Abstract
We recently described an RNA-binding protein, Y14, that binds preferentially to spliced mRNAs and persists in the cytoplasm. Y14 is part of a multi-protein complex that also contains the mRNA export factor TAP. This suggests that splicing imprints the mRNA with a unique set of proteins that communicate the history of the transcript to the cytoplasm. Here, using microinjection of pre-mRNAs into Xenopus oocyte nuclei followed by immunoprecipitation of RNase-fragmented mRNAs from the cytoplasm, we show that Y14 is stably bound to sequences immediately upstream of exon–exon junctions. This feature appears to be unique to Y14. Using monoclonal antibodies that we produced against Aly/REF, another component recently reported to be an mRNA export factor, we show that Aly/REF is associated with spliced mRNAs in the nucleus but is not detectable on mRNAs in the cytoplasm. Thus, we propose that the splicing- dependent binding of Y14 provides a position-specific molecular memory that communicates to the cytoplasm the location of exon and intron boundaries. This novel mechanism may play an important role in post-splicing events.
Keywords: Aly/mRNA/nuclear transport/splicing/Y14
Introduction
Numerous observations have documented a communication between the nucleus and cytoplasm in various aspects of gene expression, and pre-mRNA splicing appears to play an important role in this interconnection (reviewed in Shyu and Wilkinson, 2000). For example, the presence of introns enhances translation of the spliced mRNAs in some genes (Matsumoto et al., 1998). It has also been shown that splicing can generate specific mRNA–protein (mRNP) complexes that are more competent for nuclear export (Luo and Reed, 1999). Additional compelling evidence was provided by nonsense-mediated decay (NMD), the process of degradation of aberrant mRNAs that contain premature termination codons (reviewed in Li and Wilkinson, 1998; Nagy and Maquat, 1998; Culbertson, 1999; Czaplinski et al., 1999; Hentze and Kulozik, 1999; Hilleren and Parker, 1999). In mammalian systems, the position of nonsense codons relative to the last exon–exon junction is critically important (Cheng et al., 1990). If translation terminates before ∼50–55 nt upstream of the last exon–exon junction, the mRNA is subject to decay (Cheng et al., 1994), suggesting that there is a marker or a tag on the last exon–exon junction that defines a boundary of recognition and allows discrimination between premature and normal termination codons. Therefore, it was suggested that the mark is endowed by splicing in the nucleus and accompanies mRNA to the cytoplasm. Although this was an attractive hypothesis, the mechanism of marking and the identity of the marker(s) on exon–exon junctions were yet to be determined.
It has been shown by several laboratories that splicing can serve not only to remove introns, but also to change the composition of mRNP complex (Luo and Reed, 1999; Kataoka et al., 2000; Le Hir et al., 2000b). Specific proteins have been found on spliced mRNAs, including RNPS1 (Mayeda et al., 1999), SRm160 (Blencowe et al., 1998; Le Hir et al., 2000b), DEK (McGarvey et al., 2000), Aly/REF (Zhou et al., 2000) and Y14 (Kataoka et al., 2000). Among these, Aly/REF and Y14 have been shown to shuttle between the nucleus and the cytoplasm (Kataoka et al., 2000; Zhou et al., 2000). Aly/REF appears to play a role in mRNA export as it interacted with TAP (Stutz et al., 2000) and enhanced export of spliced mRNA when the recombinant protein was injected into the oocyte (Zhou et al., 2000). The recently described Y14 is a promising candidate for a factor to communicate the nuclear history of mRNA to the cytoplasm. Y14 binds efficiently to mRNAs produced by splicing both in vitro and in vivo. Notably, Y14 has been shown to remain associated with newly exported mRNAs in the cytoplasm.
Because Y14 binds to different mRNAs that have no common nucleotide sequence, it is possible that its binding to mRNA, which is spliceosome dependent, is either random or position specific. In order to address this question we wished to map the position of Y14 on newly exported mRNAs in the cytoplasm by using oocyte microinjections and a monoclonal antibody against Y14 (4C4). Our studies show that Y14 is located on mRNAs in the cytoplasm approximately –20 nt upstream of exon–exon junctions. Furthermore, we demonstrate using monoclonal antibodies that we produced against Aly/REF (11G5) that unlike Y14, Aly/REF does not persist with spliced mRNA in the cytoplasm.
Thus, our results demonstrate that some of the proteins that bind the mRNA in a splicing-dependent manner are maintained at the same position after export to the cytoplasm and, importantly, that the composition of the complex changes during this process.
Results
Experimental approach to map the location of Y14 on spliced mRNAs
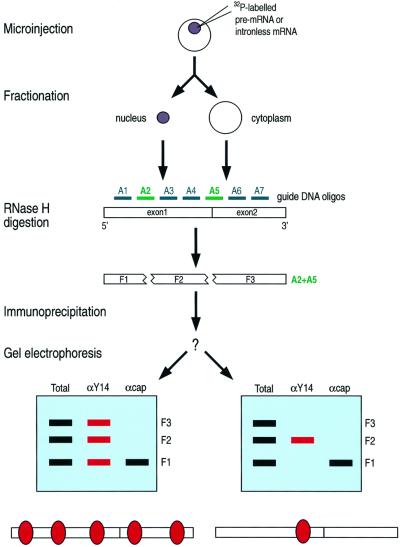
We first microinjected the radiolabeled pre-mRNA into the nuclei of oocytes, incubated the oocytes to allow splicing and export to occur, and prepared the nuclear and cytoplasmic fractions from the oocytes (Figure 1). Antisense deoxyoligonucleotides corresponding to specific mRNA sequences were added to these fractions, together with RNase H, in order to cleave the mRNA in a sequence-specific manner. We then carried out immunoprecipitation with anti-Y14 antibodies (4C4) (Kataoka et al., 2000), anti-cap antibodies (H20; to define the 5′ fragment) (Bochnig et al., 1987) and control non-immune antibodies (SP2/0). The total RNA fragments and those that co-immunoprecipitated with each of these antibodies were extracted and resolved by PAGE. If Y14 preferentially binds to a certain region, anti-Y14 antibody should immunoprecipitate the fragment that contains it. The experimental design and the two possible outcomes are shown in Figure 1.
Fig. 1. Experimental strategy to locate Y14 on mRNA in vivo. Random binding of Y14 throughout the mRNA would result in non-specific precipitation of all the fragments (bottom, left), whereas specific binding would bring about precipitation of specific fragments (bottom, right).
Preliminary experiments without immunoprecipitation were first carried out to determine which oligonucleotides are suitable for this mapping procedure, namely whether there are any regions of the mRNAs used that are not accessible to the oligonucleotides. The fragmentation pattern obtained from chicken δ-crystallin (CDC) (Kataoka et al., 2000), immunoglobulin (IgM) (Watakabe et al., 1989) and adenovirus MLP ΔIVS (Ad2) mRNA (Kataoka et al., 2000) demonstrated that most parts of these mRNAs are accessible to the oligonucleotides, except for a region corresponding to about –34 to –18 nt upstream of the exon–exon junction (data not shown). Interestingly this region was not protected when the mRNA was produced from cDNA (data not shown). This experiment showed that the oligonucleotide-guided RNase H cleavage strategy is feasible for in vivo spliced and exported mRNAs. It also suggested that splicing leads to changes in the region immediately upstream of exon–exon junction either by modifying the structure of the mRNA or by binding of factors (protein and/or RNAs) that hinder the hybridization of the oligonucleotides.
Y14 is bound immediately upstream of exon–exon junctions
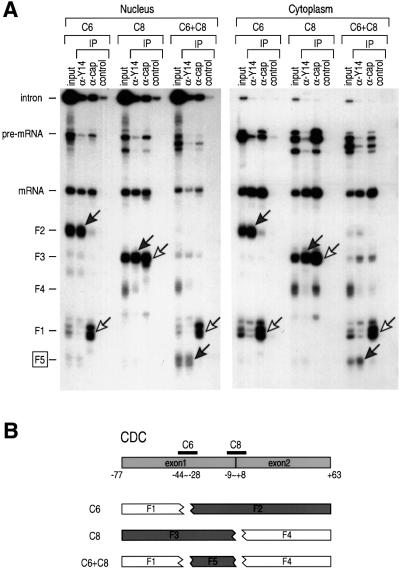
Based on the results of the experiments described above, we chose several oligonucleotides, singly or in combinations, to cleave the CDC mRNAs. The results of nuclear microinjection of the CDC pre-mRNA followed by immunoprecipitation with anti-Y14 antibody from the nucleus and the cytoplasm are shown in Figure 2A. Cleavage of the CDC with two oligonucleotides, C6 and C8, singly or together generates several fragments as shown in Figure 2B. Anti-Y14 antibody immunoprecipitates the fragments designated as F2, F3 and F5. The middle fragment, F5 (approximately –33 to –4 relative to the exon–exon junction, centered at approximately –18.5), contains the sequence that overlaps in F2 and F3, and therefore represents the minimum sequence to which Y14 is bound. Fragments F1 and F4 were not immunoprecipitated with anti-Y14 antibody. In contrast, the anti-cap antibody immunoprecipitates F1 and F3, confirming the identification of these fragments. This demonstrates that Y14 is specifically associated with sequences immediately upstream of the exon–exon junction.
Fig. 2. Y14 associates with sequences immediately upstream of the exon–exon junction. Immunoprecipitations (IP) and RNA analysis were performed following RNase H digestion with the indicated oligonucleotides. (A) 32P-labeled CDC pre-mRNA was injected in oocyte nuclei. The cytoplasmic fraction was recovered after 60 min and subjected to RNase H digestion with 7 µM oligonucleotides (C6 alone, C8 alone or C6 in combination with C8) for 25 min at 30°C. Immuno precipitations were carried out with anti-Y14 (4C4), anti-cap (H20) or control non-immune antibody (SP2/0). RNAs were analyzed on a 9% denaturing polyacrylamide gel. Gray arrows indicate the fragments bound to Y14 and white arrows indicate the 5′ fragments. (B) Positions of the oligonucleotides and the RNA fragments from the spliced CDC mRNA. Gray boxes indicate the fragments bound to Y14.
In order to determine whether the positioning of Y14 upstream of exon–exon junctions is a general phenomenon, rather than unique in the case of CDC mRNA, we carried out similar experiments with the IgM mRNA. Figure 3A shows the results of immunoprecipitation of fragments generated from the IgM mRNA from the cytoplasm. Figure 3B indicates the positions of the oligonucleotides, M2, M5 and M6, that were used in these experiments. The anti-Y14 antibody immunoprecipitates fragments F2, F4 and the partially digested fragments that contain them (F1∼F2 and F1∼F4). F4, which corresponds to sequences approximately –43 to –5 relative to the exon–exon junction, is the smallest fragment (∼38 nt) that immunoprecipitates with anti-Y14 antibodies (centered at approximately –24). In contrast, the anti-cap antibody immunoprecipitates only the F1 fragment, confirming its identification as the one derived from the 5′ terminus of the mRNA.
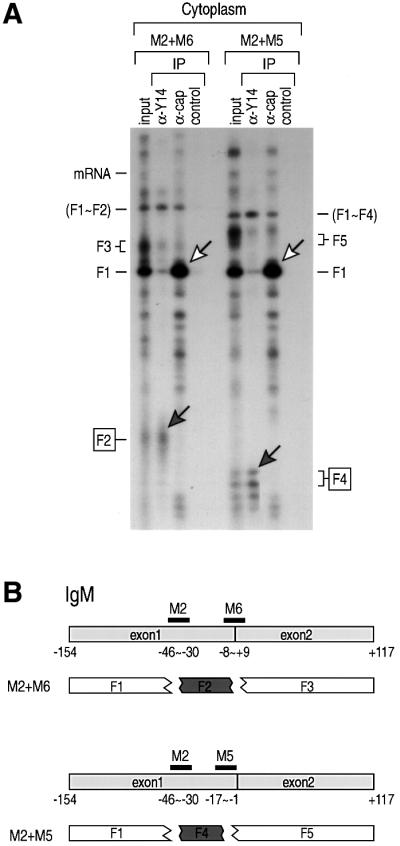
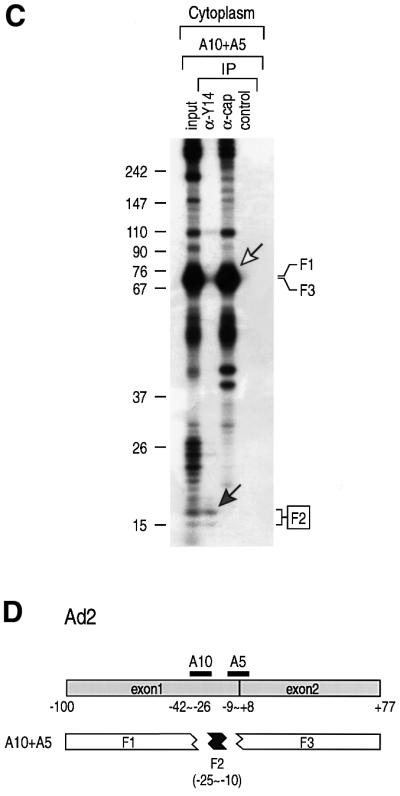
Fig. 3. Association of Y14 on spliced mRNA is position specific. Immunoprecipitations (IP) and RNA analysis were performed following RNase H digestion with the indicated oligonucleotides. (A) 32P-labeled IgM pre-mRNA was injected in oocyte nuclei. The cytoplasmic fraction was recovered after 90 min and subjected to RNase H digestion with 7 µM oligonucleotides (M2 together with M6, or M2 in combination with M5) for 25 min at 30°C. Immunoprecipitations were carried out with anti-Y14 (4C4), anti-cap (H20), or control non-immune antibody (SP2/0). RNAs were analyzed on a 9% denaturing polyacrylamide gel. Gray arrows indicate the fragments bound to Y14 and white arrows indicate the 5′ fragments. (B) Positions of the oligonucleotides and the RNA fragments from the spliced IgM mRNA. Gray boxes indicate the fragments bound to Y14. (C) Ad2 pre-mRNA was used in an identical experiment to that in (A) except that oligonucleotides A10 and A2 were used for digestion. (D) Positions of the oligonucleotides and the RNA fragments from the spliced Ad2 mRNA.
Another pre-mRNA, Ad2, was used to verify the position specificity of binding as well as to define better the minimal binding site of Y14. Different sets of oligonucleotides were tested to generate the smallest fragment that can be immunoprecipitated with anti-Y14 antibody. Among these, A10 and A5 produced an ∼16 nt fragment encompassing –25 to –10 relative to the exon–exon junction (centered at approximately –17.5) (Figure 3), which was specifically immunoprecipitated by anti-Y14 antibody. Putting these results together (Figures 2 and 3), the average center of the Y14 binding region is estimated to be at 20 nt upstream of the exon–exon junction.
The Y14 protein remains bound at the same position after mRNA export
Interestingly, the immunoprecipitation patterns from the nuclear and cytoplasmic fractions (Figure 2A) are essentially identical. This indicates that the splicing-dependent Y14 binding that is established in the nucleus persists in the cytoplasm. The RNase H cleavage patterns from the nucleus and the cytoplasm were also identical (data not shown), supporting this conclusion. The notion that Y14 persists in the cytoplasm raises intriguing questions about what kind of alterations, if any, are being made on exon–exon junctions during the process of mRNA export, and which other factor(s) remains on mRNAs to modulate the downstream steps in mRNA metabolism.
Aly/REF, unlike Y14, does not persist on cytoplasmic mRNAs
Among the proteins that are enriched in spliced mRNAs, both Y14 and Aly/REF have been indicated to be nucleo-cytoplasmic shuttling proteins (Kataoka et al., 2000; Zhou et al., 2000). In fact, Y14 and Aly/REF have several common features; both are small RNA-binding proteins with a single RNP motif in the middle of the proteins, and they localize mainly in nuclear speckles and the nucleoplasm. To study Aly/REF further, we produced monoclonal antibodies against human Aly/REF, because efficient and specific antibodies to this protein were not available. One antibody, designated 11G5, immunoprecipitated human Aly/REF specifically (data not shown) and recognized a single band from HeLa cells in western blot analysis (Figure 4). Immunofluorescence microscopy showed bright staining of nuclear speckles and the nucleoplasm in HeLa cells (data not shown), a pattern consistent with a previous report (Zhou et al., 2000) and similar to that of Y14 (Kataoka et al., 2000). Western blotting on the nucleus and the cytoplasm of Xenopus laevis oocytes showed a single band of identical size mobility to that of human Aly/REF, indicating that 11G5 also recognizes X.laevis Aly/REF (Figure 4).
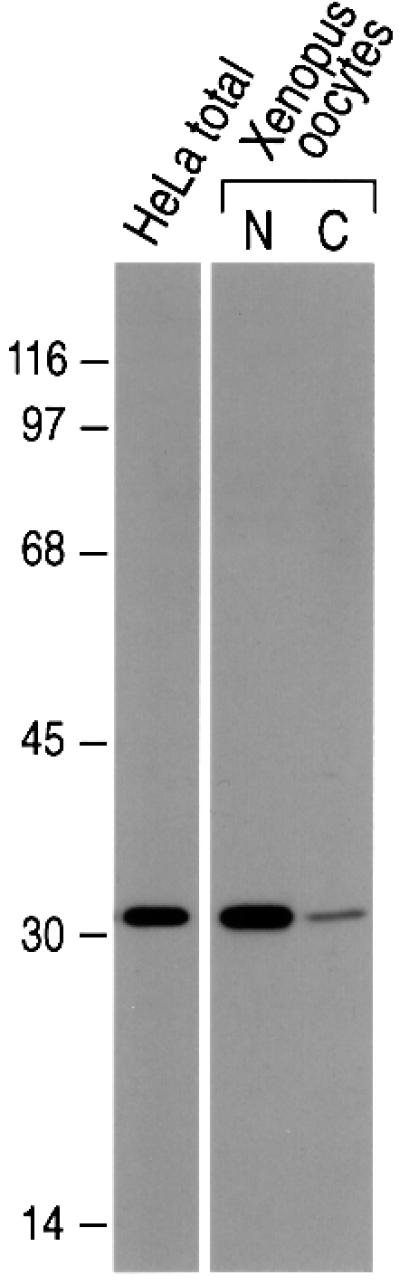
Fig. 4. Characterization of anti-Aly/REF monoclonal antibody, 11G5. Western blot analysis on total extract of HeLa cells, and the nuclear (N) or cytoplasmic (C) fractions of Xenopus oocytes. To load equal amounts of proteins in each well, the nuclear fraction from 10 oocytes and the cytoplasmic fraction from two oocytes were separated on 12.5% SDS–PAGE.
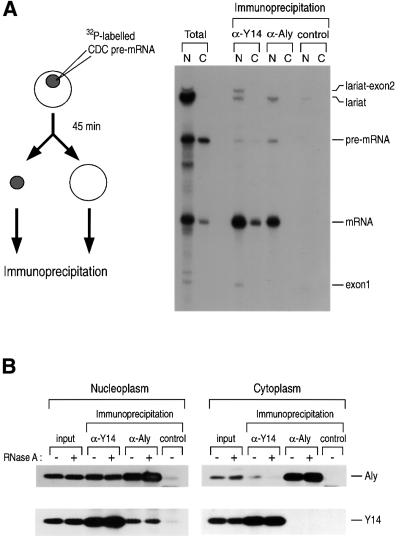
In order to compare the RNA-binding patterns of Y14 and Aly/REF, CDC pre-mRNA was injected into the nuclei of Xenopus oocytes. Following incubation for 45 min, immunoprecipitations from the nuclear and cytoplasmic fractions were carried out with anti-Y14 antibody (4C4), anti-Aly antibody (11G5) and control antibody (SP2/0) (Figure 5A). In the nucleus, both Y14 and Aly/REF are associated preferentially with spliced mRNA. Similar patterns were observed from in vitro splicing experiments using HeLa nuclear extract (data not shown).
Fig. 5. Y14 constitutes a cytoplasmic mRNP complex while Aly/REF dissociates during or immediately after export. (A) Immunoprecipi tations and RNA analysis were performed following nuclear injection of 32P-labeled Ad2 pre-mRNA. Oocytes were incubated for 45 min before dissection. Anti-Y14 (4C4), anti-Aly (11G5) and control antibodies (SP2/0) were used for immunoprecipitation. (B) Immunoprecipitations and western blotting were carried out on the nucleoplasm and cytoplasm of HeLa cells. The fractions were pretreated with 10 µg/ml RNase A for 20 min before immunoprecipitation. Anti-Y14 (4C4), anti-Aly (11G5) and control non-immune antibodies (SP2/0) were used for immunoprecipitations. For western blotting, anti-Aly (11G5) and anti-Y14 antibodies (1F12) were used.
Importantly, antibodies to Aly/REF, a reportedly shuttling protein, did not co-immunoprecipitate mRNA from the cytoplasm (Figure 5A). This indicates that Aly/REF dissociates from the mRNA immediately prior to, during or immediately after export, whereas Y14 stays on the mRNA. The same result was obtained with anti-Aly/REF polyclonal serum, making it unlikely that this is simply the result of epitope inaccessibility in the cytoplasm (data not shown). Therefore, the mechanism and timing of dissociation of Aly/REF from the mRNA must be different from that of Y14.
Next, we examined whether Y14 and Aly/REF are present on the same mRNP complexes, rather than being on separate mRNA molecules, by performing immunoprecipitations followed by western blotting (Figure 5B). Y14 co-immunoprecipitates with Aly/REF from the nucleus, demonstrating that they are present in the same complexes (Figure 5B, left panel). This complex is resistant to RNase A treatment, suggesting that Y14 and Aly/REF bind to each other through protein–protein interaction, although it is not yet clear whether they interact directly. This complex, however, does not persist in the cytoplasm as the same proteins do not co-immunoprecipitate from the cytoplasmic fraction (Figure 5B, right panel), although we can not exclude the possibility that a small fraction of Aly/REF is still on the Y14–mRNA complex. These results show that both Y14 and Aly/REF associate with spliced mRNA in the nucleus but that only Y14 persists on mRNAs in the cytoplasm. Thus, Y14, unlike Aly/REF, can serve to provide the splicing-generated positional information that defines the location of exon–exon junctions on mRNAs in the cytoplasm.
Discussion
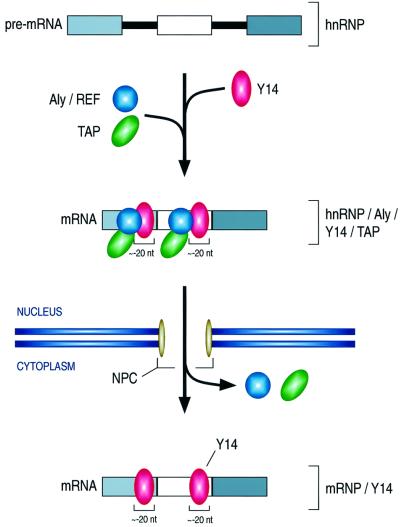
From these experiments we conclude that Y14, which is recruited to the mRNAs by the spliceosome, is associated with the mRNAs in a position-specific manner such that it is bound to sequences immediately upstream of the exon–exon junctions (Figure 6). This association, which persists on newly exported mRNAs in the oocyte cytoplasm, can therefore serve to communicate to the cytoplasm the position of the introns, even though they had been removed earlier in the nucleus. Thus, Y14 binding essentially documents and retains the structure of the gene from which the mRNA was transcribed. How long this ‘scar’ persists in the cytoplasm and what removes Y14 from the mRNA so that it can shuttle back to the nucleus is not yet known.
Fig. 6. Schematic diagram of the formation and transport of the Y14 complex. See text for details.
What might the function of Y14 binding near exon– exon junctions be? In the nucleus, Y14 binding may have a role in mRNA export along with Aly/REF, as an indicator that splicing has taken place and as a means of recruiting export factors such as TAP (Izaurralde and Adam, 1998; Kataoka et al., 2000) (Figure 6). The binding of Y14 near exon–exon junctions may also be a way of marking that region as one that has already been worked on by the splicing machinery and thereby preventing any further unnecessary processing of the RNA. In the cytoplasm, several possible functions for Y14 can be considered, including a role in translation, mRNA turnover, transport and localization in the cytoplasm. We find the possibility of a role for the position-specific Y14 binding in NMD particularly intriguing.
In higher eukaryotes, the termination codon is generally found in the last exon of the mRNA. Mutations that create premature termination codons are potentially deleterious as they can lead to the formation of C-terminal truncated proteins, which often results in severe human diseases (Frischmeyer and Dietz, 1999). Premature termination codons trigger a process of NMD that is designed to destroy the mRNA and thus prevent the accumulation of mutant proteins (Li and Wilkinson, 1998; Nagy and Maquat, 1998; Culbertson, 1999; Czaplinski et al., 1999; Hentze and Kulozik, 1999; Hilleren and Parker, 1999). Previous studies have demonstrated a link between splicing and NMD (Zhang et al., 1998) and showed the requirement for translation (Muhlrad and Parker, 1994; Zhang et al., 1997; Thermann et al., 1998). Our findings suggest that splicing imprints (or marks) exon–exon junctions by the binding of the Y14 complex. The Y14 complex, however, appears not to be directly on the junction itself but immediately upstream of it, in a region that we could only delineate to within approximately –25 to –10 nt 5′ to the exon–exon junction (the average center of the binding estimated from three different mRNAs is at –20 nt). By virtue of its consistent position, strict dependence on splicing and persistence on newly exported mRNAs in the cytoplasm, we suggest that Y14 has the necessary attributes to serve as the marker that signals premature termination codons and can thus trigger NMD. According to this model, translating ribosomes (or factors closely associated with the translation machinery) displace Y14 complexes from the mRNA as part of the first round of translation in the cytoplasm. Thus, in the course of translation of a normal mRNA, all of its Y14 complexes will be removed by the time the first translating ribosome reaches the termination codon. If, however, a nonsense mutation occurs more than ∼50 nt upstream of the last exon–exon junction, the ribosomes will dissociate from the mRNA as they reach that premature termination codon and fail to displace the remaining Y14 complex(es). This remaining Y14 complex, perhaps together with a component of the termination complex, may serve as the trigger of NMD by recruiting the factors that degrade this mRNA (Czaplinski et al., 1998).
Over the past several years, evidence has accumulated that shuttling RNA-binding proteins interconnect the nuclear and cytoplasmic aspects of the life cycle of mRNA (Pinol-Roma and Dreyfuss, 1992; Shyu and Wilkinson, 2000). The observation of position-specific Y14 binding on the cytoplasmic mRNA provides an interesting example of a novel mode of regulation that can serve as molecular memory, and of the transmission of information from one cellular compartment to another. While this manuscript was in the final stage of preparation, a report by Le Hir et al. was published describing several complementary observations to those we report here (Le Hir et al., 2000a). Using the 4C4 antibody as well as several additional antibodies, these authors showed that in vitro spliced pre-mRNAs give rise to mRNAs that contain a complex of proteins, including Y14 and Aly/REF, which assembles immediately upstream of exon–exon junctions (–24/20 nt). They showed further that nuclear mRNA, produced by splicing of nucleus-injected pre-mRNA, also contains a complex at the same position, although the composition of this in vivo generated nuclear complex was not investigated in that study. Our findings are consistent with these observations but extend them in several significant ways. Importantly, the splicing-dependent binding of Y14 upstream of exon–exon junctions persists in the cytoplasm, suggesting that this protein can serve to communicate the position of exon–exon junctions to the cytoplasm. In contrast, Aly/REF is not found on the cytoplasmic mRNAs and thus is not likely to serve such a role. Current observations on Aly/REF suggest that it plays a role in mRNA export only. The property of Y14 makes it the most promising candidate for the hypothesized ‘marker’ on exon–exon junctions.
Materials and methods
In vitro transcription
To prepare templates for in vitro transcription, pCDC (previously referred to as pSP14–15), pIgM (previously named as pµC3-C4) and pAd2 were linearized with SmaI (Kataoka et al., 2000), HindIII (Watakabe et al., 1989) or XbaI, respectively. Preparation of RNAs was carried out as described previously (Kataoka et al., 2000).
Xenopus oocyte microinjections
Injections were carried out as described previously (Kataoka et al., 2000). Briefly, in vitro transcribed RNAs were injected into the nuclei of Xenopus oocytes. After a 45–90 min incubation (see figure legends), oocytes were manually dissected to the nucleus and cytoplasm. Each fraction was homogenized in RSB-100, centrifuged and the supernatants were used for RNase H digestion.
RNase H digestion and gel electrophoresis
Nuclear or cytoplasmic fractions were treated with 1 U of RNase H (Promega) and 5–7 µM oligonucleotide in a total 50 µl reaction for 20–25 min (see figure legends) at 30°C. Oligonucleotides for CDC RNAs are C6 (5′-AACCTGGAGCACAGCAG-3′) and C8 (5′-TTGTTGACCTGGAGGGT-3′). Oligonucleotides used for digestion of IgM RNAs are M2 (5′-CCTGTGAGTCACAGTAC-3′), M5 (5′-CATTGGGTTTTGAGATG-3′) and M6 (5′-GTGCACCTCATTGGGTT-3′). For digestion of Ad2 mRNA, A10 (5′-CGATGCGGAAGAGATGA-3′) and A2 (5′-GGCGGCCGTTACTAGTG-3′) were used.
Antibodies, immunoprecipitations and western blot analyses
Anti-Y14 antibody, 4C4, was described elsewhere (Kataoka et al., 2000). Additional anti-Y14 antibody, 1F12, was used for western blotting. Anti-Aly/REF monoclonal antibody, 11G5, was generated against recombinant human Aly/REF protein fused to glutathione S-transferase. The anti-mono-methyl guanosine cap antibody, H20 (Bochnig et al., 1987), was a generous gift from Dr R.Lührmann. Following RNase H digestion, nucleoplasmic or cytoplasmic fractions were incubated for 30 min with the antibodies immobilized on protein A–Sepharose CL-4B (Amersham Pharmacia). After thorough washing, RNAs were extracted with phenol and analyzed on denaturing 9% polyacrylamide gels. Subcellular fractionation of HeLa cells, RNase A treatment and western blot analyses were carried out as described previously (Siomi et al., 1997).
Acknowledgments
Acknowledgements
We are grateful to members of our laboratory especially to Drs Westley Friesen, Livio Pellizzoni and Josée Dostie, for critical reading of and comments on this manuscript, and to Sue Kelchner for secretarial assistance. We also thank Dr Reinhard Lührmann for the H20 antibody and Dr Peter Klein for generously providing Xenopus laevis. This work was supported by grants from the National Institutes of Health and the Human Frontier of Science Program Organization. G.D. is an Investigator of the Howard Hughes Medical Institute.
References
- Blencowe B.J., Issner,R., Nickerson,J.A. and Sharp,P.A. (1998) A coactivator of pre-mRNA splicing. Genes Dev., 12, 996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochnig P., Reuter,R., Bringmann,P. and Luhrmann,R. (1987) A monoclonal antibody against 2,2,7-trimethylguanosine that reacts with intact, class U, small nuclear ribonucleoproteins as well as with 7-methylguanosine-capped RNAs. Eur. J. Biochem., 168, 461–467. [DOI] [PubMed] [Google Scholar]
- Cheng J., Fogel-Petrovic,M. and Maquat,L.E. (1990) Translation to near the distal end of the penultimate exon is required for normal levels of spliced triosephosphate isomerase mRNA. Mol. Cell. Biol., 10, 5215–5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Belgrader,P., Zhou,X. and Maquat,L.E. (1994) Introns are cis effectors of the nonsense-codon-mediated reduction in nuclear mRNA abundance. Mol. Cell. Biol., 14, 6317–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson M.R. (1999) RNA surveillance. Unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet., 15, 74–80. [DOI] [PubMed] [Google Scholar]
- Czaplinski K., Ruiz-Echevarria,M.J., Paushkin,S.V., Han,X., Weng,Y., Perlick,H.A., Dietz,H.C., Ter-Avanesyan,M.D. and Peltz,S.W. (1998) The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev., 12, 1665–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K., Ruiz-Echevarria,M.J., Gonzalez,C.I. and Peltz,S.W. (1999) Should we kill the messenger? The role of the surveillance complex in translation termination and mRNA turnover. BioEssays, 21, 685–696. [DOI] [PubMed] [Google Scholar]
- Frischmeyer P.A. and Dietz,H.C. (1999) Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet., 8, 1893–1900. [DOI] [PubMed] [Google Scholar]
- Hentze M.W. and Kulozik,A.E. (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell, 96, 307–310. [DOI] [PubMed] [Google Scholar]
- Hilleren P. and Parker,R. (1999) mRNA surveillance in eukaryotes: kinetic proofreading of proper translation termination as assessed by mRNP domain organization? RNA, 5, 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E. and Adam,S. (1998) Transport of macromolecules between the nucleus and the cytoplasm. RNA, 4, 351–364. [PMC free article] [PubMed] [Google Scholar]
- Kataoka N., Yong,J., Kim,V.N., Velazquez,F., Perkinson,R.A., Wang,F. and Dreyfuss,G. (2000) Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell, 6, 673–682. [DOI] [PubMed] [Google Scholar]
- Le Hir H., Izaurralde,E., Maquat,L.E. and Moore,M.J. (2000a) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J., 19, 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Moore,M.J. and Maquat,L.E. (2000b) Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon–exon junctions. Genes Dev., 14, 1098–1108. [PMC free article] [PubMed] [Google Scholar]
- Li S. and Wilkinson,M.F. (1998) Nonsense surveillance in lymphocytes? Immunity, 8, 135–141. [DOI] [PubMed] [Google Scholar]
- Luo M.J. and Reed,R. (1999) Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl Acad. Sci. USA, 96, 14937–14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Wassarman,K.M. and Wolffe,A.P. (1998) Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J., 17, 2107–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A., Badolato,J., Kobayashi,R., Zhang,M.Q., Gardiner,E.M. and Krainer,A.R. (1999) Purification and characterization of human RNPS1: a general activator of pre-mRNA splicing. EMBO J., 18, 4560–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey T. et al. (2000) The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon–product complexes. J. Cell Biol., 150, 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D. and Parker,R. (1994) Premature translational termination triggers mRNA decapping. Nature, 370, 578–581. [DOI] [PubMed] [Google Scholar]
- Nagy E. and Maquat,L.E. (1998) A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci., 23, 198–199. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S. and Dreyfuss,G. (1992) Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature, 355, 730–732. [DOI] [PubMed] [Google Scholar]
- Shyu A.B. and Wilkinson,M.F. (2000) The double lives of shuttling mRNA binding proteins. Cell, 102, 135–138. [DOI] [PubMed] [Google Scholar]
- Siomi M.C., Eder,P.S., Kataoka,N., Wan,L., Liu,Q. and Dreyfuss,G. (1997) Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J. Cell Biol., 138, 1181–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F., Bachi,A., Doerks,T., Braun,I.C., Seraphin,B., Wilm,M., Bork,P. and Izaurralde,E. (2000) REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA, 6, 638–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermann R., Neu-Yilik,G., Deters,A., Frede,U., Wehr,K., Hagemeier,C., Hentze,M.W. and Kulozik,A.E. (1998) Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J., 17, 3484–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watakabe A., Inoue,K., Sakamoto,H. and Shimura,Y. (1989) A secondary structure at the 3′ splice site affects the in vitro splicing reaction of mouse immunoglobulin µ chain pre-mRNAs. Nucleic Acids Res., 17, 8159–8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Sun,X., Qian,Y., LaDuca,J.P. and Maquat,L.E. (1998) At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol. Cell. Biol., 18, 5272–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Welch,E.M., Hogan,K., Brown,A.H., Peltz,S.W. and Jacobson,A. (1997) Polysome-associated mRNAs are substrates for the nonsense-mediated mRNA decay pathway in Saccharomyces cerevisiae. RNA, 3, 234–244. [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Luo,M.J., Straesser,K., Katahira,J., Hurt,E. and Reed,R. (2000) The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature, 407, 401–405. [DOI] [PubMed] [Google Scholar]