteins. Endonuclease cleavage, which is activated by the subsequent binding of the 3′ terminal sequence of vRNA, resides in a PB1 sequence that contains three essential acidic amino acids, similar to the active sites of other enzymes that cut polynucleotides to produce 3′-OH ends. These results, coupled with those of our previous study, provide a molecular map of the five known essential active sites of the influenza viral polymerase.
Keywords: cap binding/capped RNA primers/endonuclease cleavage/influenza virus polymerase
Introduction
The influenza virus RNA polymerase catalyzes the synthesis of viral mRNAs and the replication of genome RNAs in infected cells (reviewed in Krug et al., 1989). During viral mRNA synthesis the polymerase carries out two functions: (i) the production of the novel primers that are required for the initiation of viral mRNA synthesis, i.e. capped RNA fragments cleaved from host cell nuclear RNAs; and (ii) the subsequent initiation and elongation of viral mRNA chains. Capped primers are produced only after the polymerase is activated by the sequential binding of the 5′ and 3′ terminal sequences of virion RNA (vRNA) (Hagen et al., 1994; Cianci et al., 1995; Li et al., 1998). Consequently, vRNA molecules function not only as templates for mRNA synthesis, but also as essential cofactors which activate the catalytic functions of the polymerase that produces capped RNA primers.
The influenza virus polymerase is comprised of three viral polymerase (P) proteins, PB1, PB2 and PA (Krug et al., 1989). The 5′ and 3′ terminal sequences of vRNA bind to different amino acid sequences of the PB1 polymerase subunit, which is part of functional polymerase complexes (Li et al., 1998). Such functional complexes are formed only when all three P proteins are coexpressed (Hagen et al., 1994; Cianci et al., 1995; Li et al., 1998). The 5′ terminal sequence of vRNA (5′ vRNA) binds to the PB1 sequence centered around two arginine residues at positions 571 and 572 (Li et al., 1998). This binding activates two new functions of the polymerase: the PB2 protein acquires the ability to bind the 5′ capped ends of RNAs and the PB1 protein acquires an RNA-binding site that is specific for the 3′ terminal sequence of vRNA (3′ vRNA), an RNP1-like motif extending from amino acids 249 to 256 (Li et al., 1998). Subsequent binding of the 3′ vRNA to this PB1 sequence activates the endonuclease that cleaves the bound capped RNAs 10–13 nucleotides from their 5′ ends (Hagen et al., 1994; Li et al., 1998). This enzyme produces 3′-OH ends, thereby enabling the capped fragments that are generated to serve as primers to initiate viral mRNA synthesis.
In contrast to the two sites that bind 5′ and 3′ vRNA, less is known about other functional sites of the polymerase complex. Cap-binding activity has been shown to reside in the PB2 subunit (Ulmanen et al., 1981; Blass et al., 1982), but the specific PB2 sequence that binds cap structures has not been identified. It has been postulated that the PB2 subunit also catalyzes endonuclease cleavage (Licheng et al., 1995), but this has not been clearly established, so that the amino acid sequence that functions in endonuclease cleavage has not been identified. The PB1 protein, which also catalyzes nucleotide addition during mRNA chain elongation (Krug et al., 1989), contains the four consensus motifs for nucleic acid polymerases, including the sequence SDD in motif C (Argos, 1988; Poch et al., 1989; Biswas and Nayak, 1994). This SDD sequence in the PB1 protein has been shown to be required for viral mRNA synthesis (Biswas and Nayak, 1994), but it has not been established whether this sequence is specifically required for the catalysis of nucleotide addition rather than for another PB1 function.
In the present study, we use ultraviolet (UV) light cross-linking and mutagenesis to identify the specific polymerase sequences that acquire cap-binding and endonuclease cleavage activity as a result of the binding of 5′ and 3′ vRNA. We show that cap-binding activity resides in a tryptophan-rich PB2 sequence and that at least one of these tryptophans functions in cap binding, indicating that the PB2 protein probably interacts with the m7G of the cap, like other cap-binding proteins whose structures are known (Hodel et al., 1997; Marcotrigiano et al., 1997; Matsuo et al., 1997). Furthermore, we demonstrate that endonuclease cleavage activity resides in the PB1 subunit rather than the PB2 subunit. This active site contains three essential acidic amino acids, and is thus similar to the active sites of other enzymes that also cut polynucleotides (RNA or DNA) to produce 3′-OH ends (Kanaya et al., 1990; Davies et al., 1991; Dyda et al., 1994; Yang and Steitz, 1995; Rice et al., 1996).
Consequently, we can conclude that the sequential binding of the two influenza genome RNA sequences to the PB1 subunit induces the formation of new active sites in the PB2 and PB1 subunits that are similar, respectively, to that of other cap-binding proteins and to that of the class of nucleases that produce 3′-OH ends. Finally, we show that the consensus SDD sequence in the PB1 subunit specifically functions in the catalysis of nucleotide addition and not in the other PB1 functions required for the production of capped RNA primers. Hence, these results, coupled with those of our previous study (Li et al., 1998), provide a molecular map of the five known essential active sites of the influenza viral polymerase.
Results
The active site for the binding of the 5′ terminal cap of RNA resides in a tryptophan-rich sequence in the PB2 protein subunit
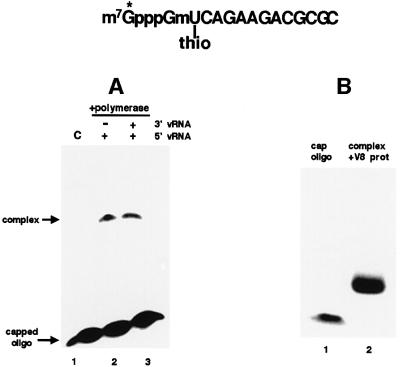
To identify the cap-binding site of the influenza virus polymerase, we used a short, capped RNA oligonucleotide that contains 32P only in its 5′ terminal cap structure and an unlabeled thio U residue at the nucleotide adjacent to the m7GpppGm cap structure (Figure 1). The thio U residue affords very high efficiency of cross-linking with amino acids after exposure to UV light (Favre et al., 1998; Wang and Rana, 1998). This labeled oligonucleotide was added to a preparation of recombinant influenza virus RNA polymerase, namely the nuclear extract from HeLa cells infected with three vaccinia virus vectors expressing the PB1, PB2 and PA proteins (Hagen et al., 1994; Cianci et al., 1995; Li et al., 1998).
Fig. 1. Isolation of the polymerase peptide that cross-links to the capped RNA oligonucleotide that contains 32P only in its 5′ terminal cap structure and an unlabeled thio U residue at the nucleotide adjacent to the cap. (A) This labeled oligoribonucleotide was incubated with a nuclear extract from cells expressing the three P proteins in the presence of either 5′ vRNA alone (lane 2) or both 5′ vRNA and 3′ vRNA (lane 3). After the binding reaction and subsequent exposure to 366 nm UV light for 30 min, the reaction mixtures were electro phoresed on a 7% denaturing gel to separate the cross-linked protein–oligoribonucleotide complex from free oligoribonucleotide. Lane 1 (C), the 32P-cap-labeled oligoribonucleotide alone. (B) The radiolabeled protein–oligoribonucleotide complex [from (A), lane 2] was digested with V8 protease, and the resulting products were analyzed by electrophoresis on an 18% denaturing gel [lane 2 of (B)]. Lane 1, the 32P-cap-labeled oligoribonucleotide prior to UV cross-linking.
The cap-labeled, thio U-containing oligoribonucleotide cross-links to polymerase complexes in the HeLa cell nuclear extracts when the polymerase is activated by 5′ vRNA (Figure 1A, lane 2). No detectable cross-linking was observed in the absence of 5′ vRNA (data not shown). Further addition of 3′ vRNA does not increase the cross-linking of the oligonucleotide to the polymerase complexes (compare lanes 2 and 3), as expected for the cap-binding site of the polymerase, which is fully activated by 5′ vRNA alone (Hagen et al., 1994; Cianci et al., 1995; Li et al., 1998). Consequently, by carrying out cross-linking in the presence of only 5′ vRNA, only the cap-binding site and not the endonuclease cleavage site should be activated and available for cross-linking (see below). Cross-linking of this capped oligoribonucleotide in the presence of 5′ vRNA requires that the nuclear extracts contain all three P proteins (data not shown), in confirmation of previous results showing that a functional cap-binding site requires that the complex contains all three P proteins (Hagen et al., 1994; Cianci et al., 1995; Li et al., 1998).
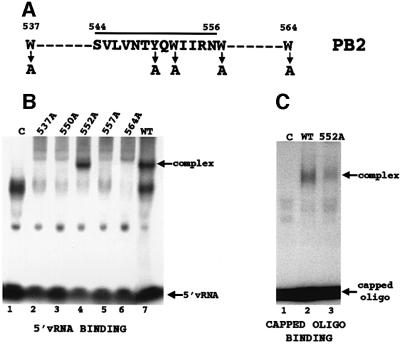
To identify the binding site of the capped oligonucleotide, the single protein species that is cross-linked to the cap-labeled oligoribonucleotide in the presence of 5′ vRNA was isolated by gel electrophoresis (Figure 1A, lane 2) and then digested with V8 protease. The labeled digestion product migrates slower than the labeled oligoribonucleotide, consistent with the presence of a small peptide covalently attached to the oligoribonucleotide (Figure 1B, compare lanes 1 and 2). Sequencing of this peptide shows that it matches a sequence in the PB2 protein subunit, extending from amino acid 544 to amino acid 556 (Figure 2A). This sequence contains a W (tryptophan) and a Y (tyrosine), two aromatic amino acids that in other cap-binding proteins have been shown to interact with the m7G of the cap (Hodel et al., 1997; Marcotrigiano et al., 1997; Matsuo et al., 1997). This PB2 peptide is part of a larger PB2 sequence (amino acids 533–564) that is tryptophan rich, containing additional Ws at positions 537, 557 and 564.
Fig. 2. Identification of the polymerase sequence that binds the 5′ terminal cap structures of RNAs. (A) Amino acid sequence of the peptide (amino acids 544–556) that cross-links to the capped, thio U-containing oligonucleotide containing 32P only in its 5′ cap structure. The positions of neighboring tryptophans (Ws) are also shown. The four tryptophans and the single tyrosine that were individually replaced with alanine are denoted by arrows. (B) Binding of 5′ vRNA (labeled at its 3′ end with 32pCp) to polymerase complexes containing a wild-type PB2 protein or one of the mutant PB2 proteins. The same amount (2.5 µg) of each nuclear extract was added. Polymerase–5′ vRNA complexes were separated from unbound 5′ vRNA by electrophoresis on 4% non-denaturing gels. Lane 1 (C), labeled 5′ vRNA incubated in the presence of a nuclear extract from cells infected with a vaccinia virus vector expressing T7 RNA polymerase. The virus-specific complex is denoted. The species that migrate faster than the virus-specific complex, which are also found in the absence of polymerase complexes, i.e. in lane 1 (C), vary in amount in different experiments. (C) Binding of a capped RNA fragment to polymerase complexes containing a wild-type PB2 protein or the mutant PB2 protein with an alanine substitution at position 552. The same amount (2.5 µg) of each nuclear extract was added. Subsequent to the binding of unlabeled 5′ vRNA to the polymerase complexes, the 13-nucleotide-long capped AlMV RNA 4 RNA fragment containing an m7G32pppGm 5′ end (10 ng, 1 × 105 c.p.m.) was incubated with the wild-type (lane 2) or the mutant polymerase complexes (lane 3). The capped RNA–polymerase complexes were separated from free capped RNA fragments by electrophoresis on 4% non-denaturing gels. The arrows denote the positions of free capped RNA fragments and of the capped RNA–polymerase complexes. The species that migrate faster than the virus-specific complex, which are also found in the absence of polymerase complexes (lane 1), vary in amount in different experiments. The amounts of the cap-binding complexes were quantitated using a phosphoimager. Lane 1 (C), capped RNA fragment incubated with a nuclear extract from cells infected with a vaccinia virus vector expressing T7 RNA polymerase.
To determine which aromatic acids in the larger PB2 sequence (amino acids 533–564) function in cap binding, we individually replaced each of its tryptophans (positions 537, 552, 557 and 564) and its tyrosine (position 550) with the non-aromatic amino acid alanine. Each of the mutated PB2 genes was inserted into a vaccinia virus vector (Elroy-Stein and Moss, 1996), which was used to infect HeLa cells together with the two vaccinia virus vectors expressing the wild-type PB1 and PA proteins. Western analysis showed that similar amounts of the mutant and wild-type PB2 proteins were synthesized (data not shown). To determine whether the mutant PB2 proteins form functional polymerase complexes, we assayed the nuclear extracts for the presence of polymerase complexes that bind 5′ vRNA (Figure 2B). Only one of the alanine substitutions, at position 552, yielded a PB2 protein that is capable of forming such functional polymerase complexes (see Discussion). Consequently, the polymerase complexes containing the PB2 protein with this alanine substitution (W552A) are the only ones that we can assay for cap-binding activity in the presence of 5′ vRNA.
The cap-binding activity of the W552A mutant polymerase complexes in the presence of 5′ vRNA was compared with that of wild-type polymerase complexes, using as substrate the 13-nucleotide-long capped RNA fragment that had been cleaved from alfalfa mosaic virus (AlMV) RNA 4 by the influenza virion endonuclease itself (Li et al., 1998). The W552A mutant polymerase complexes are defective in cap-binding activity: they possess only ∼25% of the activity of wild-type polymerase complexes (Figure 2C, compare lanes 2 and 3). In contrast, these polymerase complexes retain wild-type activity for the binding of 3′ vRNA in the presence of 5′ vRNA, a step that is activated concomittantly with cap binding (data not shown). Consequently, these results indicate that the W552A mutation in the PB2 protein results in polymerase complexes that possess wild-type 5′ vRNA binding activity but are defective in the subsequent step of cap binding (see Discussion).
The active site for the endonucleolytic cleavage of capped RNAs is located in the PB1 subunit and contains several essential acidic amino acids
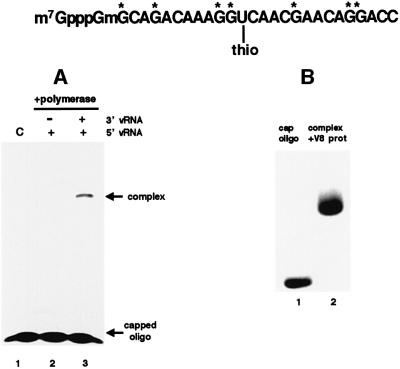
To identify the active site for endonucleolytic cleavage of capped RNAs, we used a 26-nt-long capped RNA oligonucleotide in which thio U was placed close to the presumed site of cleavage—the 3′ phosphodiester bond of the A residue at position 14 (Figure 3). The G residues of this oligonucleotide were labeled. Our rationale was that during the cleavage reaction the thio U residue in the resulting 14-nt-long capped RNA oligonucleotide would cross-link with the nuclease active site. As an important control, we demonstrated that this labeled oligonucleotide does not cross-link to the polymerase in the presence of 5′ vRNA alone (Figure 3A, lane 2), conditions under which cap binding but not endonucleolytic cleavage is activated (Hagen et al., 1994; Cianci et al., 1995; Li et al., 1998). These results indicate that the distance of the thio U from the cap structure is large enough to prevent it from cross-linking to the cap-binding site of the PB2 protein.
Fig. 3. Isolation of the polymerase peptide that cross-links to the capped RNA oligonucleotide that contains thio U close to the site of endonucleolytic cleavage. (A) This oligoribonucleotide, which was labeled internally with 32P (labeled Gs are denoted by asterisks), was incubated with a nuclear extract from cells expressing the three P proteins in the presence of either 5′ vRNA alone (lane 2) or both 5′ vRNA and 3′ vRNA (lane 3). After binding and subsequent exposure to UV light, the reaction mixtures were electrophoresed on a 7% denaturing gel to separate the cross-linked protein–oligoribonucleotide complex from free oligoribonucleotide. Lane 1 (C), the 32P-labeled oligoribonucleotide alone. (B) The radiolabeled protein–oligoribo nucleotide complex [from (A), lane 3] was digested with V8 protease, and the resulting products were analyzed by electrophoresis on an 18% denaturing gel [lane 2 of (B)]. Lane 1, the 32P-labeled oligoribo nucleotide prior to UV cross-linking.
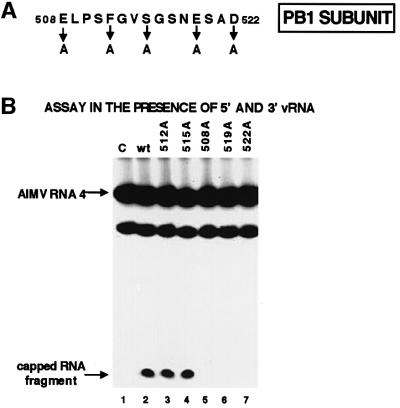
When the polymerase is activated by both 5′ vRNA and 3′ vRNA, thereby activating the endonucleolytic cleavage of the labeled oligonucleotide (Hagen et al., 1994; Cianci et al., 1995; Li et al., 1998), the thio U in the oligo nucleotide cross-links to the polymerase (Figure 3A, lane 3). Because cross-linking is dependent on endonucleolytic cleavage, we concluded that this oligonucleotide probably cross-links with the endonuclease active site and, therefore, proceeded with the identification of the cross-linked peptide. The cross-linked protein species was digested with V8 protease, yielding a product that migrates slower than the labeled oligoribonucleotide (Figure 3B, lane 2). Sequencing of the small peptide covalently attached to this oligoribonucleotide established that it matches a sequence in the PB1 subunit extending from amino acid 508 to 522 (Figure 4A). This sequence contains three acidic amino acids, which are prime candidates for essential residues because specific acidic amino acids have been shown to be essential for the activity of other enzymes that cleave polynucleotides to produce 3′-OH ends (Kanaya et al., 1990; Davies et al., 1991; Dyda et al., 1994; Yang and Steitz, 1995; Rice et al., 1996).
Fig. 4. Identification of the polymerase active site that is required for the catalytic function of the endonuclease. (A) Amino acid sequence of the region in the PB1 protein subunit that cross-links to the capped RNA oligonucleotide containing thio U close to the site of endo nucleolytic cleavage. The positions of the five amino acids that were individually replaced with an alanine are denoted. (B) Cap-dependent endonuclease activities of wild-type and mutant polymerases. After binding 5′ vRNA and 3′ vRNA to the polymerase complexes, wild-type (lane 2) or mutant polymerases (lanes 3–7) were incubated with full-length AlMV RNA 4 containing an m7G32pppGm 5′ end. The RNA products were separated on a denaturing 20% gel. The positions of full-length ALMV RNA 4 and of the specific 13-nt-long cleavage product are indicated by arrows. The species migrating faster than intact ALMV RNA 4 is a non-specific breakdown product. Lane 1 (C), full-length AlMV RNA 4 incubated in the absence of a polymerase complex.
To determine whether these three PB1 acidic amino acids (at positions 508, 519 and 522) are essential for the endonucleolytic cleavage of capped RNAs, each of these amino acids was individually replaced with alanine. As controls, we also replaced the phenylalanine at position 512 or the serine at position 515 with alanine. Each of the mutated PB1 genes was inserted into a vaccinia virus vector (Elroy-Stein and Moss, 1996), which was used to infect HeLa cells together with the two vaccinia virus vectors expressing the wild-type PB2 and PA proteins. Western analysis showed that similar amounts of the mutant and wild-type PB1 proteins were synthesized (data not shown). The polymerase complexes in the nuclear extracts from these cells were assayed for their cap-dependent endonuclease activity: full-length AlMV RNA 4 containing 32P only in its 5′ cap was incubated with each polymerase complex in the presence of both 5′ and 3′ vRNA, and the amount of the specific 13-nt-long capped cleavage product was determined (Plotch et al., 1981; Li et al., 1998) (Figure 4B).
Polymerase complexes that contain a PB1 protein in which the phenylalanine at position 512 or the serine at position 515 is replaced with alanine possess essentially the same endonuclease activity as wild-type polymerase complexes (Figure 4B, lanes 2–4). The polymerase complexes containing such altered PB1 proteins also catalyze the next step in viral mRNA synthesis, nucleotide addition (data not shown). In contrast, replacement of alanine for any of the acidic residues, at positions 508, 519 or 522, leads to a complete loss of endonuclease activity (lanes 5–7), indicating that each of these three acidic residues is required for endonuclease activity.
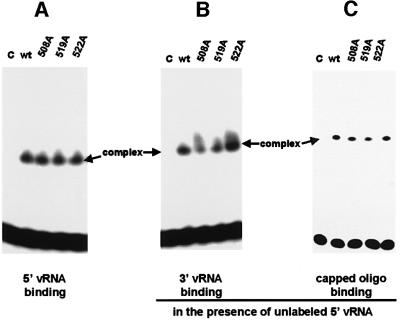
To rule out the possibility that the latter mutant polymerase complexes are defective in a step prior to endonuclease cleavage, we assayed their ability to bind 5′ vRNA (Figure 5A), 3′ vRNA in the presence of 5′ vRNA (Figure 5B) and capped RNA in the presence of 5′ vRNA (Figure 5C). The polymerase complexes containing alanine replacements of the acidic amino acids at positions 508, 519 or 522 of the PB1 subunit are not defective in these binding reactions. We conclude that these three mutant polymerases are specifically defective in the endonuclease reaction per se.
Fig. 5. The mutant polymerase complexes that lack endonuclease activity possess wild-type activities for binding (A) 5′ vRNA, (B) 3′ vRNA and (C) capped RNA. Wild-type (lane 2) or mutant (508A, 519A, 522A) polymerases (lanes 3–5) were incubated with (A) 5′ vRNA (labeled with 32P at its 5′ end), (B) labeled 3′ vRNA in the presence of unlabeled 5′ vRNA or (C) the 13-nt-long capped AlMV RNA 4 RNA fragment containing an m7G32pppGm 5′ end in the presence of unlabeled 5′ vRNA. Polymerase–RNA complexes were separated from unbound RNA by electrophoresis on 4% non-denaturing gels. Lane 1 (C), each labeled RNA incubated in the absence of a polymerase complex.
The polymerase consensus sequence in the PB1 subunit, SDD, specifically functions in the catalysis of nucleotide addition
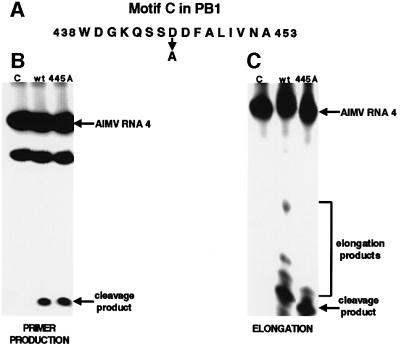
The PB1 subunit, which catalyzes nucleotide addition, contains the four consensus motifs for nucleic acid polymerases, including the sequence SDD at amino acids 444–446, which is in motif C (Figure 6A) (Argos, 1988; Poch et al., 1989; Biswas and Nayak, 1994). To determine whether this SDD sequence is specifically required for nucleotide addition and not another PB1-specific function, we prepared mutant polymerase complexes in which the PB1 subunit contains alanine instead of aspartic acid at position 445. In the presence of both 5′ and 3′ vRNA such mutant polymerase complexes possess essentially the same cap-dependent endonuclease activity as wild-type polymerase complexes (Figure 6B). To assay RNA chain elongation, the three ribonucleoside triphosphates (ATP, CTP and GTP) that are complementary to bases in the 3′ vRNA sequence were added during the cleavage reaction. Under these conditions, wild-type polymerase complexes synthesize, albeit inefficiently, RNA products that contain ∼2, 5 or 15 nt linked to the 13-nt-long capped cleavage product produced by the endonuclease (Figure 6C). In contrast, the polymerase complexes containing the PB1 protein with the alanine substitution at position 445 do not synthesize detectable amounts of the two longer elongation products. We conclude that the SDD sequence in the PB1 subunit functions in the catalysis of nucleotide addition.
Fig. 6. The SDD sequence (nt 444–446) in the PB1 subunit specifically functions in the catalysis of nucleotide addition. (A) Motif C containing the SDD sequence. The D at position 445 was replaced with A. (B) Production of capped RNA primers: cap-dependent endonuclease activity of the mutant polymerase in which the D at position 445 of the PB1 subunit has been replaced with an alanine. After binding 5′ vRNA and 3′ vRNA to the wild-type (lane 2) or mutant (lane 3) polymerase complexes, they were incubated with full-length AlMV RNA 4 containing an m7G32pppGm 5′ end. The RNA products were separated on a denaturing 20% gel. The positions of full-length AlMV RNA 4 and of the specific 13-nt-long cleavage product are indicated by arrows. The species migrating faster than intact AlMV RNA 4 is a non-specific breakdown product. Lane 1 (C), full-length AlMV RNA 4 incubated in the absence of a polymerase complex. (C) RNA chain elongation activity of the mutant and wild-type polymerases. Endonuclease assays contained 5′ and 3′ vRNA and three ribonucleoside triphosphates (ATP, CTP and GTP, each at 1 mM). The positions of the endonuclease cleavage product and the partially elongated RNA chains are denoted. Lane 1 (C), full-length AlMV RNA 4 incubated in the absence of a polymerase complex. No additional elongation products were detected in lane 3 even when the gel was exposed 20 times longer.
Discussion
After activation by two specific vRNA sequences the influenza virus polymerase acquires the cap-dependent endonuclease activity that produces the capped primers needed for the initiation of viral mRNA synthesis (Hagen et al., 1994; Cianci et al., 1995; Li et al., 1998). In the present study we demonstrate that this enzyme is comprised of two active sites that are on two different subunits of the polymerase. The cap-binding site, which is activated by the binding of 5′ vRNA to the polymerase, resides on the PB2 subunit, whereas the active site of the endonuclease, which is activated by the subsequent binding of 3′ vRNA, is located on the PB1 subunit.
The cap-binding site on the PB2 subunit
Our strategy for identifying the cap-binding site of the influenza virus RNA polymerase has several salient features. First, we used a short, capped RNA oligonucleotide that contains 32P only in its 5′ terminal cap structure and an unlabeled thio U residue at the nucleotide adjacent to the m7GpppGm cap structure. Secondly, we carried out the UV cross-linking of this thio U residue to the polymerase in the presence of only 5′ vRNA, so that only the cap-binding site and not the endonuclease cleavage site of the polymerase is activated and available for cross-linking. The peptide that cross-links under these conditions is the sequence in the PB2 protein subunit that extends from amino acid 544 to 556: SVLVNTYQWIIRN. This sequence, which is completely conserved in the PB2 protein of all influenza A virus strains sequenced, contains a Y (position 550) and W (position 552), two aromatic amino acids that in other cap-binding proteins have been shown to interact with the m7G of the cap (Hodel et al., 1997; Marcotrigiano et al., 1997; Matsuo et al., 1997). The PB2 sequence containing these two aromatic amino acids, NTYQW, is also conserved in all influenza B virus strains that have been sequenced. The influenza A virus PB2 peptide that cross-links to the cap-labeled oligonucleotide is part of a larger, tryptophan-rich PB2 sequence that extends from amino acids 533 to 564. A study carried out with polymerase complexes isolated from virions has also shown that this region of the PB2 protein cross-links to cap-labeled RNA (Honda et al., 1999). The larger PB2 sequence, which contains Ws at positions 537, 552, 557 and 564, is completely conserved in all influenza A virus strains. Only the Ws at positions 537 and 552 are conserved in the PB2 protein of influenza B virus.
Our aim was to verify the cross-linking results by mutagenesis, i.e. to determine whether substitution of alanine for individual aromatic amino acids in the PB2 sequence extending from amino acids 533 to 564 leads to a loss of the cap-binding activity of the viral polymerase. However, this aim could not be accomplished in its entirety because substitution of an alanine for individual Ws at positions 537, 557 or 564, or for the Y at position 550, renders the PB2 protein incapable of forming functional polymerase complexes. Presumably these aromatic amino acids are crucial to the correct folding of the PB2 protein and/or to its interaction with the PB1 and PA proteins. In contrast, the PB2 protein that contains a substitution of alanine for the W (position 552), which is the W that is in the peptide which cross-links to the cap-labeled oligonucleotide, forms functional polymerase complexes that bind 5′ vRNA. These mutant polymerase complexes have cap-binding activity that is only ∼25% of that of wild-type polymerase complexes. It is therefore likely that the PB2 protein in functional polymerase complexes interacts with the cap structure like other cap-binding proteins.
Structural studies of two other cap-binding proteins, the cellular eIF4E translation initiation factor and the vaccinia virus VP39 protein, have established that the m7G of the cap is sandwiched between two aromatic amino acids (Hodel et al., 1997; Marcotrigiano et al., 1997; Matsuo et al., 1997). With eIF4E, both of the aromatic amino acids are tryptophan (Marcotrigiano et al., 1997; Matsuo et al., 1997). Substitution of either of these tryptophans with a non-aromatic amino acid (leucine) eliminates cap-binding activity, and such a substitution of neighboring tryptophans either eliminates or substantially reduces cap-binding activity (Morino et al., 1996). Because replacement of the PB2 tryptophan at position 552 with a non-aromatic amino acid (alanine) substantially reduces, but does not eliminate, cap-binding activity, this tryptophan may not directly interact with the m7G of the cap. Rather, one of the neighboring aromatic amino acids (three tryptophans and one tyrosine) may serve this function. However, as discussed above, it was not possible to test this possibility.
Cap binding does not occur prior to the binding of 5′ vRNA to the PB1 subunit of the polymerase (Hagen et al., 1994; Cianci et al., 1995; Li et al., 1998). One possibility is that 5′ vRNA binding causes an allosteric change in the folding of the PB2 protein that enables two aromatic amino acids in the tryptophan-rich PB2 sequence (amino acids 533–564) to be appropriately positioned to sandwich the m7G of the cap. Alternatively, 5′ vRNA binding may expose a pre-existing cap-binding site on the PB2 protein by changing the quaternary structure of the polymerase complex.
Other than the presence of aromatic amino acids, the sequence homology between the eIF4E and VP39 cap-binding proteins is limited, in contrast to their structural homology (Hodel et al., 1997; Marcotrigiano et al., 1997; Matsuo et al., 1997). It was therefore not surprising that the tryptophan-rich PB2 peptide that cross-links to the cap does not align, without significant gaps, to any sequence in either eIF4E or VP39. However, the PB2 cap-binding peptide does align without any gaps with a sequence (amino acids 194–206) in the 80 kDa subunit of the nuclear cap-binding complex (CBC), which binds to the 5′ caps of cellular pre-mRNAs in the nucleus (Visa et al., 1996). In this alignment, six of the 13 amino acids are identical or similar. Although it is not clear whether this alignment is significant, it should be noted that at least some of the functions of CBC and the PB2 protein may be similar: the 5′ caps of influenza viral mRNAs in the nucleus are bound to the PB2 subunit of viral polymerase complexes rather than to CBC, suggesting that the PB2 protein may provide CBC-like functions for the viral mRNAs (Shih and Krug, 1996).
The endonuclease cleavage site on the PB1 subunit
The endonuclease active site on the PB1 protein contains three acidic amino acids (E508, E519, D522) that are essential for activity, whereas two nearby non-acidic amino acids are not essential. The sequence constituting this active site is completely conserved in the PB1 protein of all influenza A virus strains that have been sequenced. Also, except for three conservative amino acid changes (e.g. V instead of I), this sequence, specifically including the three acidic amino acids, is conserved in the PB1 protein of influenza B virus. Other enzymes that cleave polynucleotides (RNA or DNA) to produce 3′-OH ends also contain three or four essential acidic amino acids in their active sites (Kanaya et al., 1990; Davies et al., 1991; Dyda et al., 1994; Yang and Steitz, 1995; Rice et al., 1996). These enzymes include not only nucleases, such as RNase H, but also polynucleotidyl transferases, such as retroviral integrases and the Mu transposase. These enzymes share at least two properties with the influenza virus PB1 endonuclease activity: (i) the active sites bind one or more divalent metal ions that are required for activity (Davies et al., 1991; Yang and Steitz, 1995; Rice et al., 1996; Goldgur et al., 1998; Keck et al., 1998; Doan et al., 1999); and (ii) cleavage in vivo occurs preferentially at the 3′ end of a CA sequence (Beaton and Krug, 1981; Shaw and Lamb, 1984; Vink et al., 1991; Mizuuchi, 1992; Balakrishnan and Jonsson, 1997; Esposito and Craigie, 1998; Mahillon and Chandler, 1998).
However, unlike the influenza virus PB1 endonuclease, the acidic residues that constitute the active sites of these other enzymes that produce 3′-OH ends are separated from each other by as many as 50–60 amino acids (Davies et al., 1991; Dyda et al., 1994; Yang and Steitz, 1995; Rice et al., 1996; Goldgur et al., 1998). X-ray crystallography has established that protein folding results in the appropriate positioning of these amino acids to form the active sites for endonuclease cleavage. In contrast, the three acidic amino acids that are required for endonuclease cleavage by the PB1 protein are separated by only 10 and 2 amino acids, suggesting that only minimal protein folding is needed to position these amino acids to form an active site. However, it can not be ruled out that one or more of the other acidic amino acids that are located in the surrounding PB1 sequence are also required for PB1-catalyzed endonucleolytic cleavage. We presume that the sequential binding of 5′ vRNA and 3′ vRNA to two other sequences in the PB1 protein results in allosteric changes in this protein that are needed both to form the endonuclease active site and to position this active site so that it is at the appropriate location relative to the cap-binding site of the PB2 subunit, i.e. at a distance corresponding to the length of the capped fragments produced by cleavage, 10–13 nt plus the 5′ cap structure. Because of this distance constraint, it is possible that the cap-binding site of the PB2 protein is situated close to the interface with the PB1 subunit, thereby explaining why many of the alanine substitutions in the cap-binding region of the PB2 protein result in PB2 proteins that are incapable of forming functional polymerase complexes that bind 5′ vRNA.
Molecular map of five essential active sites of the influenza viral polymerase
Comparison of the amino acid sequences of RNA-dependent polymerases of many RNA viruses has identified four conserved sequence motifs, one of which (motif C) usually contains the sequence XDD (Poch et al., 1989). The location of this motif has been identified in the three-dimensional structure of two other viral RNA polymerases (Hansen et al., 1997; Lesburg et al., 1999). The structure of these viral RNA polymerases, like that of both DNA-dependent polymerases and reverse transcriptase (Joyce and Steitz, 1995), resembles a right-hand structure (Hansen et al., 1997; Lesburg et al., 1999). Motif C is located in the palm subdomain. The PB1 subunit of the influenza virus RNA polymerase contains a sequence that fits motif C, including the core SDD sequence that is located at amino acids 444–446. A previous study showed that mutation of this sequence inactivated the polymerase (Biswas and Nayak, 1994), indicating that this sequence is essential for one or more PB1 functions. Here we demonstrate that the SDD sequence of the PB1 subunit is not required for the production of capped RNA primers, but rather is part of the active site for nucleotide addition.
The influenza virus PB1 protein has two active sites that are similar to those in retroviral reverse transcriptases. Both the PB1 endonuclease and the retroviral RNase H active sites contain several essential acidic amino acids, and both enzymes cleave RNA to produce primer RNAs containing 3′-OH ends (Kanaya et al., 1990; Davies et al., 1991; Dyda et al., 1994; Yang and Steitz, 1995; Rice et al., 1996; this study). In addition, as is the case for reverse transcriptase (Kohlstaedt et al., 1992; Jacobo-Molina et al., 1993; Joyce and Steitz, 1995), the PB1 catalytic site for nucleotide addition is likely to be in the palm subdomain of the right-hand structure.
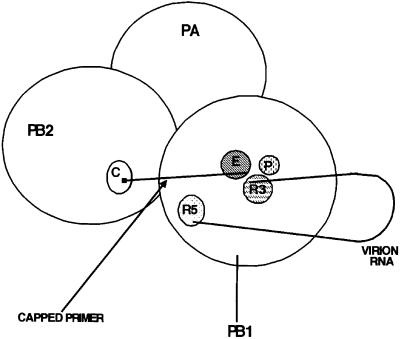
However, the interactions between these two active sites are probably not the same for the two enzymes. During the reverse transcriptase-catalyzed synthesis of second strand DNA, one of the functions of RNase H is to produce RNA primer(s) (Luo et al., 1990; Wohrl and Moelling, 1990; Randolph and Champoux, 1994). This RNA primer remains hydrogen-bonded with the template DNA, and the polymerase functions to juxtapose the resulting double-stranded region relative to the polymerase site (Kohlstaedt et al., 1992; Jacobo-Molina et al., 1993; Gao et al., 1998). The RNase H catalytic site itself apparently does not have a large role in this process, because it is separated from the polymerase active site by 18–20 base pairs (Kohlstaedt et al., 1992; Jacobo-Molina et al., 1993; Suo and Johnson, 1997). In contrast, we propose that during the initiation of influenza viral mRNA synthesis the activated endonuclease site (amino acids 508–522) and the binding site for 3′ vRNA (amino acids 249–256) are juxtaposed (Figure 7). Because the capped RNA primer produced by the endonuclease does not form hydrogen bonds with the 3′ vRNA template (Krug et al., 1989), the polymerase alone would have to be responsible for juxtaposing the primer and template. This could be achieved by the juxtaposition of the endonuclease and 3′ vRNA binding sites. In addition, both of these PB1 sequences should be close to the catalytic site for nucleotide addition (including amino acids 444–446) that is located in the palm subdomain (Figure 7). Conse quently, three non-contiguous amino acid sequences in the PB1 subunit would be close to each other in three dimensions during the initiation of viral mRNA synthesis (Figure 7).
Fig. 7. Schematic representation of the interactions between the catalytic sites of the influenza virus RNA polymerase after activation by the sequential binding of 5′ vRNA and 3′ vRNA. C indicates the location of the tryptophan-rich cap-binding site in the PB2 subunit; P indicates the location of the polymerase active site in the PB1 subunit, which includes the SSD sequence at amino acids 444–446; E indicates the location of the endonuclease active site in the PB1 subunit (amino acids 508–522); R3 denotes the binding site of 3′ vRNA in the PB1 subunit (amino acids 249–256); and R5 denotes the binding site of 5′ vRNA in the PB1 subunit (centered around two arginines at positions 571 and 572). A small black square denotes the 5′ cap of the primer RNA.
Our mapping of the five known active sites of the influenza virus polymerase has provided intriguing predictions about the structure of the activated form of the polymerase. Clearly, these predictions need to be confirmed by structural studies. The protein subunits of the polymerase only attain these active structures after the sequential binding of two specific vRNA sequences to the PB1 subunit. The challenge will be to determine precisely how this RNA binding induces the protein subunits to refold to form the active sites that are needed for the production of capped RNA primers and for the ensuing synthesis of viral mRNA.
Materials and methods
Preparation of recombinant influenza virus polymerase complexes
Recombinant vaccinia virus vectors individually encoding the influenza virus PB1, PB2 and PA proteins (Vac-PB1, Vac-PB2 and Vac-PA) were grown in HeLa cells (Smith et al., 1987; Cianci et al., 1995). Recombinant vaccinia virus vectors containing a mutant PB1 or PB2 gene were constructed as described previously (Li et al., 1998). HeLa cells were triply infected with Vac-PB2 (mutant or wild type), Vac-PA and Vac-PB1 (mutant or wild type) (Smith et al., 1987; Cianci et al., 1995; Li et al., 1998). At 20–24 h after infection, nuclear extracts were prepared and used as the source of the influenza virus polymerase complex (Fiering et al., 1990; Hagen et al., 1994; Li et al., 1998).
Preparation of labeled oligoribonucleotides and RNAs
The oligoribonucleotides used for the identification of the cap-binding and endonuclease active sites were synthesized using T7 RNA polymerase and synthetic DNA templates (Milligan et al., 1987). Transcriptions were carried out in the presence of thio-UTP. The oligoribonucleotide used to identify the cap-binding site was synthesized using unlabeled ribonucleoside triphosphates, and was then incubated with the vaccinia virus capping enzyme (guanyltransferase and 7-methyl transferase) and 2′-O-methyltransferase in the presence of [α-32P]GTP to produce an m7G32pppGm 5′ end (Plotch et al., 1981). The oligoribonucleotide used to identify the endonuclease active site was synthesized in the presence of [α-32P]GTP and m7GpppGm. The RNA substrate for endonuclease assays was AlMV RNA 4 containing an m7G32pppGm 5′ end (Plotch et al., 1981). The substrate for capped RNA-binding was prepared by incubating full-length AlMV RNA 4 (containing a labeled 5′ cap) with the polymerase complexes in detergent-treated influenza virions (Plotch et al., 1981; Ulmanen et al., 1981). The 13-nt-long capped RNA fragment produced by the virion endonuclease was isolated by gel electrophoresis. Oligoribonucleotides containing the 5′ or 3′ terminal sequence of vRNA were synthesized using an oligonucleotide synthesizer, and were then labeled at their 5′ ends using polynucleotide kinase and [γ-32P]ATP (Li et al., 1998). In some experiments, the 5′ vRNA sequence was labeled at its 3′ end using 32pCp and RNA ligase.
UV cross-linking
A nuclear extract containing wild-type polymerase complexes (750 µg) was incubated for 25 min at 25°C with either the 32P-cap-labeled oligoribonucleotide containing thio U (80 ng, 1 × 107 c.p.m.) or the capped oligoribonucleotide containing thio U and labeled internal G residues (80 ng, 1 × 107 c.p.m.) under conditions described previously (Li et al., 1998). The reactions contained either 5′ vRNA alone (80 ng) or both 5′ and 3′ vRNA (each at 80 ng). After the binding reaction, the mixtures were put on ice and exposed to 366 nm UV light at a distance of 2 cm for 30 min (Favre et al., 1998; Li et al., 1998; Wang and Rana, 1998). The cross-linked protein–oligoribonucleotide complexes were separated from free oligoribonucleotide by electrophoresis on 7% denaturing gels. The gel slices containing the cross-linked complexes were dialysed overnight to achieve equilibration with the buffer used for V8 protease digestion. The proteins in the gel slices were then digested with this protease (0.3 µg) for 18 h at 25°C, and the gel slices were placed on the top of an 18% denaturing gel. After electrophoresis, the labeled band was cut out and blotted onto PVDF membranes. This material was microsequenced using automated Edman degradation by the Protein Chemistry Section of the HHMI Biopolymer Facility and W.M.Keck Foundation Biotechnology Resource Laboratory at Yale University.
Assays for the activities of viral polymerase complexes
Assays for the binding of labeled 5′ vRNA, 3′ vRNA and the capped RNA fragment (13 nt long) to the polymerase complex were carried out as described previously (Li et al., 1998). The assay for the cap-dependent endonuclease assay: following binding of 5′ and 3′ vRNA, full-length AlMV RNA 4 containing an m7G32pppGm 5′ end (0.25 ng, 1 × 105 c.p.m.) was added, along with 0.1 µg of transfer RNA as an additional inhibitor of non-specific nucleases, and the mixture was incubated for 60 min at 30°C (Plotch et al., 1981; Hagen et al., 1994; Cianci et al., 1995; Li et al., 1998). To assay for RNA chain elongation, three ribonucleoside triphosphates (ATP, CTP and GTP, each at 1 mM) were added during the cleavage reaction. For both the endonuclease and elongation assays, the RNA products were separated on a denaturing 20% polyacrylamide–7 M urea gel.
Acknowledgments
Acknowledgements
This investigation was supported by two grants to R.M.K.: AI11772 from the US National Institutes of Health and F-1468 from the Welch Foundation.
References
- Argos P. (1988) A conserved motif in many polymerases. Nucleic Acids Res., 16, 9909–9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan M. and Jonsson,C.B. (1997) Functional identification of nucleotides conferring substrate specificity to retroviral integrase reactions. J. Virol., 71, 1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaton A.R. and Krug,R.M. (1981) Selected host cell capped RNA fragments prime influenza viral RNA transcription in vivo. Nucleic Acids Res., 9, 4423–4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S.K. and Nayak,D.P. (1994) Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J. Virol., 68, 1819–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blass D., Patzelt,E. and Keuchler,E. (1982) Identification of the cap binding protein of influenza virus. Nucleic Acids Res., 10, 4803–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianci C., Tiley,L. and Krystal,M. (1995) Differential activation of the influenza virus polymerase via template RNA binding. J. Virol., 69, 3995–3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J.F.I., Hostomska,Z., Hostomsky,Z., Jordan,S.R. and Matthews,D.A. (1991) Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science, 252, 88–95. [DOI] [PubMed] [Google Scholar]
- Doan L., Handa,B., Roberts,N.A. and Klumpp,K. (1999) Metal ion catalysis of RNA cleavage by the influenza virus endonuclease. Biochemistry, 38, 5612–5619. [DOI] [PubMed] [Google Scholar]
- Dyda F., Hickman,A.B., Jenkins,T.M., Engelman,A., Craigie,R. and Davies,D.R. (1994) Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotide transferases. Science, 266, 1981–1986. [DOI] [PubMed] [Google Scholar]
- Elroy-Stein O. and Moss,B. (1996) Expression of proteins in mammalian cells using vaccinia viral vector. Curr. Protocols Mol. Biol., 16.15.11–16.19.19. [Google Scholar]
- Esposito D. and Craigie,R. (1998) Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein–DNA interaction. EMBO J., 17, 5832–5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre A., Saintome,C., Fourrey,J.-L., Clivio,P. and Laugaa,P. (1998) Thionucleobases as intrinsic photoaffinity probes of nucleic acid structure and nucleic acid–protein interactions. J. Photochem. Photobiol. B, 42, 109–124. [DOI] [PubMed] [Google Scholar]
- Fiering S., Northrop,J.P., Nolan,G.P., Mattila,P.S., Crabtree,G.R. and Herzenberg,L.A. (1990) Single cell assay of a transcription factor reveals a threshold in transcription activation by signals emanating from the T-cell antigen receptor. Genes Dev., 4, 1823–1834. [DOI] [PubMed] [Google Scholar]
- Gao H.-Q., Boyer,P.L., Arnold,E. and Hughes,S.H. (1998) Effects of mutations in the polymerase domain on the polymerase, RNase H and strand transfer activities of human immunodeficiency virus type 1 reverse transcriptase. J. Mol. Biol., 277, 559–572. [DOI] [PubMed] [Google Scholar]
- Goldgur Y., Dyda,F., Hickman,A.B., Jenkins,T.M., Graigie,R. and Davies,D.R. (1998) Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium. Proc. Natl Acad. Sci. USA, 95, 9150–9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen M., Chung,T.D.Y., Butcher,A. and Krystal,M. (1994) Recombinant influenza virus polymerase: requirement of both 5′ and 3′ viral ends for endonuclease activity. J. Virol., 68, 1509–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J.L., Long,A.M. and Schultz,S.C. (1997) Structure of the RNA-dependent RNA polymerase of poliovirus. Structure, 5, 1109–1122. [DOI] [PubMed] [Google Scholar]
- Hodel A.E., Gershon,P.D., Shi,X.N., Wang,S.M. and Quiocho,F.A. (1997) Specific protein recognition of an mRNA cap through its alkylated base. Nature Struct. Biol., 4, 350–354. [DOI] [PubMed] [Google Scholar]
- Honda A., Mizumoto,K. and Ishihama,A. (1999) Two separate sequences of PB2 subunit constitute the RNA cap-binding site of influenza virus RNA polymerase. Genes Cells, 4, 475–485. [DOI] [PubMed] [Google Scholar]
- Jacobo-Molina A. et al. (1993) Crystal structure of human immunodeficiency virus type-1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl Acad. Sci. USA, 90, 6320–6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce C.M. and Steitz,T.A. (1995) Polymerase structures and function: variations on a theme? J. Bacteriol., 177, 6321–6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya S., Kohara,A., Miura,Y., Sekiguchi,A., Iwai,S., Inoue,H., Ohtsuka,E. and Ikehara,M. (1990) Identification of the amino acid residues involved in an active site of Escherichia coli ribonuclease H by site-directed mutagenesis. J. Biol. Chem., 265, 4615–4621. [PubMed] [Google Scholar]
- Keck J.L., Goedken,E.R. and Marqusee,S. (1998) Activation/attenuation model for RNase H. J. Biol. Chem., 273, 34128–34133. [DOI] [PubMed] [Google Scholar]
- Kohlstaedt L.A., Wang,J., Friedman,J.M., Rice,P.A. and Steitz,T.A. (1992) Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science, 256, 1783–1790. [DOI] [PubMed] [Google Scholar]
- Krug R.M., Alonso-Caplen,F.V., Julkunen,I. and Katze,M. (1989) Expression and replication of the influenza virus genome. In Krug,R.M. (ed.), The Influenza Viruses. Plenum Press, New York and London, pp. 89–152.
- Lesburg C.A., Cable,M.B., Ferrari,E., Hong,Z., Mannarino,A.F. and Weber,P.C. (1999) Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nature Struct. Biol., 6, 937–943. [DOI] [PubMed] [Google Scholar]
- Li M.-L., Ramirez,C. and Krug,R.M. (1998) RNA-dependent activation of primer RNA production by the influenza virus polymerase: different regions of the same protein subunit constitute the two required RNA-binding sites. EMBO J., 17, 5844–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licheng S., Summers,D.F., Peng,Q. and Galarza,J.M. (1995) Influenza A virus polymerase subunit PB2 is the endonuclease which cleaves host cell mRNA and functions only as the trimeric enzyme. Virology, 208, 38–47. [DOI] [PubMed] [Google Scholar]
- Luo G.-X., Sharmeen,L. and Taylor,J. (1990) Specificities involved in the initiation of retroviral plus-strand DNA. J. Virol., 64, 592–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahillon J. and Chandler,M. (1998) Insertion sequences. Microbiol. Mol. Biol. Rev., 62, 725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J., Gingras,A.C., Sonenberg,N. and Burley,S.K. (1997) Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell, 89, 951–961. [DOI] [PubMed] [Google Scholar]
- Matsuo H., Li,H., McGuire,A.M., Fletcher,C.M., Gingras,A.C., Sonenberg,N. and Wagner,G. (1997) Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nature Struct. Biol., 4, 717–724. [DOI] [PubMed] [Google Scholar]
- Milligan J.F., Groebe,D.R., Witherell,G.W. and Uhlenbeck,O.C. (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res., 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K. (1992) Transpositional recombination: mechanistic insights from Mu and other elements. Annu. Rev. Biochem., 61, 1011–1051. [DOI] [PubMed] [Google Scholar]
- Morino S., Hazama,H., Ozaki,M., Teraoka,Y., Shibata,S., Doi,M., Ueda,H., Ishida,T. and Uesugi,S. (1996) Analysis of the mRNA cap-binding ability of human eukaryotic initiation factor-4E by use of recombinant wild-type and mutant forms. Eur. J. Biochem., 239, 597–601. [DOI] [PubMed] [Google Scholar]
- Plotch S.J., Bouloy,M., Ulmanen,I. and Krug,R.M. (1981) A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell, 23, 847–858. [DOI] [PubMed] [Google Scholar]
- Poch O., Sauvaget,I., Delarue,M. and Tordo,N. (1989) Identification of four conserved motifs among the RNA-dependent polymerase encoding element. EMBO J., 8, 3867–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph C. and Champoux,J. (1994) The use of DNA and RNA oligonucleotides in hybrid structures with longer polynucleotide chains to probe the structural requirements for Moloney murine leukemia virus plus strand priming. J. Biol. Chem., 269, 19207–19215. [PubMed] [Google Scholar]
- Rice P., Craigie,R. and Davies,D.R. (1996) Retroviral integrases and their cousins. Curr. Opin. Struct. Biol., 6, 76–83. [DOI] [PubMed] [Google Scholar]
- Shaw M.W. and Lamb,R.A. (1984) A specific sub-set of host-cell mRNAs prime influenza virus mRNA synthesis. Virus Res., 1, 455–467. [DOI] [PubMed] [Google Scholar]
- Shih S.R. and Krug,R.M. (1996) Surprising function of the three influenza viral polymerase proteins: selective protection of viral mRNAs against the cap-snatching reaction catalyzed by the same polymerase proteins. Virology, 226, 430–435. [DOI] [PubMed] [Google Scholar]
- Smith G., Levin,J.Z., Palese,P. and Moss,B. (1987) Synthesis and cellular location of the ten influenza polypeptides individually expressed by recombinant vaccinia viruses. Virology, 160, 336–345. [DOI] [PubMed] [Google Scholar]
- Suo Z. and Johnson,K.A. (1997) Effect of RNA secondary structure on RNA cleavage catalyzed by HIV-1 reverse transcriptase. Biochemistry, 36, 12468–12476. [DOI] [PubMed] [Google Scholar]
- Ulmanen I., Broni,B.A. and Krug,R.M. (1981) The role of two of the influenza virus core P proteins in recognizing cap 1 structures (m7GpppNm) on RNAs and in initiating viral RNA transcription. Proc. Natl Acad. Sci. USA, 78, 7355–7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink C., van Gent,D.C., Elgersma,Y. and Plasterk,R.H. (1991) Human immunodeficiency virus integrase protein requires a subterminal position of its viral DNA recognition sequence for efficient cleavage. J. Virol., 65, 4636–4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N., Izaurralde,E., Ferreira,J., Daneholt,B. and Mattaj,I.W. (1996) A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particles during nuclear export. J. Cell Biol., 133, 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. and Rana,T.M. (1998) RNA–protein interactions in the Tat-trans-activation response element complex determined by site-specific photo-cross-linking. Biochemistry, 37, 4235–4243. [DOI] [PubMed] [Google Scholar]
- Wohrl B.M. and Moelling,K.M. (1990) Interactions of HIV-1 ribonuclease H with polypurine tract containing RNA–DNA hybrids. Biochemistry, 29, 10141–10147. [DOI] [PubMed] [Google Scholar]
- Yang W. and Steitz,T.A. (1995) Recombining the structures of HIV integrase, RuvC and RNase H. Structure, 3, 131–134. [DOI] [PubMed] [Google Scholar]