Abstract
Centromere protein A (CENP-A) is an essential centromere-specific histone H3 homologue. Using combined chromatin immunoprecipitation and DNA array analysis, we have defined a 330 kb CENP-A binding domain of a 10q25.3 neocentromere found on the human marker chromosome mardel(10). This domain is situated adjacent to the 80 kb region identified previously as the neocentromere site through lower-resolution immunofluorescence/FISH analysis of metaphase chromosomes. The 330 kb CENP-A binding domain shows a depletion of histone H3, providing evidence for the replacement of histone H3 by CENP-A within centromere-specific nucleosomes. The DNA within this domain has a high AT-content comparable to that of α-satellite, a high prevalence of LINEs and tandem repeats, and fewer SINEs and potential genes than the surrounding region. FISH analysis indicates that the normal 10q25.3 genomic region replicates around mid-S phase. Neocentromere formation is accompanied by a replication time lag around but not within the CENP-A binding region, with this lag being significantly more prominent to one side. The availability of fully sequenced genomic markers makes human neocentromeres a powerful model for dissecting the functional domains of complex higher eukaryotic centromeres.
Keywords: CENP-A/centromere/chromatin/neocentromere/replication timing
Introduction
The centromere is an essential structure that coordinates the correct segregation of chromosomes during cell division. Human centromeres generally contain large arrays of the 171 bp, tandemly repeated α-satellite (or alphoid) DNA (reviewed in Choo, 1997a). Human neocentromeres represent a new class of centromere lacking alphoid DNA, thought to originate through the epigenetic modification of normal, often euchromatic, genomic DNA in response to chromosomal rearrangements that result in the loss of a normal centromere (Choo 1997b; Depinet et al., 1997; Warburton et al., 2000). Immunolocalization studies of >20 functionally important centromere proteins have confirmed the structural and functional similarity of neocentromeres and normal centromeres (Saffery et al., 2000).
Studies in yeast have shown recently that eukaryotic centromeres are made up of a number of structurally and functionally distinct molecular domains (Megee et al., 1999; Partridge et al., 2000; Pinto and Winston, 2000). Similar studies in higher eukaryotes have been hampered by the presence of large tandem arrays of amorphous repetitive DNA. Neocentromeres, which lack such highly repetitive sequences, carry unique genomic markers that should provide an unparalleled model system for the study of the detailed organization of the complex centromeres of higher eukaryotes.
The nucleosomal structure of the higher eukaryotic centromere is poorly understood. CENP-A is a 17 kDa protein that associates with all active centromeres including neocentromeres (Palmer et al., 1987; Warburton et al., 1997; Saffery et al., 2000). Molecular analyses of CENP-A homologues in yeast (Cse4p in Saccharomyces cerevisiae and SpCENP-A in Schizosaccharomyces pombe) have suggested a strong association of these proteins with the specialized centromere chromatin structure (Meluh et al., 1998; Glowczewski et al., 2000; Takahashi et al., 2000). In vitro reconstitution experiments have demonstrated the capacity of CENP-A to replace histone H3 for the packaging of α-satellite DNA into the familiar ‘beads-on-a-string’ primary chromatin structure (Yoda et al., 2000). Cenpa gene disruption in mice has demonstrated an essential role of the protein for CENP-C localization and assembly of kinetochores in early embryonic cells (Howman et al., 2000). Thus, CENP-A plays an important role in mediating the formation of specialized nucleosomes at the centromere and is, therefore, a prime candidate for a centromere-specific epigenetic marker (Choo, 2000).
The timing of CENP-A expression has suggested that late replication of centromere DNA may underlie the epigenetic regulation of centromere formation (Shelby et al., 1997; Csink and Henikoff, 1998). However, most evidence suggests that higher eukaryotic centromeres replicate at different times throughout the second half of S phase (Camargo and Cervenka, 1982; Ten Hagen et al., 1990; Vig et al., 1990; O’Keefe et al., 1992; Haaf and Ward, 1994; Haaf, 1997). Further detailed replication study has been difficult due to the paucity of genomic markers within these centromeres.
We have described previously the characterization of a neocentromere at the 10q25.3 region of a marker chromosome mardel(10) (Voullaire et al., 1993; du Sart et al., 1997; Barry et al., 1999, 2000). Here, we have employed a novel strategy involving chromatin immunoprecipitation and genomic array analysis to define the CENP-A binding DNA domain of this neocentromere. The binding profile of histone H3, as well as the replication and sequence properties of the CENP-A and surrounding DNA domains are presented.
Results
Construction of a BAC contig containing the 10q25.3 neocentromere
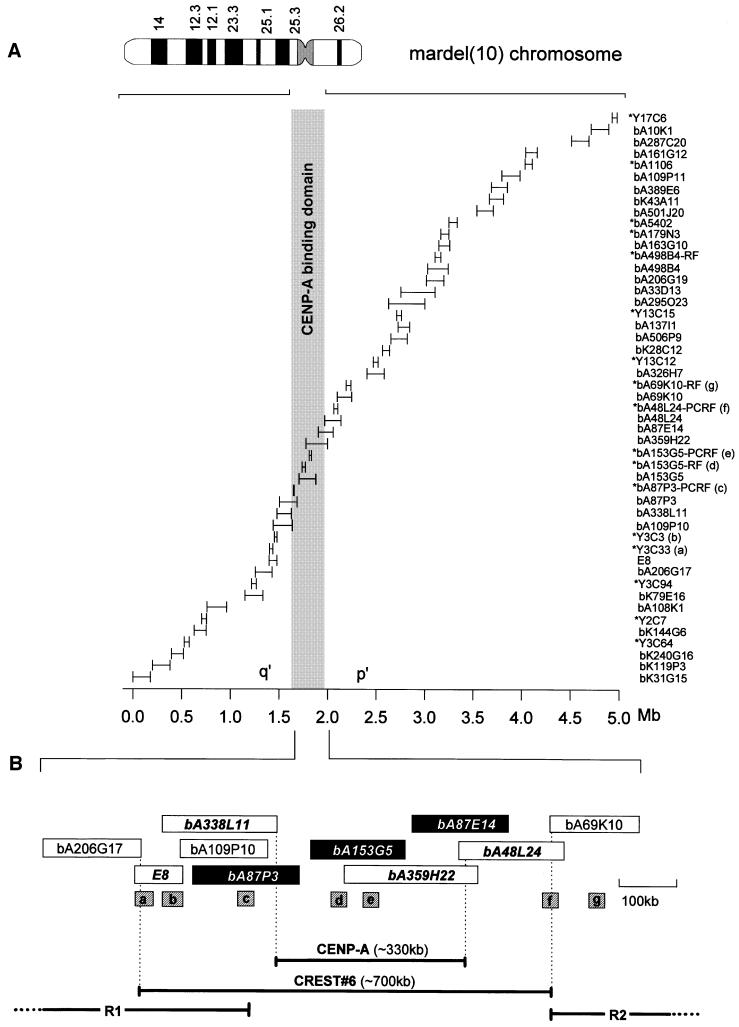
A sequence contig spanning a region of ∼5 Mb and containing the mardel(10) neocentromere was constructed (Figure 1A). This contig encompasses the entire chromosome band 10q25.3 (originally 10q25.2; Voullaire et al., 1993) and contains all the bacterial artificial chromosomes (BACs) and other probes used in this study.
Fig. 1. Contig map of the 10q25.3 neocentromere and surrounding DNA on the mardel(10) chromosome. (A) Ideogram of the G-banded mardel(10) chromosome and arrangement of the 5 Mb sequence contig. Positions of the BACs used for CIA analysis are shown, together with the probes used for replication timing analysis (indicated with an asterisk). The map was constructed using an overlapping BAC contig, a subset of which is used in this study, with additional mapping data from the Sanger Centre and sequence data derived from the Celera Corporation. The sizes of the bars are proportional to the size of each BAC or FISH probe. The shaded region represents the CENP-A binding domain. The distal p′ BAC bA287C20 was used as a control in the CIA experiments. (B) Detailed relative positions of the BACs used to delineate the CENP-A and CREST#6 binding regions. CENP-A binding BACs are shown in black and CREST#6 binding BAC names are in bold and italics. BAC bA109P10 did not show significant binding to CENP-A and was not used in CREST#6 experiments. Determination of the boundaries of the CENP-A and CREST#6 binding domains (shown as horizontal bars below) takes into account the overlapping regions (vertical dotted lines) of flanking non-binding BACs. a–g represent probes within this region used for replication timing experiments, where probes c and f define the inner boundaries of two regions of delayed replication (R1 and R2) immediately flanking the CENP-A binding domain (see Figure 3).
A 700 kb CREST#6 binding domain on mardel(10)
Chromatin immunoprecipitation and array (CIA) analysis was performed initially on the somatic cell hybrids 1f and 5f, containing the normal chromosome 10 and mardel(10), respectively, using an anti-centromere antiserum, CREST#6 (du Sart et al., 1997). Oligonucleosomes obtained from micrococcal nuclease digestion of nuclei were used as the input fraction in chromatin immunoprecipitation. Immunoprecipitated chromatin was captured by protein A–Sepharose (bound fraction). DNA extracted from both input and bound fractions was randomly amplified by degenerate oligonucleotide-primed (DOP) PCR for use as probes on an array of dot-blotted BACs mapped to the 10q25.3 sequence contig (Figure 1). As a positive control, the hybridization signals for α-satellite DNA were compared for the bound and input fractions in 1f and 5f. The results indicated that for both cell lines, the bound fraction yielded 200–300 times enhancement of the α-satellite signals over the input fraction (n = 4), verifying the effectiveness of this procedure in substantially enriching for centromere-specific chromatin (data not shown).
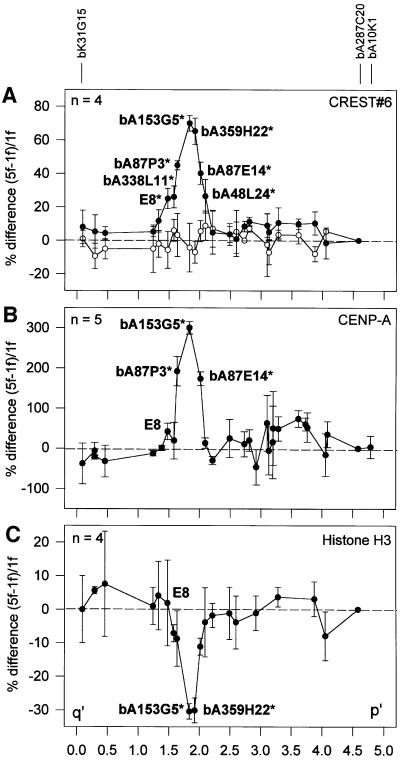
When the relative hybridization signals for the bound and input fractions in 1f and 5f across the 10q25.3 array were determined, a major peak was observed in the bound fraction of 5f over that of 1f, spanning BACs E8, bA338L11, bA87P3, bA153G5, bA359H22, bA87E14 and bA48L24 (p <0.01; Figure 2A, filled circles; see also Supplementary data available at The EMBO Journal Online). The remaining BACs on the array gave only baseline values. The minimal CREST#6 binding region spanned ∼700 kb, centring around the two overlapping BACs bA153G5 and bA359H22 (Figures 1B and 2A). Similar results were not observed when 1f was compared with another somatic hybrid cell line, GM10926D, containing a normal chromosome 10 (Figure 2A, open circles).
Fig. 2. Distribution of CREST#6 antigens (A), CENP-A (B) and histone H3 (C) along the 10q25.3 BAC contig as determined by the CIA procedure. The y-axis represents the percentage difference between the normalized bound/input ratio of 5f and 1f (filled circles) or the difference between 1f and the somatic cell hybrid GM10926D (open circles in A). Each data point is the mean of four or five independent experiments and is shown with standard errors. BAC locations are represented on the x-axis by their midpoints relative to the start of the contig. Positions of BACs with signals significantly deviating from the baseline (p <0.01) are indicated on the graphs with an asterisk. The position of E8 is also indicated on (B) and (C), reference BACs are indicated above the graphs, and the positions of all remaining BACs can be obtained from their relative positions in Figure 1A.
CENP-A binds to a 330 kb domain
CIA analysis of 1f and 5f was repeated using a polyclonal anti-mouse CENP-A antibody (Figure 2B). For both cell lines, hybridization of the immunoprecipitated chromatin DNA on to the α-satellite spot yielded 250- to 435-fold enhancement (n = 4) of the signals for the bound fraction over the input fraction (data not shown), again establishing the effectiveness of this antibody for the immunoprecipitation of centromeric chromatin. Such a significant enrichment of α-satellite DNA has also been described previously using standard chromatin immunoprecipitation (similar to that used in the first part of our CIA technique) with an anti-CENP-A antibody (Vafa and Sullivan, 1997).
When the relative hybridization profiles for the bound and input fractions in 1f and 5f were compared across the 10q25.3 contig (Figure 2B; see also Supplementary data), three different BACs in the 5f sample (bA87P3, bA153G5 and bA87E14) showed significantly elevated hybridization signals (p <0.01). The peak for these signals was approximately four times greater than that obtained with the CREST#6 antibody, probably reflecting the greater specificity of the anti-CENP-A antibody. Other BACs within the contig did not show a significant difference between 1f and 5f. Note that while the region showing the highest CENP-A binding (bA153G5) coincided with that seen for CREST#6, E8 and bA48L24, which both mapped to the shoulders of the CREST#6 peak, did not produce specificities significantly above the baseline. Taking into account the non-CENP-A binding status of flanking overlapping BACs (bA338L11 and bA48L24 on the q′ and p′ sides, respectively), the minimal CENP-A binding domain was estimated to be ∼330 kb (Figure 1B).
Depletion of histone H3 within the CENP-A binding domain
Since CENP-A is a histone H3-related protein, we have determined the status of normal histone H3 within the CENP-A binding domain using an anti-histone H3 antibody in the CIA procedure. The results indicated a significant depletion of histone H3 in the region where CENP-A was observed to bind maximally, represented by the two overlapping BACs bA153G5 and bA359H22 (p <0.01; Figure 2B and C; see also Supplementary data). The magnitude of the decrease in H3 binding relative to the surrounding genomic regions, however, was not as marked as the increase seen for CREST#6 and CENP-A. This could reflect differences in antibody affinities and/or the much greater abundance of total genomic histone H3 that would be expected to be immunoprecipitated.
Replication timing changes around the neocentromere
An initial low-resolution analysis using BrdU (5-bromo-2′-deoxyuridine) incorporation before Colcemid arrest indicated that the mardel(10) neocentromere replicated just after the middle of S phase (J.M.Craig, E.Earle and K.H.A.Choo, in preparation). To obtain a higher-resolution analysis, a fluorescence in situ hybridization (FISH) assay was employed (Kitsberg et al., 1993; Bickmore and Carothers, 1995; Boggs and Chinault, 1997). In this assay, a length of DNA is judged to have replicated when it can be detected as a double spot using FISH. It relies on the fact that the probes used are of appropriate size to produce only a single spot before replication and two after. Replication time is inferred from the relative number of S-phase nuclei in an asynchronous cell population with single or double FISH signals, with the time expressed as a percentage of S phase elapsed by the time the probe region completes replication. This elapsed time is inversely proportional to the percentage of double FISH signals (Bickmore and Carothers, 1995). Since the ascertainment of individual alleles within diploid nuclei is known to be troublesome, due to replication asynchrony and technical artefacts (Haaf and Ward, 1994; Bickmore and Carothers, 1995; Smith and Higgs, 1999), we chose to measure replication timing in 1f and 5f cells. This was a valid approach given that other studies have shown that replication times are maintained when human chromosomes and genes are transferred to heterologous vertebrate systems (Smith and Higgs, 1999 and references therein).
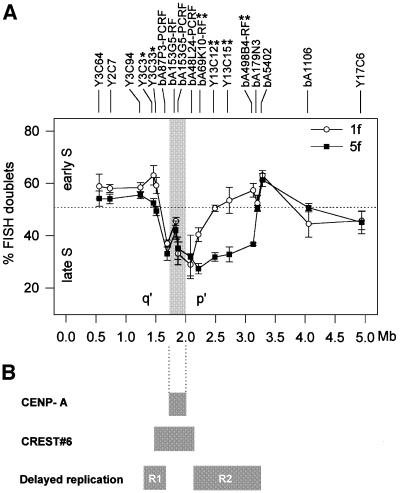
Seventeen probes distributed across the 10q25.3 contig (Figures 1A and 3A) were hybridized to S-phase nuclei from 1f and 5f cells. Hybridization efficiency for these experiments was >95%. Two hundred nuclei were scored for each of four separate experiments. The proportion of nuclei with FISH signal doublets for the normal chromosome 10 in 1f cells fell between 37 and 63%, indicating that the whole contig region on a normal chromosome 10 replicated around the middle of S phase (Figure 3A, open circles; see also Supplementary data). Use of an additional chromosome 10-containing somatic cell hybrid, GM10926D, produced statistically identical results to that of 1f, indicating a reproducible replication pattern in two independently derived cell lines.
Fig. 3. Replication timing at the 10q25.3 neocentromere. (A) Results from the FISH replication assay. The average percentages of double signals for each probe (inversely proportional to replication time), along with error bars (±1 SD), are plotted for the 1f (open circles) and 5f (filled squares) somatic cell hybrids carrying a normal chromosome 10 and the mardel(10) chromosome, respectively. The CENP-A binding domain is indicated by the shaded box. Data points with statistically significant differences between 1f and 5f are highlighted with either * (p <0.05) or ** (p <0.01). The horizontal dotted line defines mid S phase and arbitrarily separates early (above the line) from late (below the line) S phase. (B) Relationship of the replication pattern with the minimal CENP-A and CREST#6 binding domains at the mardel(10) neocentromere. R1 and R2 represent the two domains of delayed replication that have accompanied neocentromere formation. R1, a relatively small region of 80–200 kb, residing q′ of the CENP-A binding region, replicates ∼10% later (p <0.05), while R2, which spans a significantly larger domain of ∼1 Mb, to the p′ side of the CENP-A binding region, replicates ∼20% later (p <0.01) on the neocentromeric chromosome.
When the replication time was determined for the same region on the mardel(10) chromosome in 5f cells, two regions showing significantly delayed replication times were observed (Figure 3A, black squares). The first region, defined by Y3C3 and Y3C33 and spanning a maximum of 200 kb, was located q′ of the CENP-A binding domain and overlapped with the CREST#6 binding domain (Figures 1B, 3A and B). This region replicated ∼10% later in mardel(10), bringing its replication time to ∼50% through S phase. Note that at p = 0.40 and 0.026 for Y3C33 and Y3C3, respectively, this difference was only just significant. In contrast, the second region immediately p′ of the CENP-A binding domain, and spanning ∼1 Mb (Figure 3A and B), showed a more pronounced and highly significant delay in replication timing on the mardel(10) chromosome compared with the same region on normal chromosome 10 (by 18–22%; 0.000075 < p < 0.0095). Thus, the replication time of this p′ region has changed to around two-thirds of the way through S phase (27–37%D, Figure 3A). Interestingly, sandwiched between these two regions of delayed replication, four probes, bA87P3-PCRF, bA153G5-RF, bA153G5-PCRF and bA48L24-PCRF, showed no significant change in replication time. Two of these probes (bA153G5-RF, bA153G5-PCRF) were localized within the CENP-A binding domain (Figures 1B and 3A).
Sequence analysis around the neocentromere
To investigate possible DNA sequence features that may be associated with neocentromere formation, the sequence of the 330 kb minimal CENP-A binding domain was compared with that of the surrounding 3.17 Mb, the human genome in general, and for AT-rich genomic regions (Table I). The CENP-A binding region demonstrated a significantly higher average AT content (65.4%) than the surrounding DNA (58.3%) or the rest of the genome (58%). This places the CENP-A binding region amongst the most AT-rich euchromatic regions in the genome (Bernardi, 1995; Smit, 1999). Within this region, there were two AT-rich islands of a size comparable to that of the AT28 region found in E8 (Barry et al., 1999) containing >500 bp of >80% AT. Within each of these regions were ∼65% perfect AT-rich tandem repeats of unit length 27 and 30 bp, repeated 10 and seven times, respectively. Indeed, the overall content of tandem repeats (two or more copies of unit length of >2 bp) was significantly higher within the CENP-A binding region.
Table I. Summary of the sequence analysis of the 10q25.3 BAC contig.
| Sequence motifs | CENP-A binding region | Non-binding region | p value | Genome average | (>64%) AT-rich average | |
|---|---|---|---|---|---|---|
| Base composition | AT, % | 65.4 | 58.3 | <0.001 | 58.0a | n/a |
| Repeats | tandem repeatsb, % | 5.5 | 2.3 | <0.05 | n/a | n/a |
| satellitesc, % | 0 | 0 | 1 | n/a | n/a | |
| LINEs, % | 43.7 | 14.7 | <0.05 | 18.5a | 26.2a | |
| L1, % | 40.6 | 11.0 | <0.05 | 15.0a | 23.2a | |
| SINEs, % | 6.3 | 12.9 | <0.001 | 14.1a | 6.3a | |
| Alus, % | 5.1 | 10.0 | <0.001 | 11.8a | 4.8a | |
| LTRs, % | 9.8 | 6.1 | >0.05 | 7.8a | 7.2a | |
| DNA elements, % | 2.9 | 2.9 | >0.05 | n/a | n/a | |
| Protein binding motifs | HMG1d,e | 25.6 | 16.2 | <0.001 | 17.7f | n/a |
| topo IIe,g | 9.3 | 6.2 | >0.05 | n/a | n/a | |
| CENP-Be,h | 0 | 1 | n/a | n/a | n/a | |
| Genes | predicted gene densityg,i | 0.30 | 1.1 | n/a | 2.6j | 1.0j |
| CpG island densityg,i | 0 | 0.53 | n/a | 1.4j | 0.37j |
To facilitate statistical analysis of differences between the two regions, the 330 kb CENP-A binding region was split into four equal segments of 82.5 kb, which were analysed separately. Similarly, the non-binding regions were split into 32 segments of 100 kb and one of 70 kb. Results are averages for each region unless otherwise stated and p values from t-tests comparing both regions are also given. Where known, values for average genomic sequences and genomic AT-rich sequences are quoted in the final two columns.
aFrom Smit (1999).
bTandem repeats were classed as being two or more copies of any repeat of unit length ≥2 bp.
cSatellite repeats (with % homology to consensus): α-satellite (80%); β-satellite (100%); γ-satellite (100%); 48 bp repeat (100%); classical satellites Ia and Ib (100%); classical satellite III (100%, two or more copies in tandem).
dPer kb.
e100% homology to consensus.
fFrom Barry et al. (1999).
gPer 100 kb.
hNumber of matches.
iFigures were calculated for whole binding and non-binding regions only.
jPredicted gene density for the whole genome and for AT-rich G bands (Craig, 1995).
n/a, not applicable/attempted.
No tandem repeats normally found in and around human centromeres (Choo, 1997a; Barry et al., 1999) were detected in the contig. Of the interspersed repeats analysed, the CENP-A binding region contained a significantly higher level of long interspersed nuclear elements (LINEs) and a significantly lower level of short interspersed nuclear elements (SINEs) than the surrounding DNA. SINEs and LINES are the most common types of retrotransposon in the human genome. An average of 43.7% LINEs was seen in the CENP-A binding region, compared with 14.8% for the rest of the contig, 18.5% for the rest of the genome and 26.2% for AT-rich (>64% AT) genomic sequences. The density of the L1 family of elements, which represents the majority of LINEs (Smit, 1999), within the CENP-A binding region (40.6%) was also much greater than in the non-binding DNA (11%), the genome as a whole (15%) and AT-rich regions (23.2%). There are, however, a small number of genomic regions with similar LINE densities to those of the CENP-A binding region, including AT-rich regions on the X chromosome (Smit, 1999).
The CENP-A binding region also contained a lower percentage of SINEs, of which Alus are the largest subgroup. This region contained an average of 6.3% SINEs and 5.1% Alus, lower than that of the surrounding DNA (12.9% SINEs and 10% Alus) and the genome average (14.1% SINEs and 11.8% Alus). However, the SINE and Alu densities were similar to that expected for such an AT-rich region (6.3 and 4.8%, respectively; Smit, 1999).
Levels of long terminal repeats (LTRs) and DNA retrotransposon elements were slightly but not significantly higher than those found elsewhere within the 10q25.3 contig. When the binding sites for three centromere-associated proteins, HMG1, DNA topoisomerase II (topo II) and CENP-B, were analysed, only that for HMG1 was significantly more abundant within the CENP-A binding region. However, as the recognition sequence for HMG1 is AT rich, this may simply reflect the AT richness of the region. Only one CENP-B binding motif was found in the contig, outside the CENP-A binding domain. Finally, the CENP-A binding domain showed a much lower density of predicted genes and CpG islands (associated with 50–60% of all genes) than the non-binding BACs. Only one putative gene was predicted within the CENP-A binding domain, which had no CpG islands. However, the predicted gene density is similar to that found within AT-rich DNA (Bernardi, 1995; Craig, 1995). Thus, the CENP-A binding region of the mardel(10) neocentromere appears to have a higher AT content, significantly more L1s and fewer Alu repeats, and is genetically less active, than the surrounding DNA and the genome as a whole.
A similar analysis was performed on the regions exhibiting significantly delayed replication after neocentromere formation. No major differences were observed between these regions and the rest of the contig, only a lower Alu repeat density and a slightly lower gene density (see Supplementary data).
Discussion
CENP-A binds to an ∼330 kb domain on the mardel(10) neocentromere
Previous mapping of the 10q25.3 neocentromere of mardel(10) was performed on chromosomes using combined immunofluorescence and FISH (du Sart et al., 1997). Within the limitations of fluorescence microscopy and the resolution afforded by condensed, mechanically stretched chromosomes, we defined a 640 kb YAC clone (YAC-3) that spanned the CREST#6 binding region. This region was further localized to an 80 kb BAC (E8) within YAC-3, but a definitive boundary of this domain could not be ascertained by the approach used (du Sart et al., 1997; Cancilla et al., 1998). In the present study, we have re-investigated the CREST#6 binding profile using the higher-resolution CIA method. The results confirm that E8 resides within the CREST#6 binding domain (∼700 kb in size), but is located at the edge of this domain. CIA analysis using a CENP-A-specific antibody showed strong binding to the same CREST#6 binding region, although the width of the binding peak was noticeably narrower than that of CREST#6, covering ∼330 kb and not including E8. The difference between the position of E8 relative to CREST#6 in the present and earlier studies (du Sart et al., 1997) is possibly due to non-uniform stretching of chromosomes employed in the initial study (e.g. Heiskanen et al., 1996), the differences in the topology of DNA packaged into metaphase chromosomes, and/or the more accurate and greater molecular resolution offered by the present approach.
The reasons for the observed variation between the widths of the CREST#6 and CENP-A peaks is at present unclear. It is possible that differences in the affinities of the CREST#6 and CENP-A antibodies may account for this variation. However, a more plausible explanation may be related to the potentially greater antigenic heterogeneity of the autoimmune patient-derived CREST antiserum. As we have shown earlier (Voullaire et al., 1993; du Sart et al., 1997), CREST#6 contains, in addition to the anti-CENP-A antibody, a second characterized antibody, to CENP-B, which is absent at the neocentromere (du Sart et al., 1997). We therefore cannot exclude the possibility that antibodies against other centromeric antigens may also be present. Such antigens may bind to the regions within and/or peripheral to the critical CENP-A-associated domain and thus give the expanded width of the CREST#6 peak in the CIA analysis. As will be discussed below in relation to the replication data, it is possible that the mammalian centromere may contain multiple structurally distinct chromatin domains, of which the CENP-A binding domain is only one. The extent of a functional mammalian centromere has not been previously defined. Our data suggest that the extent to which the 10q25 neocentromere, as defined by CENP-A binding, spans ∼330 kb of DNA. However, in view of the above discussion, it is probably premature at present to assume that the CENP-A binding domain is the only important component for consideration when attempting to define the functional extent of a centromere or a neocentromere. Future studies including the use of specific antibodies against other proteins, such as CENP-C or those that are involved in cohesion or heterochromatin, will be necessary to address the ‘simple’ question of how big a centromere really is. The CIA method described here should facilitate such studies.
In our earlier study, we compared extensively the restriction map of the mardel(10) neocentromere region with its progenitor normal chromosomal region (du Sart et al., 1997). Within a domain of up to 1 Mb, which includes the entire CENP-A binding domain defined in the present study, no significant difference was seen between the two chromosomes. This suggests that the DNA underlying the CENP-A binding domain was not significantly altered between the mardel(10) and its progenitor chromosome, at least at the level of restriction mapping. Thus it is likely that the formation of the ∼330 kb CENP-A binding domain on the mardel(10) is epigenetically controlled, although final confirmation will await direct sequence analysis of DNA derived from mardel(10) and its progenitor chromosome, as was carried out previously with E8 (Barry et al., 1999, 2000).
Sequence analysis indicates that the CENP-A binding region coincides with a peak in AT, tandem repeat and LINE retrotransposon content and a trough in SINE content. It is possible that this AT-rich region was competent for neocentromere formation because of its relative genetic inertness, which would result in a minimal disruption to cellular physiology. It is also possible that neocentromeres are assembled preferentially within regions of high AT content or LINE density for structural or conformational advantages. Interestingly, a similar sequence analysis of a second neocentromere at chromosome 20p12 identified through the CIA procedure has revealed that although a similar trough in SINE density was found, no extremes of LINE elements or gene density were present within the CENP-A binding region (Lo et al., 2001). A common feature shared between these two neocentromeres and with α-satellite DNA appears to be a high AT content (65% for the 10q25.3 neocentromere, 61.1% for the 20p12 neocentromere and 62.6% for α-satellite DNA, compared with a genome average of 58%). As AT-rich DNA has a narrower minor groove than GC-rich DNA, it is possible that these regions of DNA may have a greater affinity for factor(s) involved in the maintenance of centromere structure. The application of the CIA strategy described for the rapid identification of the CENP-A binding domains of other neocentromeres should allow more extensive sequence comparison and the identification of common features that might permit or predispose to neocentromere formation.
Histone H3 is depleted at the site of CENP-A binding
CENP-A is a histone H3-like protein that has been proposed to play a primary role in the formation of centromere-specific chromatin (Choo, 2000; Maggert and Karpen, 2000). Studies in yeast have provided evidence for the involvement of the CENP-A homologue Cse4p in the organization of specialized centromeric chromatin (Meluh et al., 1998; Keith et al., 1999; Glowczewski et al., 2000). The complementary distribution profile observed for CENP-A and histone H3 in this study demonstrates that histone H3 is significantly depleted in the CENP-A binding domain, thus providing evidence for replacement of H3 by CENP-A in centromeric nucleosomes. This conclusion is supported by studies showing that CENP-A co-purifies with core histones and with nucleosome core particles (Palmer et al., 1987), can replace histone H3 in nucleosome reconstitution in vitro (Yoda et al., 2000) and self-associates in vivo (Chen et al., 2000). Furthermore, CENP-A binding, α-satellite-containing chromatin was shown to exhibit phased nucleosomal arrays (Vafa and Sullivan, 1997). In addition, preliminary CIA analysis using antibodies to other core histones has shown no difference in nucleosomal density around the 10q25 neocentromere (A.W.I.Lo, unpublished data). However, it remains unclear whether the observed CENP-A binding domain is completely devoid of histone H3 or whether only a subset of centromeric nucleosomes contains CENP-A.
Replication timing of the neocentromere region
We have presented evidence that the CENP-A binding and surrounding regions replicate within the third quarter (50–75%) of S phase. This supports the majority of data obtained from mammalian centromeres containing repetitive DNA, showing replication in mid S phase (O’Keefe et al., 1992; Haaf, 1997) or throughout late S phase (Camargo and Cervenka, 1982; Ten Hagen et al., 1990; Vig et al., 1990). Thus, it appears that centromeres as a group replicate between the middle and the end of S phase. It also seems that the replication time of individual centromeres occurs within a particular time-span rather than at a fixed time point, as indicated by the reported levels of replication asynchrony (O’Keefe et al., 1992; Haaf and Ward, 1994; Litmanovitch et al., 1998; J.M.Craig, E.Earle and K.H.A.Choo, in preparation). It is therefore possible that the precise time at which a centromere replicates within late S phase is unimportant as long as this occurs within the period of CENP-A expression (see Introduction) or within a similar window of opportunity for some other molecular interactions. As centromeres are not the only bands to replicate in late S phase, there must be some way in which CENP-A is preferentially recruited to centromeres after replication. One possible candidate would be a homologue of the S.pombe mis6 protein, which has been shown to be essential for localizing CENP-A to centromeres (Takahashi et al., 2000).
It is not clear how extensively the observed mammalian replication profile can be extrapolated to the centromeres of other eukaryotes. Both field bean and S.cerevisiae centromeres replicate in the first half of S phase (McCarroll and Fangman, 1988; Fuchs et al., 1998; Schubert, 1998). Studies of replication timing in a greater number of eukaryotes will address this question. Replication timing has been proposed to be one possible way to epigenetically mark neocentromeres. (Steiner and Clarke, 1994; Karpen and Allshire, 1997; Csink and Henikoff, 1998; Wiens and Sorger, 1998; Choo, 2000). Our results indicate that although neocentromere formation is accompanied by a delay in the replication timing of a substantial area around the 10q25.3 neocentromere region, the replication time of the CENP-A binding domain is not significantly altered and, therefore, does not behave in concert with its surrounding DNA. One possible explanation for this is that as the region destined to bind CENP-A on the normal chromosome already replicates during late S phase, as do the normal centromeres, requirement to replicate during late S phase has perhaps already been satisfied. Alternatively, or in addition, unlike the surrounding chromatin, formation of CENP-A-associated chromatin may have little effect on replication timing (see below).
The presence of two regions of delayed replication flanking the CENP-A binding region of mardel(10) is interesting and suggests that a centromere may consist of at least two types of structurally distinct chromatin. It is possible that the regions flanking the CENP-A binding domain may be responsible for other facets of centromere function, such as sister chromatid cohesion or formation of a heterochromatic environment (Megee et al., 1999; Partridge et al., 2000), which may predispose the region to a delay in replication. It is of interest that two mammalian heterochromatin proteins, HP1 and SUV39H1, are found at human neocentromeres (Aagaard et al., 2000; Saffery et al., 2000). Thus, although neocentromeres do not exhibit the physical properties traditionally defined for heterochromatin, such as C banding, they fall within the molecular definition of heterochromatin (Wallrath, 1998; Hennig, 1999). Heterochromatin therefore could be associated with all active centromeres and perform important roles such as maintenance of a more compact higher-order chromatin structure necessary for withstanding the spindle forces and/or for sister chromatid cohesion. In future studies, it will be of interest to address whether parts or all of the domains of delayed replication defined in this study are functionally essential.
Neocentromeres provide a good model system for centromere investigation
The dissection of the structural and functional domains of normal higher eukaryotic centromeres has been hampered by the lack of definable landmarks within their highly repetitive sequences. With the sequence of the entire human genome becoming available, we have shown that it is feasible to construct a detailed molecular map across a neocentromere, which has allowed dissection of this neocentromere at a molecular resolution that has not been previously possible for normal centromeres. In addition to the studies described here, the strategy presented should allow the detailed investigation of other changes in chromatin structure and function, such as the roles of other centromere binding proteins, scaffold organization, histone acetylation and other chromatin modifications such as methylation, phosphorylation and poly(ADP) ribosylation (reviewed in Choo, 2000).
Materials and methods
Cell lines
Somatic cell hybrids containing the normal chromosome 10 (1f) and the neocentric mardel(10) were established as described elsewhere (du Sart et al., 1997). GM10926D (ATCC, Manassas, VA, USA) is a CHO somatic cell hybrid containing a single human chromosome 10. All cell lines were maintained in Ham’s KAO medium with 12% dialysed fetal calf serum (Trace Biosciences, Castle Hill, Australia).
10q25.3 BAC contig
A BAC contig containing 3.5 Mb of contiguous BACs from 10q25.2-q26.1, centred on the neocentromere within 10q25.3, was assembled by standard techniques, using BACs obtained from Genome Therapeutics Corporation (www.genomecorp.com; marked with prefixes ‘bK’ in Figure 1) and the human chromosome 10 sequencing group at the Sanger Centre (www.sanger.ac.uk/HGP/Chr10/; marked with prefixes ‘bA’). The BAC contig was verified and extended using sequence data generated through the use of Celera Discovery System and Celera’s associated databases (www.celera.com). This resulted in a 99.4% complete 5 Mb sequence contig. The positions of all DNA probes used in this study are illustrated in Figure 1A. For further details, see Supplementary data.
Antisera/antibodies
Antisera CREST#6, kindly provided by S.Wittingham and T.Kaye (The Walter and Eliza Hall Institute of Medical Research, Melbourne, Australia), was from a patient with the autoimmune disease, CREST, and contained antibodies to centromere components including CENP-A and CENP-B (du Sart et al., 1997).
For anti-CENP-A-specific antibody production, a 25-amino-acid immunogen, KPQTPRRRPSSPARGPSRQSSSVGS (residues 6–30 of the mouse CENP-A protein; Kalitsis et al., 1998), was conjugated to diphtheria toxin and used to immunize rabbits using standard procedures. The specificity of this antibody has been established (Saffery et al., 1999, 2000; Howman et al., 2000). Rabbit anti-histone H3 was obtained from Upstate Biotech, Waltham, MA, USA.
CIA analysis
Chromatin immunoprecipitation was carried out as described by Johnson et al. (1998) with modifications. Approximately 107 exponentially growing cells were incubated in TBS [0.01 M Tris–HCl pH 7.5, 3 mM CaCl2, 2 mM MgCl2 with 0.1 mM phenylmethylsulfonyl fluoride (PMSF) and proteinase inhibitors (Complete, Roche Molecular Biochemicals, Indianapolis, IN, USA)] with 0.25% Tween 40 at 4°C, on a roller for 2 h. The nuclei were released using Dounce homogenization (30–40 strokes on ice with a Type-A pestle, Wheaton, Millville, NJ, USA). Nuclei were further purified by centrifugation at 1500 g for 20 min at 4°C through a 25%/50% discontinuous sucrose gradient. Purified nuclei were then digested with micrococcal nuclease (USB Corp., Cleveland, OH, USA) in digestion buffer (0.32 M sucrose, 50 mM Tris–HCl pH 7.5, 4 mM MgCl2, 1 mM CaCl2, 0.1 mM PMSF) at a concentration of 80 U/mg DNA at 37°C for 10 min. After centrifugation at 15 000 g at 4°C, the first supernatant was kept on ice. The pellet was further incubated with lysis buffer (1 mM Tris–HCl pH 7.5, 0.2 mM EDTA, 0.2 mM PMSF and proteinase inhibitors) on ice for 1 h, centrifuged as before and the two supernatants pooled. The resulting oligonucleosome suspension was precleared by incubation with 1:1000 dilution of pre-immune serum and 1% protein A–Sepharose (AP Biotech, Uppsala, Sweden) at 4°C for 4 h, and centrifuged at 250 g for 5 min at 4°C. The supernatant (input fraction) was used immediately for immunoprecipitation. Equal volumes of the input fraction and incubation buffer (50 mM NaCl, 20 mM Tris–HCl pH 7.5, 5 mM EDTA, 0.1 mM PMSF and protease inhibitors) were incubated with antibodies at 1:500 dilution at 4°C overnight. Immune complexes were captured by incubation with 12.5% v/v protein A–Sepharose at 4°C for 2 h. At the end of the incubation, the protein A–Sepharose was washed stepwise in buffers A (50 mM Tris–HCl pH 7.5, 10 mM EDTA) containing 50, 100 and 150 mM NaCl. Bound immune complexes (bound fraction) were eluted with 2 vol of incubation buffer containing 1% SDS. DNA was extracted from both the input and bound fractions by phenol/chloroform extraction and 100 ng amplified by DOP PCR as described previously (Telenius et al., 1992).
Genomic arrays were generated by immobilizing 100 ng of BAC DNA on to Hybond N+ nylon membranes (AP Biotech) in a dot blot format (Minifold SRC-96, Schleicher & Schuell, Dassel, Germany). An identical amount of cloned α-satellite DNA (αRI; Jorgensen et al., 1987) was spotted on to membranes as a positive control. Five hundred nanograms of DOP PCR-amplified input or bound DNA from 1f and 5f were radioactively labelled by random priming and used to probe identical arrays after pre-annealing with 5 µg of human Cot-1 DNA. Hybridization and wash were performed at high stringency (0.1× SSC/0.1% SDS, 65°C). The results were quantified using a PhosphorImager system (Molecular Dynamics Storm 860 and ImageQuaNT software, AP Biotech).
Calculations for CIA experiments
The intensities of equal areas around the hybridization spot (IBAC) and nearby background (Ibkg) on the array were measured and the true hybridization intensity (I) calculated by subtracting one from the other: I = IBAC – Ibkg. This was repeated for the bound fraction-hybridized spot (Ibound) and input fraction-hybridized spot (Iinput). The ratio of enhancement (R) was calculated by dividing one by the other: R = Ibound/Iinput. In order to correct for the intra- and interexperimental variations, normalization was performed against the R values of a control BAC, bA287C20, which mapped to one end of the 10q25 contig, ∼2.5 Mb p′ of the CENP-A binding region: nR = R/RbA287C20. For each BAC within the contig, normalized R (nR) values from the normal chromosome 10 (1f cell line; nR1f) and the neocentromere (5f cell line; nR5f) were compared by expressing the difference in R values between the two cell lines as a percentage of the R value of the normal chromosome: % difference = [(nR5f – nR1f) × 100%]/nR1f. Experiments were repeated at least four times and the average percentage difference for each serum or antibody was plotted, together with standard error, against the position along the contig for each BAC. All results, from ratios of enhancements (R) onwards are provided in Supplementary data.
DNA probes used for replication timing experiments
Preliminary analysis of interphase FISH with DNA probes of various lengths indicated that probes <85 kb in size produced the expected number of FISH spots (one or two) in >95% of interphase nuclei. Three BACs fulfilled this criterion: bA179N3, bA5402 and bA1106. Other probes included cosmids, BAC fragments and PCR fragments. BAC fragments were a 24 kb EcoRV fragment of bK153G5 (bK153G5-RF), a 50 kb SmaI fragment of bA69K10 (bA69K10-RF) and a 60 kb NotI fragment of bA498B4 (bA498B4-RF). Additional probes (designated PCRF) of 8, 22 and 23 kb were generated by PCR amplification of BACs bA87P3, bA153G5 and bA48L24, respectively, using standard techniques. All probes, or clones used to generate the probes, are marked with an asterisk in Figure 1A; further details are provided in Supplementary data.
FISH
BrdU was added to cultures in log phase at a final concentration of 10–4 M and incubated for 1.5 h before harvesting (Bickmore and Carothers, 1995; Smith and Higgs, 1999). FISH was carried out using standard techniques (Craig, 1999) and digoxygenin-labelled probes were detected using sheep anti-dig-FITC (Roche; 1:50) followed by donkey anti-sheep FITC (Jackson, West Grove, USA; 1:100). BrdU was detected as follows: after 5 min incubation in 0.5% w/v bovine serum albumin (BSA)/phosphate-buffered saline (PBS), slides were incubated at room temperature with rat anti-BrdU (Harlan, Indianapolis, USA; 1:10 in 0.5% w/v BSA/PBS). Slides were washed in 0.5% w/v BSA/PBS/0.5% Tween 20 and incubated with anti-rat AMCA (Jackson; 1:100). All spot-counting was performed blind. Two hundred BrdU-positive (S-phase) nuclei were scored per hybridization experiment for the presence of single or double signals. Each probe was hybridized in four independent experiments (see Supplementary data).
Sequence analysis
DNA (3.5 Mb) from positions 0.5 to 4 Mb on Figure 1A was analysed. To facilitate data handling and the comparison of properties within the contig, the sequence was split up into smaller pieces: four segments of 82.5 kb for the CENP-A binding region, 32 segments of 100 kb and one of 70 kb for the non-binding region. These values were then averaged, and the significance of any differences quantified using Student’s t-test. DNA sequence was analysed for base composition and interspersed repeat content using RepeatMasker (ftp.genome.washington.edu/cgi-bin/RepeatMasker) with default settings. Gene density information was derived from existing and predicted genes using Genie and Emsembl via the Human Genome Browser (www.genome.ucsc.edu), and from CpG islands, using the program CPGPLOT via the EMBOSS server (bioweb.pasteur.fr/seqanal/interfaces/cpgplot.html) using default settings, apart from a CpG island length of >400 bp and GC content of >55%. All other sequence analysis was performed via the ANGIS web server (www.angis.org.au) using the programs FindPatterns and Tandem, with search criteria as outlined elsewhere (Barry et al., 1999).
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank R.Little for BACs from Genome Therapeutics Corporation, A.Stafford, B.Griffiths and A.K.Aung for assistance with BAC preparation, and L.Wong for useful discussion. A.L. was supported by Melbourne International Research Scholarship and International Postgraduate Research Scholarship. This work was funded by NH&MRC of Australia to K.H.A.C.
References
- Aagaard L., Schmid,M., Warburton,P. and Jenuwein,T. (2000) Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres. J. Cell Sci., 113, 817–829. [DOI] [PubMed] [Google Scholar]
- Barry A.E., Howman,E.V., Cancilla,M.R., Saffery,R. and Choo,K.H.A. (1999) Sequence analysis of an 80kb human neocentromere DNA. Hum. Mol. Genet., 8, 217–227. [DOI] [PubMed] [Google Scholar]
- Barry A.E., Bateman,M., Howman,E.V., Cancilla,M.R., Tainton,K.M., Irvine,D.V., Saffery,R. and Choo,K.H.A. (2000) The 10q25.2 neocentromere and its inactive progenitor have identical primary nucleotide sequence: further evidence for epigenetic modification. Genome Res., 10, 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi G. (1995) The human genome: organization and evolutionary history. Annu. Rev. Genet., 29, 445–476. [DOI] [PubMed] [Google Scholar]
- Bickmore W.A. and Carothers,A.D. (1995) Factors affecting the timing and imprinting of replication on a mammalian chromosome. J. Cell Sci., 108, 2801–2809. [DOI] [PubMed] [Google Scholar]
- Boggs B.A. and Chinault,A.C. (1997) Analysis of DNA replication by fluorescence in situ hybridization. Methods, 13, 259–270. [DOI] [PubMed] [Google Scholar]
- Camargo M. and Cervenka,J. (1982) Patterns of DNA replication of human chromosomes. II. Replication map and replication model. Am. J. Hum. Genet., 34, 757–780. [PMC free article] [PubMed] [Google Scholar]
- Cancilla M.R., Tainton,K.M., Barry,A.E., Larionov,V., Kouprina,N., Resnick,M.A., Sart,D.D. and Choo,K.H. (1998) Direct cloning of human 10q25 neocentromere DNA using transformation-associated recombination (TAR) in yeast. Genomics, 47, 399–404. [DOI] [PubMed] [Google Scholar]
- Chen Y., Baker,R.E., Keith,K.C., Harris,K., Stoler,S. and Fitzgerald-Hayes,M. (2000) The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol. Cell. Biol., 20, 7037–7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo K.H.A. (1997a) The Centromere. Oxford University Press, Oxford, UK.
- Choo K.H.A. (1997b) Centromere DNA dynamics: latent centromeres and neocentromere formation. Am. J. Hum. Genet., 61, 1225–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo K.H.A. (2000) Centromerization. Trends Cell Biol., 10, 182–188. [DOI] [PubMed] [Google Scholar]
- Craig J.M. (1995) The functional significance of mammalian chromosome banding. PhD thesis, University of Edinburgh, UK.
- Craig J.M. (1999) Isolation of vertebrate metaphase chromosomes and their analysis by FISH. In Bickmore,W.A. (ed.), Chromosome Structural Analysis: A Practical Approach. Oxford University Press, Oxford, pp. 59–80.
- Csink A.K. and Henikoff,S. (1998) Something from nothing: the evolution and utility of satellite repeats. Trends Genet., 14, 200–204. [DOI] [PubMed] [Google Scholar]
- Depinet T.W. et al. (1997) Characterization of neo-centromeres in marker chromosomes lacking detectable α-satellite DNA. Hum. Mol. Genet., 6, 1195–1204. [DOI] [PubMed] [Google Scholar]
- du Sart D. et al. (1997) A functional neo-centromere formed through activation of a latent human centromere and consisting of non-α-satellite DNA. Nature Genet., 16, 144–153. [DOI] [PubMed] [Google Scholar]
- Fuchs J., Strehl,S., Brandes,A., Schweizer,D. and Schubert,I. (1998) Molecular-cytogenetic characterization of the Vicia faba genome–heterochromatin differentiation, replication patterns and sequence localization. Chromosome Res., 6, 219–230. [DOI] [PubMed] [Google Scholar]
- Glowczewski L., Yang,P., Kalachnikova,T., Santisteban,M.S. and Smith,M.M. (2000) Histone–histone interactions and centromere function. Mol. Cell. Biol., 20, 5700–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T. (1997) Analysis of replication timing of ribosomal RNA genes by fluorescence in situ hybridization. DNA Cell Biol., 16, 341–345. [DOI] [PubMed] [Google Scholar]
- Haaf T. and Ward,D.C. (1994) Structural analysis of α-satellite DNA and centromere proteins using extended chromatin and chromosomes. Hum. Mol. Genet., 3, 697–709. [DOI] [PubMed] [Google Scholar]
- Heiskanen M., Peltonen,L. and Palotif,A. (1996) Visual mapping by high resolution FISH. Trends Genet., 12, 379–382. [DOI] [PubMed] [Google Scholar]
- Hennig W. (1999) Heterochromatin. Chromosoma, 108, 1–9. [DOI] [PubMed] [Google Scholar]
- Howman E.V., Fowler,K.J., Newson,A.J., Redward,S., MacDonald,A.C., Kalitsis,P. and Choo,K.H.A. (2000) Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl Acad. Sci. USA, 97, 1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C.A., O’Neill,L.P., Mitchell,A. and Turner,B.M. (1998) Distinctive patterns of histone H4 acetylation are associated with defined sequence elements within both heterochromatic and euchromatic regions of the human genome. Nucleic Acids Res., 26, 994–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen A.L., Bostock,C.J. and Bak,A.L. (1987) Homologous subfamilies of human alphoid repetitive DNA on different nucleolus organizing chromosomes. Proc. Natl Acad. Sci. USA, 84, 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalitsis P., MacDonald,A.C., Newson,A.J., Hudson,D.F. and Choo,K.H. (1998) Gene structure and sequence analysis of mouse centromere proteins A and C. Genomics, 47, 108–114. [DOI] [PubMed] [Google Scholar]
- Karpen G.H. and Allshire,R.C. (1997) The case for epigenetic effects on centromere identity and function. Trends Genet., 13, 489–496. [DOI] [PubMed] [Google Scholar]
- Keith K.C., Baker,R.E., ChenY., Harris,K., Stoler,S. and Fitzgerald-Hayes,M. (1999) Analysis of primary structural determinants that distinguish the centromere-specific function of histone variant Cse4p from histone H3. Mol. Cell. Biol., 19, 6130–6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitsberg D., Selig,S., Brandeis,M., Simon,I., Keshet,I., Driscoll,D.J., Nicholls,R.D. and Cedar,H. (1993) Allele-specific replication timing of imprinted gene regions. Nature, 364, 459–463. [DOI] [PubMed] [Google Scholar]
- Litmanovitch T., Altaras,M.M., Dotan,A. and Avivi,L. (1998) Asynchronous replication of homologous α-satellite DNA loci in man is associated with nondisjunction. Cytogenet. Cell Genet., 81, 26–35. [DOI] [PubMed] [Google Scholar]
- Lo A.W.I., Magliano,D.J., Sibson,M.C., Kalitsis,P., Craig,J.M. and Choo,K.H.A. (2001) A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A binding neocentromere DNA. Genome Res., 11, 448–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggert K.A. and Karpen,G.H. (2000) Acquisition and metastability of centromere identity and function: sequence analysis of a human neocentromere. Genome Res., 10, 725–728. [DOI] [PubMed] [Google Scholar]
- McCarroll R.M. and Fangman,W.L. (1988) Time of replication of yeast centromeres and telomeres. Cell, 54, 505–513. [DOI] [PubMed] [Google Scholar]
- Megee P.C., Mistrot,C., Guacci,V. and Koshland,D. (1999) The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol. Cell, 4, 445–450. [DOI] [PubMed] [Google Scholar]
- Meluh P.B., Yan,P., Glowczewski,L., Koshland,D. and Smith,M.M. (1998) Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell, 94, 607–613. [DOI] [PubMed] [Google Scholar]
- O’Keefe R.T., Henderson,S.C. and Spector,D.L. (1992) Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific α-satellite DNA sequences. J. Cell Biol., 116, 1095–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer D.K., O’Day,K., Wener,M.H., Andrews,B.S. and Margolis,R.L. (1987) A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol., 104, 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge J.F., Borgstrom,B. and Allshire,R.C. (2000) Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev., 14, 783–791. [PMC free article] [PubMed] [Google Scholar]
- Pinto I. and Winston,F. (2000) Histone H2A is required for normal centromere function in Saccharomyces cerevisiae. EMBO J., 19, 1598–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffery R., Earle,E., Irvine,D.V., Kalitsis,P. and Choo,K.H. (1999) Conservation of centromere protein in vertebrates. Chromosome Res., 7, 261–265. [DOI] [PubMed] [Google Scholar]
- Saffery R., Irvine,D.V., Griffiths,B., Kalitsis,P., Wordeman,L. and Choo,K.H.A. (2000) Human centromeres and neocentromeres show identical distribution patterns of >20 functional important kinetochore-associated proteins. Hum. Mol. Genet., 9, 175–185. [DOI] [PubMed] [Google Scholar]
- Schubert I. (1998) Late-replicating satellites: something for all centromeres? Trends Genet., 14, 385–386. [DOI] [PubMed] [Google Scholar]
- Shelby R.D., Vafa,O. and Sullivan,K.F. (1997) Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol., 136, 501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A. (1999) Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev., 9, 657–663. [DOI] [PubMed] [Google Scholar]
- Smith Z.E. and Higgs,D.R. (1999) The pattern of replication at a human telomeric region (16p13.3): its relationship to chromosome structure and gene expression. Hum. Mol. Genet., 8, 1373–1386. [DOI] [PubMed] [Google Scholar]
- Steiner N.C. and Clarke,L. (1994) A novel epigenetic effect can alter centromere function in fission yeast. Cell, 79, 865–874. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Chen,E.S. and Yanagida,M. (2000) Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science, 288, 2215–2219. [DOI] [PubMed] [Google Scholar]
- Telenius H., Carter,N., Bebb,C.E., Magnus,B., Nordenskjold,M., Ponder,B.A. and Tunnacliffe,A. (1992) Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics, 13, 718–725. [DOI] [PubMed] [Google Scholar]
- Ten Hagen K.G., Gilbert,D.M., Willard,H.F. and Cohen,S.N. (1990) Replication timing of DNA sequences associated with human centromeres and telomeres. Mol. Cell. Biol., 10, 6348–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafa O. and Sullivan,K.F. (1997) Chromatin containing CENP-A and α-satellite DNA is a major component of the inner kinetochore plate. Curr. Biol., 7, 897–900. [DOI] [PubMed] [Google Scholar]
- Vig B.K., Schroeter,D. and Paweletz,N. (1990) Centromere separation. Early replication of repetitive DNA associated with inactive centromeres. Cancer Genet. Cytogenet., 50, 57–65. [DOI] [PubMed] [Google Scholar]
- Voullaire L.E., Slater,H.R., Petrovic,V. and Choo,K.H.A. (1993) A functional marker centromere with no detectable α-satellite, satellite III, or CENP-B protein: activation of a latent centromere. Am. J. Hum. Genet., 52, 1153–1163. [PMC free article] [PubMed] [Google Scholar]
- Wallrath L.L. (1998) Unfolding the mysteries of heterochromatin. Curr. Opin. Genet. Dev., 8, 147–153. [DOI] [PubMed] [Google Scholar]
- Warburton P.E. et al. (1997) Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol., 7, 901–904. [DOI] [PubMed] [Google Scholar]
- Warburton P.E. et al. (2000) Molecular cytogenetic analysis of eight inversion duplications of human chromosome 13q that each contain a neocentromere. Am. J. Hum. Genet., 66, 1794–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiens G.R. and Sorger,P.K. (1998) Centromeric chromatin and epigenetic effects in kinetochore assembly. Cell, 93, 313–316. [DOI] [PubMed] [Google Scholar]
- Wolffe A.P. and Pruss,D. (1996) Deviant nucleosomes: the functional specialization of chromatin. Trends Genet., 12, 58–62. [DOI] [PubMed] [Google Scholar]
- Yoda K., Ando,S., Morishita,S., Houmura,K., Hashimoto,K., Takeyasu,K. and Okazaki,T. (2000) Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc. Natl Acad. Sci. USA, 97, 7266–7271. [DOI] [PMC free article] [PubMed] [Google Scholar]