Abstract
Kluyveromyces lactis killer strains secrete a zymocin complex that inhibits proliferation of sensitive yeast genera including Saccharomyces cerevisiae. In search of the putative toxin target (TOT), we used mTn3:: tagging to isolate zymocin-resistant tot mutants from budding yeast. Of these we identified the TOT1, TOT2 and TOT3 genes (isoallelic with ELP1, ELP2 and ELP3, respectively) coding for the histone acetyltransferase (HAT)-associated Elongator complex of RNA polymerase II holoenzyme. Other than the typical elp ts-phenotype, tot phenocopies hypersensitivity towards caffeine and Calcofluor White as well as slow growth and a G1 cell cycle delay. In addition, TOT4 and TOT5 (isoallelic with KTI12 and IKI1, respectively) code for components that associate with Elongator. Intriguingly, strains lacking non-Elongator HATs (gcn5Δ, hat1Δ, hpa3Δ and sas3Δ) or non-Elongator transcription elongation factors TFIIS (dst1Δ) and Spt4p (spt4Δ) cannot confer resistance towards the K.lactis zymocin, thus providing evidence that Elongator equals TOT and that Elongator plays an important role in signalling toxicity of the K.lactis zymocin.
Keywords: Elongator/killer yeast/TOT/zymocin
Introduction
In competing for limited resources, microorganisms have evolved sophisticated strategies to gain selective advantages over their competitors. One of these is the secretion of toxic compounds that results in killing or growth arrest of other species or genera. Well studied cases are the dsRNA-encoded viral KT28 and K1 killer toxins of Saccharomyces cerevisiae (Wickner, 1996), which cause sensitive yeast cells to irreversibly block DNA synthesis and arrest in S phase (Schmitt et al., 1996) or to destroy cytoplasmic membrane function by TOK1 hyperactivation and lethal ion channel formation (Ahmed et al., 1999). Killer dsDNA plasmid-carrying strains of the dairy yeast Kluyveromyces lactis also secrete a zymo-toxin (referred to as zymocin) that inhibits cellular growth of various sensitive yeast genera including S.cerevisiae (Stark et al., 1990; Schaffrath and Breunig, 2000). As judged from fluorescence-activated cell sorter (FACS) analyses, it predominantly causes sensitive budding yeast cells to arrest at the unbudded G1 stage of the cell cycle with an unreplicated (1n) DNA content (Butler et al., 1991a). Although reminiscent of pre-START arrests induced by the pheromone cascade or displayed by cdc28ts strains at restrictive growth temperatures (Leberer et al., 1997), the speculation that zymocin might act by antagonizing G1 cyclin function does not hold true: hyperactive dominant CLN3 alleles are not able to rescue sensitive cells from zymocin treatment (Butler et al., 1994). Further evidence that the zymocin might act in late G1 before START is provided by the finding that cells that have been chemically arrested in S phase by hydroxyurea, prior to zymocin treatment, are able to commit one complete round of cell division and get arrested in the new unbudded G1 cell cycle stage once they have been released from the chemical S block in the continued presence of zymocin (Butler et al., 1991a).
Despite the heterotrimeric (αβγ) structure of native holo-zymocin (Stark and Boyd, 1986), intracellular expression of its smallest subunit, the γ-toxin, alone is lethal. Thus, conditional expression of the γ gene from regulatable GAL promoters leads to a galactose-dependent G1 arrest that mimics the effect of native holo-zymocin (Tokunaga et al., 1989; Butler et al., 1991b). The α and β subunits, however, are needed for holo-zymocin to act from the cell exterior. The β subunit is predicted to form four C-terminal hydrophobic domains, suggesting cell membrane association, while the α subunit exhibits an exo-chitinase activity that is essentially required for holo-zymocin function (Stark et al., 1990; Butler et al., 1991c). Sensitive yeast cells can be rescued from zymocin action by exogenously applying crude chitin preparations (D.Jablonowski and R.Schaffrath, unpublished data) and chitin-deficient S.cerevisiae mutants are resistant towards exo-zymocin (Takita and Castilho-Valavicius, 1993) but not against endogenous γ-toxin expression. These observations suggest a role for the α and β subunits in zymocin docking to sensitive cells, whereas cytotoxicity resides solely within the γ-toxin subunit. Based on their ability to grow in the presence of the holo-form, zymocin-resistant mutants termed skt (sensitivity to K.lactis toxin), iki (insensitive to killer) and kti (K.lactis toxin insensitive), respectively, have been isolated independently (Kawamoto et al., 1990; Butler et al., 1994; Kishida et al., 1996). Sensitivity of these mutants towards intracellular, conditional expression of the γ-toxin can distinguish zymocin binding/uptake (class I) from potential γ-toxin target site mutants (class II). Mutations in the class I genes SKT5 and KTI2 (isoallelic with CHS4 and CHS3, respectively) affect chitin biosynthesis and only lead to exo-zymocin resistance (Kawamoto et al., 1993; Butler et al., 1994), whereas mutations in the class II genes KTI12, IKI1 and IKI3 confer resistance against exo-zymocin and endogenously expressed γ-toxin (Butler et al., 1994; Yajima et al., 1997). Despite these recent advances, the intracellular toxin target site (TOT) still remains elusive and earlier reports that adenylate cyclase, Cdc35p, is involved in zymocin action (Sugisaki et al., 1983) have been ruled out (White et al., 1989).
To understand the zymocin mode of action we isolated several γ-toxin target site (tot) mutants by way of mTn3 transposon mutagenesis and PCR-mediated gene disruption following conditional expression of the γ-toxin from within S.cerevisiae cells. Thus, we identified mTn3:: integrations and re-verified individual knockouts in several TOT genes. The totΔ strains are resistant to both exo-zymocin and endogenous γ-toxin expression, providing evidence for true toxin target site mutants. Moreover, loss of TOT gene function renders yeast cells additional phenotypes: slow growth, a G1 cell cycle delay and sensitivity towards the drugs caffeine, Calcofluor White and 6-azauracil (6-AU). Consistent with the latter, which implies a functional role in transcription elongation, we identified the TOT gene products as constituents of Elongator and components associated with Elongator, a multisubunit complex interacting with elongating RNA polymerase II holoenzyme.
Results
Molecular identification of TOT genes and analysis of the tot phenotypes
Among a pool of 100 000 yeast clones carrying insertions of the mini-transposon mTn3::lacZ::LEU2 (mTn3) γ-toxin-resistant clones were screened for by switching on the expression of the γ-toxin from the UASGAL1 promoter on 2% galactose plates. Three hundred and twelve clones able to grow on galactose were selected over a period of 7 days. Of these, roughly one-sixth (58 clones) showed β-galactosidase expression in a filter assay, indicating in-frame lacZ fusions. After pHMS14 plasmid rescue, restriction pattern analysis and retransformation into non-mutagenized reporter strain LS20, 41 candidates remained to be subjected to a second viability screen using inducible γ-toxin expression from another regulatable promoter (UASMET25) to eliminate promoter-specific false positives related to UASGAL1. Fifteen clones were able to grow in the absence of added methionine. These were finally subjected to a killer eclipse assay testing exo-zymocin resistance. Only three candidates passed this test. The TOT1–3 genes disrupted by mTn3 in these three candidates were identified by vectorette PCR and sequencing as YLR384c (ELP1/IKI1/TOT1), YGR200c (ELP2/TOT2) and YPL086c (ELP3/TOT3).
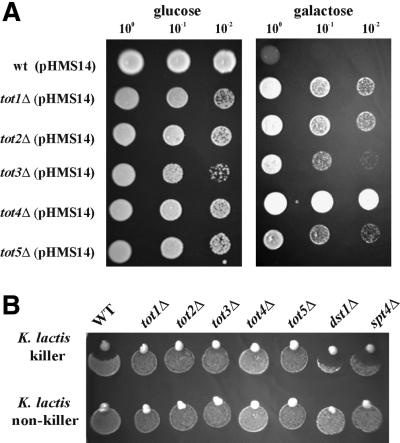
To verify that zymocin resistance caused by the mTn3 minitransposon insertion was due to inactivation of the TOT genes, individual knockouts of TOT1–3 were constructed by PCR-mediated gene disruption. In addition, two more genes reported previously as zymocin resistance determinants (Butler et al., 1994; Yajima et al., 1997) were individually disrupted and shown to display a tot phenotype: KTI12 (TOT4) and IKI1 (TOT5). All these totΔ cells showed resistance towards endogenously expressed γ-toxin (Figure 1A) as well as against extracellular holo-zymocin (Figure 1B), indicating that the tot1–5Δ strains are true toxin target site mutants. To analyse whether the tot1–5Δ mutations were dominant or recessive, the null mutants were crossed with LF20, the wild-type zymocin-sensitive strain of opposite mating type. TOT/totΔ heterozygous diploids obtained in this way were all sensitive towards exo-zymocin and intracellular γ-toxin (data not shown), showing that each of the tot1–5Δ mutations behaves recessively. Since TOT1, TOT2 and TOT3 were recently identified as ELP1, ELP2 and ELP3, structural genes encoding components of RNA polymerase II Elongator (Otero et al., 1999; Wittschieben et al., 1999; Fellows et al., 2000), mutants lacking transcription factors TFIIS (dst1Δ) (Archambault et al., 1992) and Spt4p (spt4Δ) (Hartzog et al., 1998; Wada et al., 1998), both of which affect elongation but are not components of the Elongator complex, were assayed against exo-zymocin. dst1Δ and spt4Δ cells were found to be as sensitive towards zymocin as the wild-type strain LS20 (Figure 1B), suggesting that zymocin resistance is not generally associated with elongation mutants but involves specifically ELP/TOT gene function. As shown previously by Butler et al. (1994), KTI12 (TOT4) overexpression leads to zymocin resistance, while IKI1 (TOT5) overexpression does not (Yajima et al., 1997). To test the overexpression phenotype of TOT1–TOT4, the genes were overexpressed from their native promoter on multicopy yeast episomal vectors in the wild-type background of strain LS20. TOT4 was the only gene causing γ-toxin resistance in multicopy (data not shown).
Fig. 1. The totΔ mutants are resistant towards endogenously expressed γ-toxin and exogenously applied K.lactis zymocin. (A) γ-toxin assay. Serial dilutions of toxin-sensitive (wt) and -resistant (tot1–5Δ) cells transformed with the GAL1-driven γ-toxin vector pHMS14 were replica spotted on repressing (glucose) and inducing (galactose) rich medium and grown for 2 days at 30°C. Lack of growth indicates γ-toxin sensitivity. (B) Killer eclipse assay. Wild-type, tot1–5Δ strains, dst1Δ and spt4Δ cells were spotted twice on to YPD medium. A K.lactis strain (upper row: killer strain AWJ137; lower row: non-killer strain NK40) was set on to the edge of these spots and incubated for 2 days at 30°C. Zones of inhibition around the zymocin-secreting killer strain (upper row) indicate zymocin sensitivity; lack of growth inhibition equals zymocin resistance.
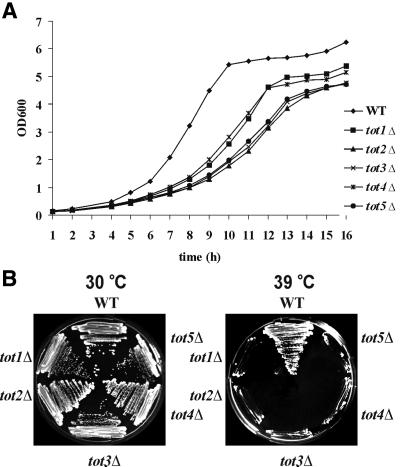
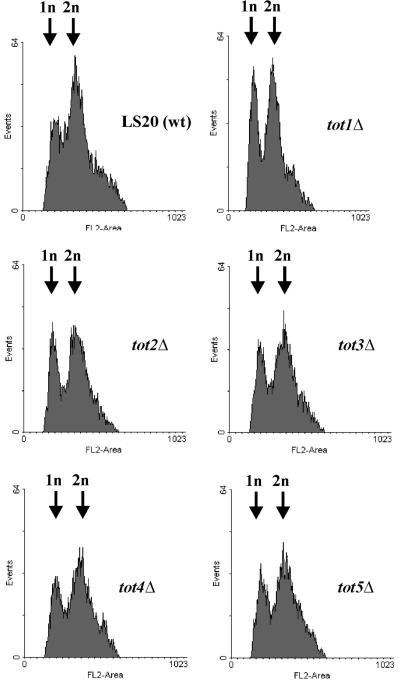
We next analysed the totΔ cells under various growth conditions. Deletion of each TOT gene renders cells a slow growth phenotype (Slg+) at 30°C (Figure 2A), which leads to thermosensitivity above 38°C (Figure 2B). Doubling times are lengthened by a factor of 1.5–2.0, resulting in growth rate reductions from µ = 0.5 (wild type) to µ = 0.25–0.3 (tot1–5Δ). To check whether the Slg+ phenotype was correlated with a delay in a specific phase of the cell cycle, FACS analysis of exponentially growing totΔ strains was performed together with wild-type strain LS20 as a control (Figure 3). This analysis showed that during exponential growth all totΔ mutant strains had a higher percentage of cells with a 1n DNA content than did wild-type cells (Figure 3). Thus, deletion of any TOT gene leads to a significant delay in the G1 phase of the cell cycle, indicating that the TOT gene products are important for normal cell cycle progression. The thermosensitive phenotype of the totΔ cells could be partially rescued by addition of 1 M sorbitol to the growth medium; however, the effect was not as striking as with a chs3Δ cell wall chitin mutant included as a positive reference strain (data not shown).
Fig. 2. Deletion of TOT1–5 genes confers slow growth and thermosensitive phenotypes. (A) Growth curve of tot1–5Δ strains. tot1–5Δ cells show a slow growth phenotype and do not reach the biomass of the wild-type reference in stationary phase. Strains were grown in YPD medium (2% glucose) and growth was measured at OD600 over a period of 16 h. (B) Ts phenotype of tot1–5Δ strains. Strains were streaked on YPD and incubated for 3 days at 30 and 39°C.
Fig. 3. The slow growth phenotype of tot1–5Δ strains correlates with a significant delay in the G1 phase of the cell cycle. FACS analyses of exponential growing tot1–5Δ cells show an extended 1n peak in comparison with the wild-type strain, indicating a delay in the G1 to S transition.
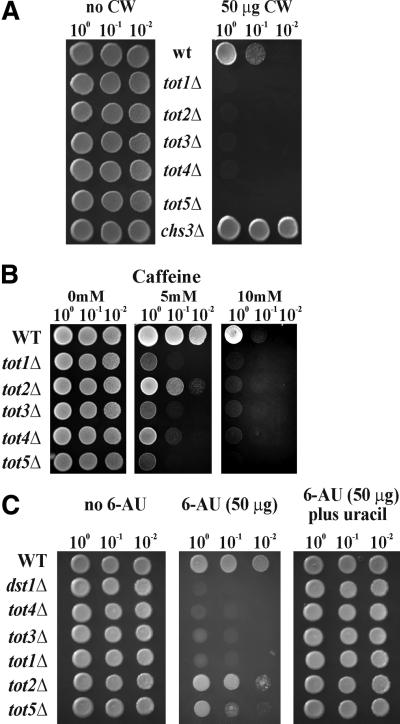
To further check whether the totΔ cells might be affected in cell wall integrity, we tested growth behaviour in the presence of the purine analogue caffeine and the fluorochrome Calcofluor White, a cell wall poison with high affinity for yeast cell wall chitin (Hampsey, 1997). As compared with wild-type strain LS20, all totΔ cells were hypersensitive towards Calcofluor White, indicative of a cell wall defect (Figure 4A). As a Calcofluor White-resistant positive control we included the chs3Δ cell wall chitin mutant (Figure 4A). Sensitivity to caffeine was also shown by all totΔ cells; however, to varying degrees (Figure 4B). Thus, the tot5Δ strain was more sensitive than tot1Δ, tot3Δ and tot4Δ cells, while the tot2Δ mutant was less sensitive to the drug. This phenotype suggests that cell wall integrity may be affected in all the totΔ cells, presumably by involvement of the Pkc1p MAP kinase signalling pathway, one of the known cellular caffeine targets (Hampsey, 1997).
Fig. 4. More tot phenotypes. (A) Calcofluor White sensitivity. Serial dilutions of strains were replica spotted on YPD plates containing no or 50 µg of Calcofluor White (CW). All totΔ strains show hypersensitivity against Calcofluor White, whereas the positive reference, chs3Δ, displays resistance towards the drug. (B) Caffeine sensitivity. Strains were spotted on YPD plates containing up to 10 mM caffeine and grown for 3 days at 30°C. All totΔ strains show more or less hypersensitivity towards caffeine. (C) 6-AU phenotype. Strains were spotted on SD plates containing no 6-AU, 50 µg/ml 6-AU or50 µg/ml 6-AU plus uracil. Except for the tot2Δ mutant, which is mildly affected by the drug, all other totΔ strains show hypersensi tivity towards 6-AU. The dst1Δ mutant served as a positive 6-AU-hypersensitive control strain.
Inspired by parallel identification of TOT1–3 as transcriptional Elongator genes ELP1–3 (Otero et al., 1999; Wittschieben et al., 1999; Fellows et al., 2000), we checked the effect of TOT gene deletion on transcriptional processes using 6-AU as indicator drug. 6-AU inhibits the nucleotide biosynthesis pathway in yeast, leading to a depletion of UTP and GTP, which in turn affects the efficacy of RNA polymerase II holoenzyme during transcriptional elongation (Exinger and Lacroute, 1992). As shown in Figure 4C, all totΔ strains showed different degrees of sensitivity to 6-AU. A dst1Δ strain lacking the elongin TFIIS was used as positive control (Archambault et al., 1992). The strains tot1Δ, tot3Δ and tot4Δ exhibited nearly the same sensitivity as the dst1Δ control. Less sensitivity was shown by tot5Δ and tot2Δ. All totΔ strains as well as dst1Δ could grow again on 6-AU when uracil was added to the media. In conclusion, this drug-induced phenotype suggests a role for the putative γ-toxin, TOT, in transcript elongation in vivo. Moreover, loss of TOT gene function has a pleiotropic effect on a yeast cell’s performance, leading to a complex tot phenotype that includes zymocin and γ-toxin resistance, slow growth, G1 cell cycle delay and thermosensitivity as well as sensitivity towards the drugs Calcofluor White, caffeine and 6-AU. None of these tot phenotypes was severely amplified on combining double tot1Δtot2Δ, tot3Δtot2Δ, tot4Δtot2Δ or tot5Δtot2Δ mutations in one genetic background (data not shown), indicating that the TOT1–5 genes are functionally related. Summing up from these phenotypic analyses, TOT can be considered to have an important role for cellular growth.
The HAT activity of Tot3p is essential for zymocin action
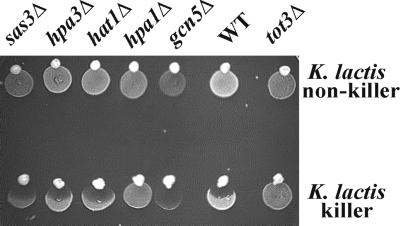
The S.cerevisiae genome encodes other histone acetyltransferase (HAT) activities than the one associated with Elp3p (Tot3p) (Wittschieben et al., 1999). To check whether histone acetylation is needed for zymocin action generally, we tested other HAT gene knockouts in GCN5, SAS3, HPA1, HPA3 and HAT1 (Kleff et al., 1995; Brownell et al., 1996; Reifsnyder et al., 1996; Brown et al., 2000) using zymocin eclipse assays. As illustrated in Figure 5, all HAT knockouts except for HPA1, which is isoallelic to TOT3/ELP3, remained zymocin sensitive. Thus, the Elp3p/Tot3p-associated HAT activity appears to be required for zymocin action, whereas other non-Elongator HATs are dispensable for zymocicity.
Fig. 5. Effect of HAT gene deletions on zymocin sensitivity. Strains deleted in the HAT-encoding genes SAS3, HPA3, HAT1, HPA1/ELP3, GCN5 and TOT3/ELP3 were subjected to a killer eclipse assay essentially as described in Figure 1. Deletion of TOT3 confers zymocin resistance, whereas the other HAT gene deletions tested are zymocin sensitive.
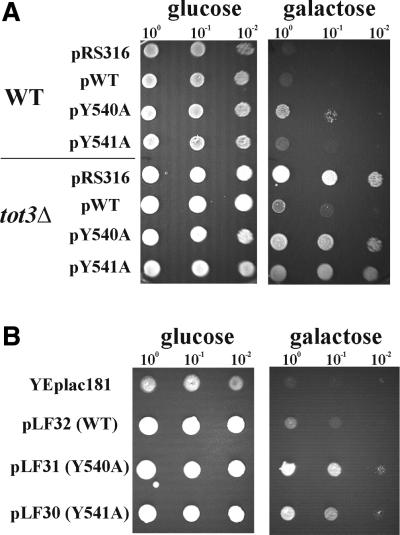
To check whether the HAT-domain activity of Elp3p (Tot3p) itself is required for γ-toxin action, point mutations in ELP3 that change two conserved tyrosine residues to alanine in the B motif of the putative catalytic HAT domain (Y540A and Y541A having <25 and ∼35% of wild-type Elp3p-HAT activity, respectively) (Wittschieben et al., 2000) were tested. These mutant alleles were expressed from their native promoters on CEN plasmids (pBOP60-14: Y540A; pBOP60-15: Y541A; and pBOP60-13: wild-type ELP3 allele) in the tot3Δ mutant and the LS20 wild-type strains co-maintaining the GAL1-driven γ-toxin vector pHMS14. The resulting strains were checked for complementation of the tot3Δ-associated γ-toxin resistance phenotype as well as suppression of γ-toxin sensitivity in wild-type LS20 (Figure 6A) using glucose to galactose shift assays.
Fig. 6. The HAT activity of Elp3p/Tot3p is essential for γ-toxin action. (A) Wild-type and tot3Δ strains containing the GAL1-driven γ-toxin vector pHMS14 were transformed with CEN plasmids carrying no insert (pRS316), the wild-type ELP3 allele (pWT) or mutated elp3 alleles (pY540A and pY541A). Serial dilutions of the resulting transformants were replica spotted on glucose-repressing and galactose-inducing medium. The mutant elp3 alleles fail to complement the tot3Δ-associated γ-toxin resistance phenotype. In the wild-type background, the Y540A allele slightly suppresses γ-toxin sensitivity. (B) When overexpressed from yeast multicopy plasmids, both the mutant elp3 alleles (pLF31: Y540A and pLF30: Y541A) suppress the γ-toxin sensitivity of the wild-type strain co-maintaining pHMS14, whereas vector without insert (YEplac181) and the wild-type ELP3 allele (pLF32) are not able to do so.
Both the Y540A and Y541A mutant ELP3 alleles failed to complement the resistance towards γ-toxin of tot3Δ cells while the wild-type ELP3 allele was able to do so (Figure 6A). Furthermore, the Y540A allele (<25% HAT activity) slightly suppressed the sensitivity of wild-type LS20 against γ-toxin, whereas the Y541A allele (∼35% HAT activity) did not (Figure 6A). When overexpressed from multicopy yeast episomal vectors, both alleles (pLF31: Y540A and pLF30: Y541A) suppressed the sensitivity of the wild-type strain LS20 towards γ-toxin, resulting in a resistance phenotype, while the wild-type ELP3 allele did not (Figure 6B). Thus, it appears that a significant reduction of activation of the HAT encoded by ELP3 phenocopies the effect of TOT3/ELP3 gene inactivation on γ-toxin sensitivity, indicating that the HAT catalytic activity of Elp3/Tot3p is required for zymocin action. Moreover, the mutant alleles are semi-dominant over wild type, indicating that the gene products are incorporated into the Elongator complex, leading to a reduction of Elongator activity.
Co-immunoprecipitation of Tot proteins
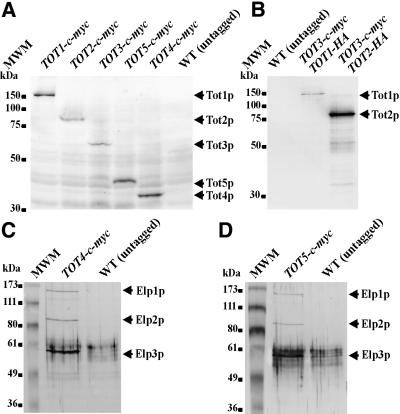
Using PCR-mediated one-step tagging in vivo (Knop et al., 1999), all five Tot proteins were C-terminally marked with the c-Myc (Figure 7A) and HA (not shown) epitope tags to analyse gene expression at the translational level and to assess protein–protein interaction using co-immuno precipitation. As judged from zymocin eclipse assays, TOT1–5 gene tagging had no effect on the biological activity of the individual Tot proteins, i.e. all tagged strains remained as zymocin sensitive as the wild-type reference strain (data not shown). Epitope-tagging identified TOT1–5 as protein-encoding structural genes. Thus, total protein extracts from exponentially grown TOT1–5-(c-myc)3 cells expressed single Tot1–5 polypeptides of estimated molecular weights consistent with the predicted ones that cross-reacted with the anti-c-Myc monoclonal antibody 9E10 (Figure 7A).
Fig. 7. Tot protein detection and co-immunoprecipitation. (A) Identification of TOT1–5 as protein-encoding structural genes. Western analysis: Tot1–5p were tagged individually by one-step in vivo tagging with a triple c-Myc tag. Total protein extracts (50 µg) were separated using 10% SDS–PAGE, electroblotted and immunoprobed with the anti-c-Myc antibody 9E10. The position of each tagged Tot protein is indicated by an arrow. (B) Tot3p interacts with Tot1p and Tot2p. Protein extracts from strains expressing c-Myc-tagged Tot3p together with HA-tagged Tot1p or HA-tagged Tot2p were immunoprecipitated using the anti-c-Myc antibody 9E10. Precipitates were then separated using 10% SDS–PAGE, electroblotted and immunoprobed with the anti-HA antibody 12CA5. The positions of HA-tagged Tot1p and Tot2p are indicated by arrows. (C and D) Tot4p (C) and Tot5p (D) interact with Elp1p (Tot1p), Elp2p (Tot2p) and Elp3p (Tot3p). Protein extracts from strains expressing c-Myc-tagged Tot4p (C) or Tot5p (D) were immunoprecipitated using the anti-c-Myc antibody 9E10, separated on a 10% SDS–PAGE gel, electroblotted and immunoprobed by western analysis using polyclonal anti-Elp1p, anti-Elp2p and anti-Elp3p antibodies. The positions of co-immuno precipitated proteins, Elp1p, Elp2p and Elp3p, are indicated by arrows.
As reported, Elp1p (Tot1p), Elp2p (Tot2p) and Elp3p (Tot3p) associate together within Elongator, a 650 kDa multisubunit complex that interacts specifically with elongating RNA polymerase II holoenzyme (Otero et al., 1999; Wittschieben et al., 1999; Fellows et al., 2000). Consistent with this, we could show by way of co-immunoprecipitation that Tot3p interacts with Tot1p and Tot2p (Figure 7B) and that Tot2p associates with Tot1p and Tot3p (data not shown). Moreover, all three Elongator constituents, Elp1p, Elp2p and Elp3p, could also be shown to interact with Tot4p (Figure 7C) and Tot5p (Figure 7D). These co-immunoprecipitations involved precipitation of c-Myc-tagged Tot4p and Tot5p with the anti-c-Myc antibody 9E10 covalently linked to protein A–Sepharose and detection by polyclonal anti-Elp1p, -Elp2p and -Elp3p antibodies (Otero et al., 1999; Wittschieben et al., 1999; Fellows et al., 2000). In conclusion, all TOT gene products were found to be associated in a multisubunit complex consisting of (at least) five polypeptides, three of which were recently identified as components of the Elongator complex. Thus, the S.cerevisiae Elongator appears to be part of or to represent the putative K.lactis γ-toxin target, TOT.
Gene transcription in response to the K.lactis zymocin
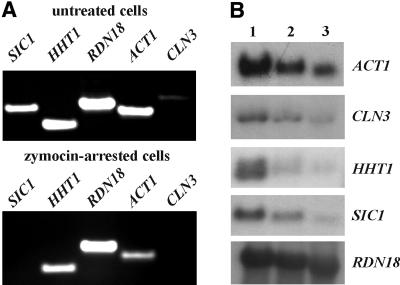
Since the Elp3p-associated HAT activity appears to be essential for Elongator function in vivo (Wittschieben et al., 2000), it is highly likely that TOT/Elongator plays a role in transcript elongation or promoter remodelling by maintaining transcriptionally competent chromatin while piggybacked to the elongating form of the RNA polymerase II holoenzyme (Travers, 1999). To investigate the effect of the K.lactis zymocin on transcriptional processes, identical amounts of total RNA prepared from untreated and zymocin-arrested cells were subjected to RT–PCR experiments using primers specific for RNA polymerase I-dependent and RNA polymerase II-driven genes. As illustrated in Figure 8A, RNA polymerase I-dependent transcription of the 18S rRNA gene (RDN18) remained largely unaffected, whereas transcription of the RNA polymerase II-dependent genes (SIC1, CLN3, HHT1 and ACT1) decreased significantly upon treating cells with the K.lactis zymocin. Similarly, this differential display was seen when using conventional northern hybridization studies (Figure 8B). Here, the effect of the K.lactis zymocin on RNA polymerase II-dependent transcription was particularly pronounced with regard to SIC1, CLN3 and HHT1 gene activation. Our observation that RNA polymerase I-dependent transcription remains largely unaffected by the K.lactis zymocin is consistent with the results of a previous study by Butler et al. (1991c) showing that bulk RNA synthesis, measured by incorporation of radiolabelled uracil, is not affected during zymocin treatment. In conclusion, the K.lactis zymocin appears to preferentially affect RNA polymerase II-dependent gene transcription.
Fig. 8. Gene transcription in response to the K.lactis zymocin. (A) Identical amounts of total RNA isolated from untreated and zymocin-arrested cells were subjected to RT–PCR experiments to study the transcription of RNA polymerase I-dependent (RDN18) and RNA polymerase II-dependent (SIC1, HHT1, ACT1 and CLN3) genes by 1% TBE–agarose gel electrophoresis. (B) Identical amounts of total RNA (10 µg) isolated from untreated cells (lane 1) and cells arrested by zymocin for 3 h (lane 2) and 6 h (lane 3) were subjected to northern blot analysis using probes specific for the RDN18, SIC1, HHT1, ACT1 and CLN3 genes.
Discussion
The K.lactis zymocin is toxic against a variety of sensitive yeast genera including S.cerevisiae. Earlier reports that the zymocin functions on S.cerevisiae by inhibiting the adenylate cyclase, and hence abolishing the roles of cAMP essential for mitotic growth and cell division (Sugisaki et al., 1983), have been disproved (White et al., 1989). Thus, its mode of action still remains unclear. Zymocin-resistant S.cerevisiae skt, iki and kti mutants have been isolated previously (Kawamoto et al., 1990; Butler et al., 1994; Kishida et al., 1996). Sensitivity of these towards intracellular, conditional expression of the γ-toxin from inducible promoters has distinguished zymocin binding/uptake (class I) from γ-toxin target site mutants (class II). The presence of as many as 10 distinct kti class II complementation groups suggests that a complex pathway transduces the zymocin’s inhibitory effect (Butler et al., 1994). While some of them may be involved in the expression of target(s) inhibited by the γ-toxin, a number of proteins could also participate in the process that is blocked by it. These might act in a biochemical pathway or, alternatively, form a putative target complex containing several components. In favour of the latter, our genetic screens involving mTn3 tagging and PCR-mediated gene disruption to search for potential γ-toxin target (tot) mutants have identified the TOT complex as a putative target. The tot phenotypes common to all TOT gene deletions provide genetic evidence that the specified Tot gene products are functionally related to one another. Thus, in addition to zymocin and γ-toxin resistance, tot1–5Δ cells commonly display slow growth and thermosensitive phenotypes as well as G1 cell cycle delay and hypersensitivity towards the drugs Calcofluor White, caffeine and 6-AU. The fact that none of these phenotypes is severely amplified on combining double totΔ mutations in one background can be genetically interpreted as TOT1–5 genes functioning in the same process. Moreover, our re-identification of ELP1/IKI3, ELP2 and ELP3, structural genes encoding Elongator complex components (Otero et al., 1999; Wittschieben et al., 1999; Fellows et al., 2000), as TOT1, TOT2 and TOT3, respectively, provides substantial evidence for the ‘putative target complex’ hypothesis. Consistent with this, our co-immunoprecipitation experiments have shown that the putative γ-toxin target complex, TOT, consists of two more constituents encoded by KTI12 (TOT4) and IKI1 (TOT5) in addition to the known Elongator components. Since tot4Δ and tot5Δ cells are phenotypically indistinguishable from tot1–3Δ (elp1–3Δ) cells, it may well be envisaged that Tot4p and Tot5p constitute further as yet uncharacterized Elongator components, suggesting that the Elongator complex represents or is part of TOT.
By introducing ELP3 HAT domain mutations into an elp3Δ strain, it has been shown recently that the HAT activity associated with Elp3p is essential for Elongator function in vivo (Wittschieben et al., 2000). Thus, in spite of being incorporated into Elongator complexes, the two mutant Elp3 proteins tested (Y540A and Y541A with <25 and ∼35% HAT activity compared with wild-type Elp3p, respectively) failed to rescue the cells from the elp3Δ-associated phenotypes. We have shown here that, similarly, both mutant Y540A and Y541A Elp3 proteins failed to confer sensitivity towards the γ-toxin when re-introduced into our toxin-resistant tot3Δ (elp3Δ) reporter strain. The point mutants behaved as dominant-negative alleles with respect to γ-toxin sensitivity. Thus, γ-toxin sensitivity apparently requires the Elongator-associated HAT activity. Moreover, they conferred γ-toxin resistance when introduced on multicopy episomal vectors into a toxin-sensitive ELP3 wild-type reporter strain. Probably, the mutant overproduced Elp3p variants compete for incorporation into Elongator with wild-type Elp3p, thus reducing the concentration of active Elongator complexes. Taken together, these results indicate that not only inactivation of Elongator by way of TOT1–5 gene deletions, which may ultimately result in non-assembly of functional Elongator complexes, but also in vivo assembly of Elongator complexes with significantly reduced HAT activity lead to the tot phenotype, including resistance towards the K.lactis zymocin.
Since all the TOT genes are non-essential, the zymocin cannot be considered to cause a G1 arrest by inhibition/inactivation, direct or otherwise, of Elongator. Instead, sensitive S.cerevisiae cells require a functional Elongator in order to be able to respond to the K.lactis zymocin. This requirement for Elongator is genetically sustained by the observation that cells lacking either non-Elongator HATs (gcn5Δ, hpa3Δ, hat1Δ and sas2Δ) (Kleff et al., 1995; Brownell et al., 1996; Reifsnyder et al., 1996; Brown et al., 2000) or non-Elongator transcription elongation factors (dst1Δ and spt4Δ) (Archambault et al., 1992; Hartzog et al., 1998; Wada et al., 1998) are not able to alter γ-toxin sensitivity in a way the tot1–5Δ strains can. This implies that the K.lactis zymocin can activate a pathway that is inhibitory to the cell cycle and of which TOT/Elongator is a part.
As for the molecular mode of action of the K.lactis zymocin, our results provide at least two possible models. First, one might envisage direct interaction of the γ-toxin with fully assembled TOT/Elongator complexes in order to promote the observed G1 cell cycle arrest. Thus, γ-toxin-bound TOT/Elongator and free Elongator would compete for a limited supply of a downstream effector molecule that is a key positive regulator of the cell cycle needed for events in late G1. Given the fact that Elongator has recently been demonstrated to associate with the CTD-hyperphosphorylated form of RNA polymerase II and proposed to be exchanged for Mediator upon CTD dephosphorylation after transcription termination (Otero et al., 1999; Wittschieben et al., 1999), it is conceivable to speculate that RNA polymerase II itself may be a prime candidate for this critical effector protein. In the absence of the γ-toxin, RNA polymerase II would associate with TOT/Elongator complexes to acquire full transcriptional activity and to promote a normal G1 to S transition by activation of START-specific genes. In the presence of the γ-toxin, RNA polymerase II would be sequestered and inactivated by the γ-toxin-bound TOT/Elongator complex, leading to a G1 cell cycle arrest, e.g. γ-toxin-bound TOT/Elongator complexes associated with RNA polymerase II may prevent exchange for Mediator during or after transcription termination, thus lowering the concentration of RNA polymerase II able to initiate transcription of START-specific genes needed for a proper G1 to S cell cycle progression. Consistent with this model, we found that transcription of RNA polymerase II-dependent genes (SIC1, CLN3, HHT1 and ACT1) was specifically down-regulated in zymocin-arrested cells, whereas RNA polymerase I-dependent transcription of the 18S rRNA gene (RDN18) remained largely unaffected. Therefore, we favour the hypothesis that the K.lactis zymocin and its cytotoxic γ subunit have the TOT/Elongator-associated RNA polymerase II holoenzyme as a putative direct target. Alternatively, one might envisage that the γ-toxin target could be a gene product whose transcript is down-regulated in strains lacking TOT/Elongator function. To address this question in more detail we are currently designing genome-wide micro-array approaches to identify genes whose transcription is particularly affected in the tot mutants.
Materials and methods
Strains, media, DNA constructs and general methods
Routine bacterial transformations used for constructing recombinant DNA, yeast plasmid shuttles and rescues as well as amplification of the mTn3-tagged yeast DNA library involved electroporation of Escherichia coli strains DH5α, XL1-Blue and TOP10F′. These were grown in Luria–Bertani medium supplemented with one or several of the following additions: ampicillin (100 µg/ml), kanamycin (80 µg/ml), X-Gal (80 µg/ml) and isopropyl-β-d-thiogalactopyranoside (IPTG) (50 µg/ml). All yeast strains used or generated in this study are described in Table I. Standard rich and minimal growth media, YEPD and SC, respectively, were prepared essentially as described by Sherman (1991). For phenotypic analysis, these media were supplemented with 6-AU (50 µg/ml), caffeine (1–20 mM) or Calcofluor White (50–1000 µg/ml). For 6-AU assays, Ura+ transformants (YCplac33) (Gietz and Sugino, 1988) of the totΔ strains were used. Yeast transformations involved electroporation (Becker and Guarente, 1991) as well as the lithium acetate method (Gietz et al., 1992). Mating type switching of LL20 (MATα, see Table I) to generate MATa cells (LF20, see Table I) was accomplished using HO gene expression from plasmid pHAL24 (YEplac181 carrying the HO gene on a 2.5 kb genomic HindIII fragment) followed by plasmid segregation. To create Ura– derivatives of strain LL20, yeast cells were transformed with a ura3Δ deleter plasmid followed by selection against URA3 on SC minimal medium containing uracil and 5-fluoroorotic acid (5-FOA). The deleter plasmid was constructed by autoligation of the YDp-U backbone fragment (Berben et al., 1991) obtained by double digestion with EcoRV and SmaI to delete an internal URA3-borne segment and reconstitute a non-functional ura3Δ allele. Among Ura– transformants (termed LS20, see Table I), ura3-linked auxotrophy was distinguished from ura5 mutants by single copy URA3 gene complementation (YCplac33). To construct the methionine-repressible γ-toxin expression vector pHAL9, the γ-toxin gene was fused as an XhoI–SalI restriction fragment from plasmid pARB1 (Butler et al., 1994) to the UASMET25 promoter of SalI-cut vector p413MET25 (Mumberg et al., 1994). The latter was obtained by deleting the GFP gene from pGFP-NFUS (Niedenthal et al., 1996) using BamHI and autoligation of the vector backbone. Galactose-inducible expression of the γ-toxin was achieved using pHMS14, a p413-based vector (Mumberg et al., 1995), carrying the UASGAL1–γ-toxin fusion on a 1.4 kb SstI–EcoRI fragment from pARB1 (Butler et al., 1994; Schaffrath et al., 1997). As a UASGAL1 vector control, plasmid pHMS22 (p413 carrying the UASGAL1 promoter only) was used. Plasmids for overexpression of Tot1p (pFF14), Tot2p (pFF10) and Tot3p (pFF9) were constructed by cloning the coding sequence with UAS and terminator into YEplac195 using SacI–SalI (pFF14), PstI–BamHI (pFF10) and HindIII–SalI (pFF9). Mutated alleles of ELP3/TOT3 (Y540A, Y541A) were cloned for overexpression in YEplac181 by using BamHI–KpnI restriction fragments of pBOP60-13, pBOP60-14 and pBOP60-15 (Wittschieben et al., 2000), generating pLF30 (Y541A), pLF31 (Y540A) and pLF32 (wild type).
Table I. Yeast strains.
| Strain | Genotype | Reference |
|---|---|---|
| K.lactis | ||
| NK40 | MATα, ade1, ade2, leu2 [k10 k2+] | Gunge et al. (1981) |
| AWJ137 | MATa, leu2, trp1 [k1+ k2+] | Kämper et al. (1991) |
| S.cerevisiae | ||
| FY1679-08A | MATa, ura3-52, leu2Δ1, trp1Δ63, his3Δ200, GAL | Euroscarf |
| FY1646 | MATα, his4-912δ, lys2-128δ, leu2Δ1, spt4Δ::HIS3 | F.Winston |
| GMY27 | MATα, ade2-101, leu2-3,-112, his3Δ200, ura3-52, lys2, gcn5Δ::hisG | Anthony Wright |
| LPY2121 | MATa, ade2-101, his3Δ200, leu2Δ1, lys2-801, TELadh4::URA3, ura3-52, trp1Δ1 sas3Δ::HIS3 | Darryl Auston |
| W303-1a | MATa, ura3-1, leu2-3, -112, his3-11, -15, trp1-1, ade2-1, can 100-1 | Anne Sutton |
| RS1236 (SK56) | as W303-1a, but hat1Δ::TRP1 | Anne Sutton |
| RS1392 (YCW2) | as W303-1a, but hpa1Δ::URA3 | Anne Sutton |
| YRP13 | as W303-1a, but hpa3Δ::HIS3 | Anne Sutton |
| LL20 | MATα leu2-3, -112, his3-11, -15, GAL | NCYC 1445 |
| LS20 | as LL20, but ura3 | this work |
| LF20 | as LL20, but MATa | this work |
| LS20′ | as LS20 plus pHMS14 (CEN4/HIS3/UASGAL1–γ-toxin) | this work |
| LFY12 | as LS20, but tot4Δ::LEU2 GAL | this work |
| FFY5 | as LS20, but tot5Δ::KlLEU2 GAL | this work |
| FFY6 | as LS20, but dst1Δ::KlLEU2 GAL | this work |
| DJY3 | as LS20, but chs3Δ::KlLEU2 GAL | this work |
| LFY1a | as LS20, but TOT4-(c-myc)3::SpHIS5 | this work |
| FFY1t | as LS20, but TOT1-(c-myc)3::SpHIS5 | this work |
| FFY2t | as LS20, but TOT2-(c-myc)3::SpHIS5 | this work |
| FFY3t | as FY1679-08A, but TOT3-(c-myc)3::SpHIS5 | this work |
| FFY5t | as FY1679-08A, but TOT5-(c-myc)3::SpHIS5 | this work |
| FFY2-1dt | as FY1679-08A, but TOT3-(c-myc)3::SpHIS5, TOT1-(HA)6::KlTRP1 | this work |
| FFY2-3dt | as FY1679-08A, but TOT3-(c-myc)3::SpHIS5, TOT2-(HA)6::KlTRP1 | this work |
Isolation and genetic analysis of tot mutants
A yeast transformant population carrying mini-transposon (mTn3) insertions randomly integrated into the genome was constructed as follows. First, strain LS20 was transformed to histidine prototrophy with pHMS14 carrying the UASGAL1–γ-toxin fusion. Several His+ candidates (termed LS20′) were checked for γ-toxin sensitivity by replica plating on to galactose SC his– medium, resulting in a Gal– phenotype. Next, LS20′ was subjected to transposon mutagenesis using electroporation-mediated transformation with the NotI-digested mTn3::yeast insertion library (Burns et al., 1994) selecting for the mTn3-based LEU2 marker on SC his–, leu– medium. His+ Leu+ yeast transformants were subsequently replica plated on galactose medium and incubated for up to 7 days to identify Gal+ toxR candidates. To distinguish genomic mTn3::integrations from plasmid-borne ones that might have caused inactivation of the UASGAL1–γ-toxin fusion on pHMS14, total DNA preparations obtained from selected Gal+ isolates were used for yeast–E.coli plasmid rescue and restriction enzyme analysis, using DNA of starting vector pHMS14 as positive control. Rescued plasmid DNAs identical to the SalI pattern of pHMS14 were retransformed into fresh recipient strain LS20 and checked for toxS by conditionally switching on γ-toxin expression on galactose SC (his–) plates. Clones that passed this test were next checked for UASGAL1-specific false positives by using a second conditional γ-toxin expression approach involving the methionine-regulated promoter UASMET25 on vector pHAL9. Using this second expression approach, clones that were able to grow under inducing conditions, i.e. in the absence of methionine, were obtained. In addition, the Gal+ Leu+ His+ toxR integrants were screened for in-frame fusions of the start-codon-less lacZ gene carried on the mTn3 portion to yeast coding regions by checking β-galactosidase production on qualitative filter assays essentially as described (Ross-Macdonald et al., 1997). Resistance towards exo-zymocin was assayed using the killer eclipse assay (Kishida et al., 1996). To identify the yeast DNA immediately adjacent to the mTn3 integration site of the mutants, the vectorette PCR approach was used (Ross-Macdonald et al., 1998). PCR products that were specifically amplified from mTn3-containing fragments were identified and directly subcloned into vector pCR2.1-TOPO using the topoisomerase cloning kit TOPO TA Version H (Invitrogen). Next, two independent subclones were sequenced for each candidate with the universal M13 reverse (5′-CAGGAAACAGCTATGAC-3′) and –20 forward primers (5′-GTAAAACGACGGCCA G-3′) and analysed using the BLAST and FASTA network services. To analyse whether the mTn3:: marked gene disruptions were recessive or dominant, the individual integrants (MATa, mTn3::LEU2, ura3, pHMS14 [HIS3]) were crossed to LF20 (MATa, leu2, his3, URA3) and diploids selected on SC medium were assayed for γ-toxin sensitivity/resistance by conditionally switching on γ expression on galactose medium. For PCR-mediated gene targeting (Wach et al., 1997) and construction of defined totΔ null alleles, the original YDp plasmid set (Berben et al., 1991) was modified with non-Saccharomyces markers to utilize YDp-KlL (K.lactis LEU2: Zhang et al., 1992), YDp-KlU (K.lactis URA3: Längle-Rouault and Jacobs, 1995) and YDp-SpH (S.pombe HIS5: Wach et al., 1997). Knockout primers (Table II) usually consisted of 50 unique nucleotides homologous to the 5′- and 3′-regions of the yeast gene of interest plus a common 21 nucleotide stretch homologous to the multiple cloning site of plasmid pUC9H-STOP (Berben et al., 1991), the backbone of the YDp plasmid set used as yeast marker templates for PCR-mediated gene targeting. For generating the tot4Δ allele, a deletion construct, pYF6 (Butler et al., 1994), was alternatively used.
Table II. Oligonucleotide primers used in this study.
| Name | Description | Sequence |
|---|---|---|
| FF1 | FW ko-primer TOT1 | 5′-AGAAACAGTACAAATGCCTAATGGCTTATGGTTGAACATGACAAGAGTGGCGACGGCCAGTGAATTCCCGG-3′ |
| FF2 | RV ko-primer TOT1 | 5′-CAATATGACTCTTAGGGAAATCATGAATCTCTGGAACAGGTATTTCTGGGAGCTTGGCTGCAGGTCGACGG-3′ |
| FF3 | FW ko-primer TOT2 | 5′-ATGGTGGAATGTATCACTCCCGAAGCCATTTTTATAGGTGCTAACAAGCACGACGGCCAGTGAATTCCCGG-3′ |
| FF4 | RV ko-primer TOT2 | 5′-CCTCAATCTTGTAATTTTGTCTGCTGGTGTTATATCCTCGTTTAGCTGCGAGCTTGGCTGCAGGTCGACGG-3′ |
| FF5 | FW ko-primer TOT3 | 5′-AGATGGCTCGTCATGGAAAAGGCCCAAAAACTAACAAAAAAAAGCTAGCACGACGGCCAGTGAATTCCCGG-3′ |
| FF6 | RV ko-primer TOT3 | 5′-CCAGAAATAACAGAAATTTTCTCTGAACCATGCTCTTCCTTGGCGATTCTAGCTTGGCTGCAGGTCGACGG-3′ |
| LF13 | FW ko-primer TOT4 | 5′-AAACTAAACAGGCAATTTAGTAAGAAGATGCCACTGGTGCTTTTTACGGGCGACGGCCAGTGAATTCCCGG-3′ |
| LF14 | RV ko-primer TOT4 | 5′-ATCTCAATTCAAGTTTTTGTTAAGATAATCAGCGAAAAGCGGACCGATCCAGCTTGGCTGCAGGTCGACGG-3′ |
| FF7 | FW ko-primer TOT5 | 5′ CTATTGCTACAGGTAGAACAAGATATAATGGCCAGTTCGTCACATAACCCCGACGGCCAGTGAATTCCCGG-3′ |
| FF8 | RV ko-primer TOT5 | 5′-AAAAGGGATCCTCATATGGATCCTCTTCATCATAATCGTCATCCTTTTCGAGCTTGGCTGCAGGTCGACGG-3′ |
| FF9 | FW ko-primer DST1 | 5′-GTAGTCAGTCCGCATAAGAGCATTCATCATGGATAGTAAGGAAGTACTGGCGACGGCCAGTGAATTCCCGG-3′ |
| FF10 | RV ko-primer DST1 | 5′-TCTGTTACCACATGCTTCACATGTACAGAAAGTGGTCAATGGTTCATCCGAGCTTGGCTGCAGGTCGACGG-3′ |
| DJ5 | FW ko-primer CHS3 | 5′-TCCGCAGGAAAGAAATTAGAATGACCGGCTTGAATGGAGATGATCCTGATCGACGGCCAGTGAATTCCCGG-3′ |
| DJ6 | RV ko-primer CHS3 | 5′-GTCTATGCAACGAAGGAGTCACTTTCCTCCTTCCGATTGAGAATATCTTCAGCTTGGCTGCAGGTCGACGG-3′ |
| S3-TOT1 | one-step in vivo tagging TOT1 | 5′-TACCTGTTCCAGAGATTCATGATTTCCCTAAGAGTCATATTGTTGATTTTCGTACGCTGCAGGTCGAC-3′ |
| S2-TOT1 | one-step in vivo tagging TOT1 | 5′-CTTTACGAGCACTATAGACAGTAATTTATATAACTAAGAAAATGGTATGCATCGATGAATTCGAGCTCG-3′ |
| S3-TOT2 | one-step in vivo tagging TOT2 | 5′-GTGTAGGAAGTAGTGATTTGTCCACCCGTATATACTCATTAGCATATGAACGTACGCTGCAGGTCGAC-3′ |
| S2-TOT2 | one-step in vivo tagging TOT2 | 5′-ATTAACTTATTATCCTCTTCTTTTCACATGAGAAATGATATAGATATTGCATCGATGAATTCGAGCTCG-3′ |
| S3-TOT3 | one-step in vivo tagging TOT3 | 5′-ATGGTAAACTAGGATATGAACTAGACGGTCCATACATGTCGAAAAGAATTCGTACGCTGCAGGTCGAC-3′ |
| S2-TOT3 | one-step in vivo tagging TOT3 | 5′-CTGCTTGGAAAACCGGCCATGTCGGCGGCACATAAAAGTTCTATTTACCTATCGATGAATTCGAGCTCG-3′ |
| S3-TOT4 | one-step in vivo tagging TOT4 | 5′-AGGATCGGTCCGCTTTTCGCTGATTATCTTAACAAAAACTTGAATCGTACGCTGCAGGTCGAC-3′ |
| S2-TOT4 | one-step in vivo tagging TOT4 | 5′-ATTTCGTCTTGCCATTTACCTTCTGATATTAATCACATGTATATCATCGATGAATTCGAGCTCG-3′ |
| S3-TOT5 | one-step in vivo tagging TOT5 | 5′-ACGAAAAGGATGACGATTATGATGAAGAGGATCCATATGAGGATCCCTTTCGTACGCTGCAGGTCGAC-3′ |
| S2-TOT5 | one-step in vivo tagging TOT5 | 5′-TAGTTTACATAATCTGGAAGCACTCACTATTTACCATCAGTTTCTACTTTATCGATGAATTCGAGCTCG-3′ |
| RDN18 FW | RT–PCR | 5′-CGCGCAAATTACCCAATCCT-3′ |
| RDN18 RV | RT–PCR | 5′-GGCAAATGCTTTCGCAGTAG-3′ |
| ACT1 FW | RT–PCR | 5′-CTTCCGGTAGAACTACTGGT-3′ |
| ACT1 RV | RT–PCR | 5′-CCTTACGGACATCGACATCA-3′ |
| HHT1 FW | RT–PCR | 5′-AGCAAGAAAGTCCACTGGTG-3′ |
| HHT1 RV | RT–PCR | 5′-GAATGGCAGCCAAGTTGGTA-3′ |
| SIC1 FW | RT–PCR | 5′-TTCACAGAACCTAGTCCCTG-3′ |
| SIC1 RV | RT–PCR | 5′-ACTCCTGGCGTCATTTTTCG-3′ |
| CLN3 FW | RT–PCR | 5′-CAATCTACGTCCCCGTTATC-3′ |
| CLN3 RV | RT–PCR | 5′-CGCTCTTTGGAGTAGTAGCA-3′ |
Flow cytometric determination of cellular DNA content
Cells from exponential growing cultures were fixed in 70% (v/v) ethanol and stored at –20°C. Aliquots (5 × 107 cells) of each sample were washed once with 1 ml of 50 mM sodium citrate solution and incubated in the dark for 30 min at 37°C in 1 ml of 1× PBS containing 1 mg of RNase A and 20 µg of propidium iodide. Each sample was analysed using a Becton-Dickinson FACS. The FACS contained a 15 mW argon laser with an excitation wavelength of 488 nm. Fluorescence was measured at 585 nm. Data were collected on 10 000 cells per sample. Under these conditions, fluorescence is considered to be proportional to DNA content (Hutter and Eipel, 1979).
Epitope tagging and immunological techniques
Epitopes were fused to genes by using PCR-based one-step in vivo epitope-tagging methods and tools as described by Knop et al. (1999). For primers used see Table II. For detection of epitope-tagged proteins, 9E10 mouse monoclonal antibody recognizing the c-Myc epitope and 3F10 rat antibody recognizing the HA epitope (Roche) were used as described (Schaffrath and Meacock, 1996). Polyclonal rabbit Elp1, Elp2 and Elp3 antibodies were kindly provided by Jesper Q.Svejstrup (ICRF, South Mimms, UK). Secondary alkaline phosphatase and peroxidase-conjugated antibodies were obtained from Jackson ImmunoResearch. Antibody cross-linking to protein A–Sepharose, preparation of protein extract and co-immunoprecipitation were carried out as described previously (Zachariae et al., 1996) using B60 buffer. Probes were then checked by western analysis. For all protein methods, proteinase inhibitors (Roche) and 0.5–1 mM phenylmethylsulfonyl fluoride were used.
Gene transcription analyses
Total RNA was isolated from equal amounts of zymocin-arrested and untreated S.cerevisiae LS20 cells using the RNAeasy midi kit (Qiagen) according to the manufacturer’s recommendations. Zymocin treatment and arrest were carried out and monitored as described (D.Jablonowski, L.Fichtner, V.J.Martin, R.Klassen, F.Meinhardt, M.J.R.Stark and R.Schaffrath, in preparation). RT–PCR experiments involved equal amounts of total RNA (4 µg) with the RevertAid™ kit (MBI Fermentas) for 1 h at 42°C in 20 µl reaction volumes. After first strand cDNA synthesis, 1/20 of the reaction was subjected to PCR (30 cycles) using Taq polymerase and oligonucleotide primers (10 µM) (Table II) to amplify fragments specific for the 18S rRNA (RDN18; 0.52 kb), the histone H3 (HHT1; 0.32 kb), the actin (ACT1; 0.44 kb), the G1 cyclin (CLN3; 0.61 kb) and the CKI (SIC1; 0.48 kb) genes. Northern blot analysis was carried out according to standard techniques (Sambrook et al., 1989).
Acknowledgments
Acknowledgements
We are indebted to Mike Stark, Fred Winston, Elmar Schiebel, Jesper Svejstrup, Mike Snyder, Rolf Sternglanz, Anne Sutton, Lorraine Pillus, Darryl Auston and Tony Wright for providing strains, plasmids, antibodies and other tools, and to T.Klapperstück for help with the FACS analysis. The project was supported by grants from the DFG (Scha 750/2) to R.S., the Martin-Luther-Universität to L.F., the Graduierten Förderung to D.J. and the AvH Foundation to R.S. (FLF-DEU/1037031).
References
- Ahmed A., Sesti,F., Ilan,N., Shih,T.M., Sturley,S.L. and Goldstein,S.A. (1999) A molecular target for viral killer toxin: TOK1 potassium channels. Cell, 99, 283–291. [DOI] [PubMed] [Google Scholar]
- Archambault J., Lacroute,F., Ruet,A. and Friesen,J.D. (1992) Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol. Cell. Biol., 12, 4142–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D.M., and Guarente,L. (1991) High-efficiency transformation of yeast by electroporation. Methods Enzymol., 194, 182–187. [DOI] [PubMed] [Google Scholar]
- Berben G., Dumont,J., Gilliquet,V., Bolle,P.A. and Hilger,F. (1991) The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast, 7, 475–477. [DOI] [PubMed] [Google Scholar]
- Brown C.E., Lechner,T., Howe,L. and Workman,J.L. (2000) The many HATs of transcription coactivators. Trends Biochem. Sci., 25, 15–19. [DOI] [PubMed] [Google Scholar]
- Brownell J.E., Zhou,J., Ranalli,T., Kobayashi,R., Edmondson,D.G., Roth,S.Y. and Allis,C.D. (1996) Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell, 84, 843–851. [DOI] [PubMed] [Google Scholar]
- Burns N., Grimwade,B., Ross-Macdonald,P.B., Choi,E.Y., Finberg,K., Roeder,G.S. and Snyder,M. (1994) Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev., 8, 1087–1105. [DOI] [PubMed] [Google Scholar]
- Butler A.R., White,J.H. and Stark,M.J. (1991a) Analysis of the response of Saccharomyces cerevisiae cells to Kluyveromyces lactis toxin. J. Gen. Microbiol., 137, 1749–1757. [DOI] [PubMed] [Google Scholar]
- Butler A.R., Porter,M. and Stark,M.J. (1991b) Intracellular expression of Kluyveromyces lactis toxin γ subunit mimics treatment with exogenous toxin and distinguishes two classes of toxin-resistant mutant. Yeast, 7, 617–625. [DOI] [PubMed] [Google Scholar]
- Butler A.R., O’Donnell,R.W., Martin,V.J., Gooday,G.W. and Stark,M.J. (1991c) Kluyveromyces lactis toxin has an essential chitinase activity. Eur. J. Biochem., 199, 483–488. [DOI] [PubMed] [Google Scholar]
- Butler A.R., White,J.H., Folawiyo,Y., Edlin,A., Gardiner,D. and Stark,M.J. (1994) Two Saccharomyces cerevisiae genes which control sensitivity to G1 arrest induced by Kluyveromyces lactis toxin. Mol. Cell. Biol., 14, 6306–6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exinger F. and Lacroute,F. (1992) 6-azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr. Genet., 22, 9–11. [DOI] [PubMed] [Google Scholar]
- Fellows J., Erdjument-Bromage,H., Tempst,P. and Svejstrup,J.Q. (2000) The Elp2 subunit of elongator and elongating RNA polymerase II holoenzyme is a WD40 repeat protein. J. Biol. Chem., 275, 12896–12899. [DOI] [PubMed] [Google Scholar]
- Gietz R.D. and Sugino,A. (1988) New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene, 74, 527–534. [DOI] [PubMed] [Google Scholar]
- Gietz D., St Jean,A., Woods,R.A. and Schiestl,R.H. (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res., 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunge N., Tamaru,A., Ozawa,F. and Sakaguchi,K. (1981) Isolation and characterization of linear deoxyribonucleic acid plasmids from Kluyveromyces lactis and the plasmid-associated killer character. J. Bacteriol., 145, 382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M. (1997) A review of phenotypes in Saccharomyces cerevisiae. Yeast, 13, 1099–1133. [DOI] [PubMed] [Google Scholar]
- Hartzog G.A., Wada,T., Handa,H. and Winston,F. (1998) Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev., 12, 357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter K.J. and Eipel,H.E. (1979) Microbial determinations by flow cytometry. J. Gen. Microbiol., 113, 369–375. [DOI] [PubMed] [Google Scholar]
- Kämper J., Esser,K., Gunge,N. and Meinhardt,F. (1991) Heterologous gene expression on the linear DNA killer plasmid from Kluyveromyces lactis. Curr. Genet., 19, 109–118. [DOI] [PubMed] [Google Scholar]
- Kawamoto S., Arai,N., Kobayashi,M., Kawahara,K., Iwaheshi,H., Tanabe,C., Hatori,H., Ohno,T. and Nakamura,T. (1990) Isolation and characterization of mutants of Saccharomyces cerevisiae resistant to killer toxin of Kluyveromyces lactis. J. Ferment. Bioeng., 4, 222–227. [Google Scholar]
- Kawamoto S., Sasaki,T., Itahashi,S., Hatsuyama,Y. and Ohno,T. (1993) A mutant allele skt5 affecting protoplast regeneration and killer toxin resistance has double mutations in its wild-type structural gene in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem., 57, 1391–1393. [DOI] [PubMed] [Google Scholar]
- Kishida M., Tokunaga,M., Katayose,Y., Yajima,H., Kawamura-Watabe, A. and Hishinuma,F. (1996) Isolation and genetic characterization of pGKL killer-insensitive mutants (iki) from Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem., 60, 798–801. [DOI] [PubMed] [Google Scholar]
- Kleff S., Andrulis,E.D., Anderson,C.W. and Sternglanz,R. (1995) Identification of a gene encoding a yeast histone H4 acetyltransferase. J. Biol. Chem., 270, 24674–24677. [DOI] [PubMed] [Google Scholar]
- Knop M., Siegers,K., Pereira,G., Zachariae,W., Winsor,B., Nasmyth,K. and Schiebel,E. (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast, 15, 963–972. [DOI] [PubMed] [Google Scholar]
- Längle-Rouault F. and Jacobs,E. (1995) A method for performing precise alterations in the yeast genome using a recyclable selectable marker. Nucleic Acids Res., 23, 3079–3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E., Thomas,D.Y. and Whiteway,M. (1997) Pheromone signalling and polarized morphogenesis in yeast. Curr. Opin. Genet. Dev., 7, 59–66. [DOI] [PubMed] [Google Scholar]
- Mumberg D., Muller,R. and Funk,M. (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res., 22, 5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D., Muller,R. and Funk,M. (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene, 156, 119–122. [DOI] [PubMed] [Google Scholar]
- Niedenthal R.K., Riles,L., Johnston,M. and Hegemann,J.H. (1996) Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast, 12, 773–786. [DOI] [PubMed] [Google Scholar]
- Otero G., Fellows,J., Li,Y., de Bizemont,T., Dirac,A.M., Gustafsson,C.M., Erdjument-Bromage,H., Tempst,P. and Svejstrup,J.Q. (1999) Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell, 3, 109–118. [DOI] [PubMed] [Google Scholar]
- Reifsnyder C., Lowell,J., Clarke,A. and Pillus,L. (1996) Yeast SAS silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nature Genet., 14, 42–49. [DOI] [PubMed] [Google Scholar]
- Ross-Macdonald P., Sheehan,A., Roeder,G.S. and Snyder,M. (1997) A multipurpose transposon system for analyzing protein production, localization, and function in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 94, 190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Macdonald P., Sheehan,A., Friddle,C., Roeder,G.S. and Snyder,M. (1998) Transposon tagging I: A novel system for monitoring protein production, function and localization. Methods Microbiol., 26, 161–179. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schaffrath R. and Breunig,K.D. (2000) Genetics and molecular physiology of the yeast Kluyveromyces lactis. Fungal Genet. Biol., 30, 173–190. [DOI] [PubMed] [Google Scholar]
- Schaffrath R. and Meacock,P.A. (1996) A cytoplasmic gene-shuffle system in Kluyveromyces lactis: use of epitope tagging to detect a killer plasmid-encoded gene product. Mol. Microbiol., 19, 545–554. [DOI] [PubMed] [Google Scholar]
- Schaffrath R., Stark,M.J.R. and Struhl,K. (1997) Toxin-mediated cell cycle arrest in yeast: the killer phenomenon of Kluyveromyces lactis. BIOforum Int., 1, 83–85. [Google Scholar]
- Schmitt M.J., Klavehn,P., Wang,J., Schonig,I. and Tipper,D.J. (1996) Cell cycle studies on the mode of action of yeast K28 killer toxin. Microbiology, 142, 2655–2662. [DOI] [PubMed] [Google Scholar]
- Sherman F. (1991) Getting started with yeast. Methods Enzymol., 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Stark M.J. and Boyd,A. (1986) The killer toxin of Kluyveromyces lactis: characterization of the toxin subunits and identification of the genes which encode them. EMBO J., 5, 1995–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark M.J., Boyd,A., Mileham,A.J. and Romanos,M.A. (1990) The plasmid-encoded killer system of Kluyveromyces lactis: a review. Yeast, 6, 1–29. [DOI] [PubMed] [Google Scholar]
- Sugisaki Y., Gunge,N., Sakaguchi,K. and Tamura,G. (1983) Kluyveromyces lactis killer toxin inhibits adenylate cyclase of sensitive yeast cells. Nature, 304, 464–466. [DOI] [PubMed] [Google Scholar]
- Takita M.A. and Castilho-Valavicius,B. (1993) Absence of cell wall chitin in Saccharomyces cerevisiae leads to resistance to Kluyveromyces lactis killer toxin. Yeast, 9, 589–598. [DOI] [PubMed] [Google Scholar]
- Tokunaga M., Kawamura,A. and Hishinuma,F. (1989) Expression of pGKL killer 28K subunit in Saccharomyces cerevisiae: identification of 28K subunit as a killer protein. Nucleic Acids Res., 17, 3435–3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A. (1999) Chromatin modification by DNA tracking. Proc. Natl Acad. Sci. USA, 96, 13634–13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A., Brachat,A., Alberti-Segui,C., Rebischung,C. and Philippsen,P. (1997) Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast, 13, 1065–1075. [DOI] [PubMed] [Google Scholar]
- Wada T. et al. (1998) DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev., 12, 343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.H., Butler,A.R. and Stark,M.J.R. (1989) Kluyveromyces lactis toxin does not inhibit yeast adenylyl cyclase. Nature, 341, 666–668. [Google Scholar]
- Wickner R.B. (1996) Prions and RNA viruses of Saccharomyces cerevisiae. Annu. Rev. Genet., 30, 109–139. [DOI] [PubMed] [Google Scholar]
- Wittschieben B.O. et al. (1999) A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell, 4, 123–128. [DOI] [PubMed] [Google Scholar]
- Wittschieben B.O., Fellows,J., Du,W., Stillman,D.J. and Svejstrup,J.Q. (2000) Overlapping roles for the histone acetyltransferase activities of SAGA and elongator in vivo. EMBO J., 19, 3060–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima H., Tokunaga,M., Nakayama-Murayama,A. and Hishinuma,F. (1997) Characterization of IKI1 and IKI3 genes conferring pGKL killer sensitivity on Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem., 61, 704–709. [DOI] [PubMed] [Google Scholar]
- Zachariae W., Shin,T.H., Galova,M., Obermaier,B. and Nasmyth,K. (1996) Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science, 274, 1201–1204. [DOI] [PubMed] [Google Scholar]
- Zhang Y.P., Chen,X.J., Li,Y.Y. and Fukuhara,H. (1992) LEU2 gene homolog in Kluyveromyces lactis. Yeast, 8, 801–804. [DOI] [PubMed] [Google Scholar]