Abstract
Pathogen-induced remodelling of the host cell actin cytoskeleton drives internalization of invasive Salmon ella by non-phagocytic intestinal epithelial cells. Two Salmonella actin-binding proteins are involved in internalization: SipC is essential for the process, while SipA enhances its efficiency. Using purified SipC and SipA proteins in in vitro assays of actin dynamics and F-actin bundling, we demonstrate that SipA stimulates substantially SipC-mediated nucleation of actin polymerization. SipA additionally enhances SipC- mediated F-actin bundling, and SipC–SipA collaboration generates stable networks of F-actin bundles. The data show that bacterial SipC and SipA cooperate to direct efficient modulation of actin dynamics, independently of host cell proteins. The ability of SipA to enhance SipC-induced reorganization of the actin cytoskeleton in vivo was confirmed using semi- permeabilized cultured mammalian cells.
Keywords: actin nucleation/bacterial invasion/Salmonella/Sips
Introduction
Invasion of intestinal epithelial cells is an essential early event in Salmonella pathogenesis, and is induced by the delivery of an array of bacterial virulence effector proteins, including the four Salmonella invasion proteins (Sips A–D). These Sips are exported across the bacterial envelope and are translocated into the mammalian cell plasma membrane or cytosol, from where they subvert host cell signal transduction pathways and stimulate cytoskeletal rearrangements at the point of bacterial contact. This drives internalization of the pathogen into a membrane-bound vacuole (Finlay and Cossart, 1997; Galán, 1999).
Actin polymerization is essential for Salmonella cell entry. Actin nucleation foci are induced beneath invading bacteria and initiate the generation of sub-membranous actin filaments (F-actin), which in turn condense to form intracellular networks and cables. Cellular actin-binding proteins (ABPs), microtubules and cell surface markers are recruited as bacterial entry progresses (Finlay et al., 1991; Garcia-Del Portillo et al., 1994). The pivotal modulation of the mammalian cell cytoskeleton at the initiation of this process is directed by two Salmonella invasion proteins, SipC and SipA (Hayward and Koronakis, 1999; Zhou et al., 1999a). SipC is essential for pathogen internalization (Kaniga et al., 1995). SipA is not essential for uptake, but it enhances the efficiency of the entry process (Zhou et al., 1999a).
Although SipC and SipA share no primary sequence similarity to each other or to known eukaryotic ABPs, each binds directly to actin and independently influences filament dynamics. In vitro characterization of the purified SipC protein has revealed that distinct C- and N-terminal domains accelerate (nucleate) actin polymerization and bundle (cross-link) F-actin, respectively. This is achieved directly, i.e. independently of host cell components (Hayward and Koronakis, 1999). These activities seem likely to be directed from the host cell plasma membrane, since SipC associates with liposomes in vitro and inserts into the plasma membrane of cultured mammalian cells during Salmonella infection (Hayward and Koronakis, 1999; Scherer et al., 2000). SipA enhances the F-actin bundling activity of the host ABP fimbrin (T-plastin), which is recruited to Salmonella-induced membrane ruffles (Zhou et al., 1999b), and a C-terminal 226-amino- acid SipA fragment (SipA226) binds to F-actin in vitro, decreasing the critical actin concentration required for polymerization and inhibiting F-actin depolymerization (Zhou et al., 1999a).
There has nevertheless been no assessment of possible collaboration between the SipC and SipA ABPs in modulating actin dynamics. A role for such interplay might be suggested by the relatively weak actin-nucleating activity of SipC. The SipC C-terminal domain (SipC-C, amino acids 200–409) nucleates actin polymerization at a 1:10 protein:actin ratio (i.e. for 5 µM actin: 500 nM > minimum [SipC-C] > 50 nM; Hayward and Koronakis, 1999). This is similar to the activity of the purified eukaryotic Arp2/3 complex (for 5 µM actin: 150 nM > minimum [Arp2/3] > 37 nM), which is considered to be a relatively weak nucleator in vitro (Mullins et al., 1998). It is striking that Arp2/3 activity is stimulated by members of the WASP and Scar protein families (Machesky and Insall, 1998; Machesky et al., 1999), and also by the Listeria ActA surface protein, which is required for actin-based intracellular motility (Welch et al., 1998). To investigate whether the Salmonella ABPs SipA and SipC do cooperate, perhaps in stimulating the actin-nucleating activity of SipC, full-length Salmonella typhimurium SipC and SipA proteins were separately overexpressed in laboratory Escherichia coli and affinity purified. Their effects on actin dynamics were initially examined using standard in vitro assays.
Results
SipA stimulates actin nucleation by SipC
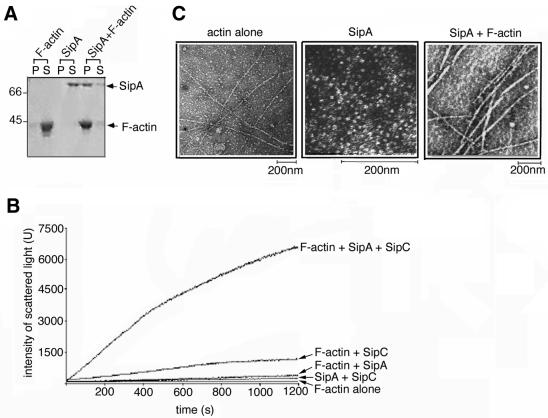
In vitro F-actin binding activity of the full-length 685-residue SipA protein was initially confirmed by routine co-sedimentation assays (100 000 g). Following incubation with F-actin, SipA partitioned predominantly to the pellet fraction with F-actin as expected, whereas SipA alone remained soluble (Figure 1A). Co-sedimentation using a range of SipA concentrations with a fixed concentration of F-actin showed that binding of SipA to F-actin was saturable at a 1:1 molar ratio (Figure 1B). The effect of SipA on actin depolymerization kinetics was monitored using pyrene–actin, a fluorescent actin derivative that has a higher fluorescence intensity in the F-actin form than as monomeric G-actin (i.e. when the pyrene labels are in close proximity rather than spatially separated, respectively). SipA inhibited the rate of F-actin depolymerization; the percentage of pyrene-labelled F-actin was constant over 60 min, in comparison with the control F-actin alone where only 30% remained (Figure 1C). F-actin stabilization by SipA was greater than that induced by the fungal toxin phalloidin, known to stabilize actin filaments (Cooper, 1987). SipA also reduced the critical concentration of G-actin required for filament assembly in vitro by one order of magnitude (data not shown). Purified full-length SipA therefore bound F-actin and modulated actin dynamics in vitro to a degree similar to the previously characterized SipA fragment (Zhou et al., 1999a). Full-length SipA was therefore used for all subsequent in vitro assays.
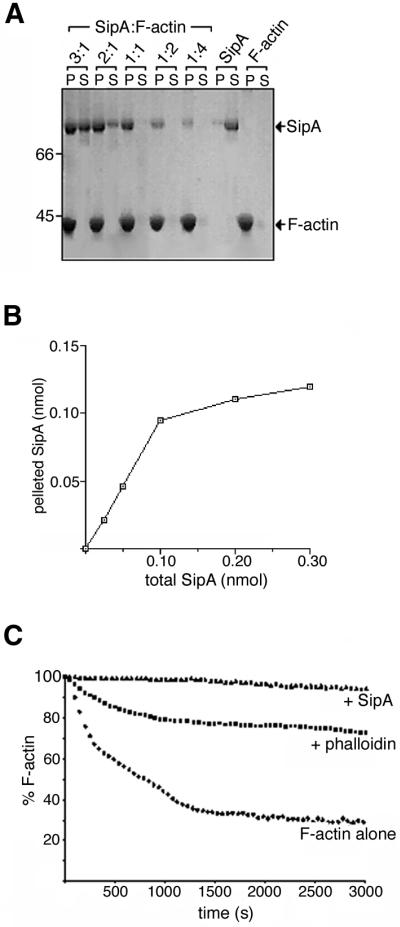
Fig. 1. Actin binding by full-length SipA. (A) Co-sedimentation assays (100 000 g, 25 min) of F-actin (2 µM) incubated with varied concentrations of SipA (indicated by molar ratios of SipA:F-actin). Pellets (P) and supernatants (S) analysed by Coomassie Blue-stained SDS–PAGE. (B) Pelleted and total amount of SipA present in each of the SipA–F-actin co-sedimentation assays shown in (A), quantified by densitometry. (C) Depolymerization assay of F-actin alone or in the presence of phalloidin or full-length SipA (each component at 0.1 µM). The percentage of F-actin remaining is plotted over incubation time.
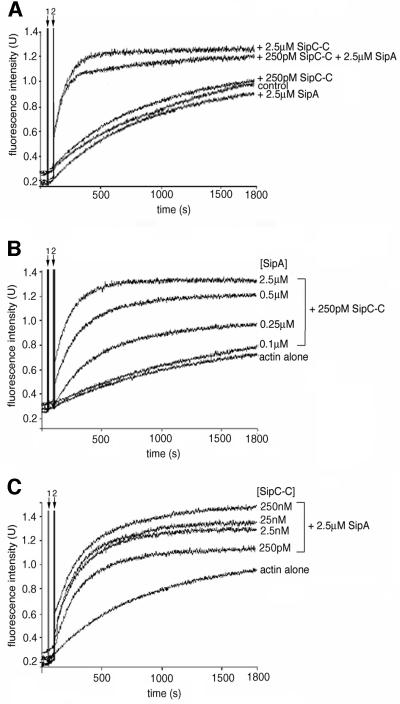
Since SipC and SipA both interact directly with actin and can modulate actin dynamics independently, either by accelerating filament assembly or inhibiting disassembly, respectively, we initially investigated the influence of SipA on the nucleation of actin polymerization directed by the SipC C-terminal domain (SipC-C; Hayward and Koronakis, 1999). Following the addition of initiation buffer to purified G-actin (2.5 µM), typical assembly kinetics were observed in pyrene–actin polymerization assays: an initial lag-phase reflecting the kinetic barrier to ‘nucleation’ (the formation of G-actin trimers or tetramers), followed by a rapid, linear phase during which filaments elongate by monomer addition, reaching steady state after 30 min (Figure 2A, control). When either purified SipA alone (Figure 2A, +2.5 µM SipA), or SipC-C at concentrations below the threshold required to accelerate the kinetics of polymerization (Figure 2A, compare +2.5 µM and +250 pM SipC-C; Hayward and Koronakis, 1999) were included, similar kinetics were observed, although consistent with its F-actin binding activity, SipA induced limited quenching of the pyrene fluorescence signal. However, when in combination with SipA (2.5 µM), picomolar concentrations of SipC-C were sufficient to stimulate spontaneous assembly of G-actin (2.5 µM), as the lag phase of polymerization was eliminated and a high rate of filament extension observed. In contrast, when purified SipD, SipC-N (amino acids 1–120; Hayward and Koronakis, 1999), SipB (Hayward et al., 2000) or purified full-length Shigella IpaA (Tran Van Nhieu and Sansonetti, 1999), a putative SipA homologue (expressed and purified in an identical manner to SipA), were similarly added in combination with SipC-C, no comparable increase in the rate of actin polymerization was observed (data not shown). Addition of a range of SipA concentrations (0.1–2.5 µM) to fixed concentrations of SipC-C and actin (250 pM and 2.5 µM, respectively) induced a dose-dependent acceleration of actin polymerization (Figure 2B). Comparable data were obtained from reciprocal analyses in which SipC-C concentration was varied (250 pM–0.25 µM) with fixed SipA and actin concentrations (both 2.5 µM) (Figure 2C). These data indicate that SipA and SipC-C collaborate to nucleate actin polymerization efficiently.
Fig. 2. Effect of SipA on SipC-mediated nucleation of actin polymer ization. (A) Pyrene–actin assay demonstrating effects of the SipC C-terminal domain (SipC-C, 250 pM or 2.5 µM) alone and in the presence of SipA (2.5 µM) on the kinetics of actin polymerization. Each component was added to 2.5 µM actin following initiation of polymerization. Fluorescence intensity (excitation 365 nm, emission 385 nm) is plotted against incubation time, with spike 1 indicating addition of initiation buffer and spike 2 the addition of test proteins. (B) Pyrene–actin assay demonstrating the actin nucleation activity of afixed concentration of SipC-C (250 pM) in the presence of 2.5 µM actin and varied concentrations of SipA (0.1–2.5 µM), following the initiation of polymerization. Fluorescence intensity is plotted against incubation time as described in (A). (C) Pyrene–actin assay demonstrating the actin nucleation activity of SipC-C using fixed concentrations of SipA and actin (both 2.5 µM) and varied concentrations of SipC-C (250 pM–250 nM), following the initiation of polymerization. Fluorescence intensity is plotted against incubation time as described in (A).
SipA enhancement of SipC-mediated F-actin bundling
We further investigated whether SipA influenced the second SipC activity, i.e. actin bundling (Hayward and Koronakis, 1999). Surprisingly, although neither F-actin nor SipA alone sedimented at low speed (12 000 g), a SipA–F-actin complex sedimented from mixtures of SipA and F-actin (Figure 3A). This suggested that full-length SipA might itself cross-link (bundle) F-actin, since co-sedimentation of an ABP with F-actin at low g force is characteristic of actin bundling activity (Goode et al., 1999; Hayward and Koronakis, 1999). This was shown not to be the case by electron microscopy and light scattering, which is positively correlated with actin filament length and is further enhanced by condensation to filament bundles (Goode et al., 1999). A mixture of SipA and F-actin showed no increase in light scattering in comparison with either control F-actin or purified SipA alone (Figure 3B, compare F-actin alone with F-actin + SipA), in contrast to the increase observed when purified SipC, shown previously to bundle F-actin (Hayward and Koronakis, 1999), was added as a control (Figure 3B, compare F-actin alone with F-actin + SipC). Transmission electron microscopy of mixtures of SipA and F-actin confirmed that the F-actin remained unpaired. This did, however, show that the SipA-coated filaments were significantly thickened (17 ± 1 nm; Figure 3C, right panel) in comparison with control F-actin alone (6.5 ± 0.5 nm; Figure 3C, left panel). Full-length SipA alone in F-buffer formed monodispersed globular structures 5 ± 0.5 nm in diameter (Figure 3C, centre panel). These SipA–F-actin structures were clearly distinct from the actin bundles induced by mixing purified SipC with F-actin (Figure 4B; Hayward and Koronakis, 1999). The low-speed sedimentation of the SipA–F-actin complex is therefore due to an increase in filament diameter due to SipA binding, and not indicative of SipA-induced filament bundling. Nevertheless, the formation of SipA–F-actin complexes precluded the use of quantitative co-sedimentation assays to study cooperation of SipA and SipC. Light scattering and electron microscopy were therefore used in subsequent experiments.
Fig. 3. Effect of SipA on SipC-mediated F-actin bundling. (A) Bundling assays (12 000 g, 15 min) with F-actin alone, SipA alone and SipA + F-actin mixtures (each at 2 µM). Pellets (P) and supernatants (S) were analysed by Coomassie Blue-stained SDS–PAGE. (B) Light scattering (U, intensity at 520 nm) over 1200 s of combinations of SipA, SipC and F-actin in a stoichiometric ratio of 1:1:1, SipC and F-actin, SipA and F-actin, SipA and SipC and F-actin alone, each component at 2.5 µM in F-buffer. (C) Transmission electron micrographs showing F-actin alone (left panel), purified full-length SipA (centre panel) and F-actin incubated with equimolar full-length SipA (right panel), each component at 2.5 µM in F-buffer.
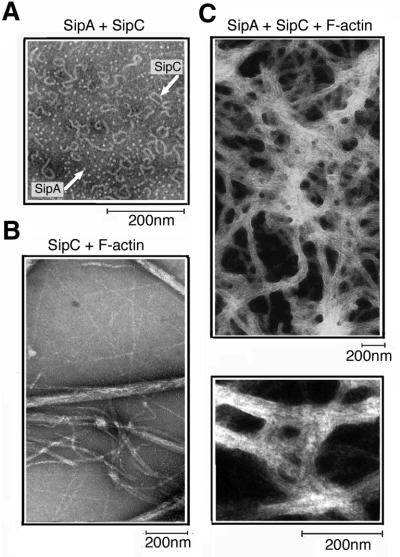
Fig. 4. Networks of bundled actin filaments generated by SipA and SipC. Transmission electron micrographs of the end-point (1200 s) corresponding to light scattering traces shown in Figure 3B. The mixture of SipA, SipC and F-actin (1:1:1; each at 2.5 µM) (C), compared with the mixture of SipC and F-actin (1:1; 2.5 µM) (B). A mixture of SipA and SipC alone (1:1; 2.5 µM) is also shown (A).
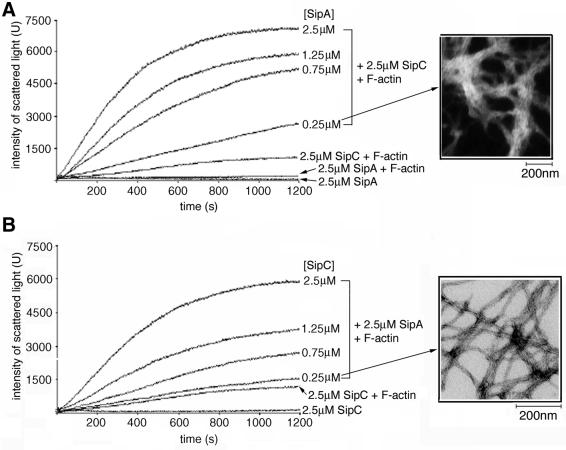
When equimolar mixtures of SipA and SipC were added to F-actin, there was a marked increase in light scattering in comparison with an equimolar mixture of SipC and F-actin (Figure 3B, compare F-actin + SipA + SipC with F-actin + SipC; all 2.5 µM). A constant light scattering trace was obtained for control SipA and SipC mixtures and for each protein independently in F-buffer (Figures 3B, SipA + SipC; and 5A and B, SipA and SipC, respectively), confirming that the purified recombinant proteins were soluble alone and in combination. Transmission electron microscopy revealed that combinations of SipA and SipC induced the formation of a cross-linked network of F-actin bundles, distinct from the parallel F-actin bundles induced by the SipC protein alone (compare Figure 4B and C). Only SipC filaments as shown by Hayward and Koronakis (1999), and SipA globular structures (Figure 3C, centre panel), were evident in mixtures of SipA and SipC (Figure 4A). Adding a range of SipA concentrations (0.25–2.5 µM) to fixed concentrations of SipC and F-actin (both 2.5 µM) induced concentration-dependent increases in light scattering (Figure 5A), and comparable data were obtained from reciprocal analyses in which the SipC concentration was varied (0.25–2.5 µM) and SipA and F-actin concentrations were fixed (both 2.5 µM) (Figure 5B). Transmission electron microscopy confirmed that these observed increases were due to the cross-linking of F-actin bundles (Figure 5A and B, insets). As would be expected, indistinguishable networks were formed when SipC and SipA were added to G-actin during polymerization (data not shown).
Fig. 5. Cooperation of SipA and SipC in F-actin bundling. (A) Light scattering (U, intensity at 520 nm) over 1200 s using a range of concentrations of SipA (0.25–2.5 µM) incubated with a fixed concentration of SipC and F-actin (each 2.5 µM). Control traces with SipA alone (SipA, 2.5 µM), SipC and F-actin (both 2.5 µM), and SipA and F-actin (both 2.5 µM) are shown for comparison. Inset shows a transmission electron micrograph of F-actin (2.5 µM), incubated with SipA (2.5 µM) and SipC (0.25 µM). (B) Light scattering as in (A) using a range of concentrations of SipC (0.25–2.5 µM) incubated with a fixed concentration of SipA and F-actin (each 2.5 µM). Control traces with SipC alone (2.5 µM) and SipC and F-actin (both 2.5 µM) are also shown. Inset shows a transmission electron micrograph of F-actin (2.5 µM), incubated with SipA (0.25 µM) and SipC (0.25 µM).
SipA enhances SipC-induced cytoskeletal rearrangements in vivo
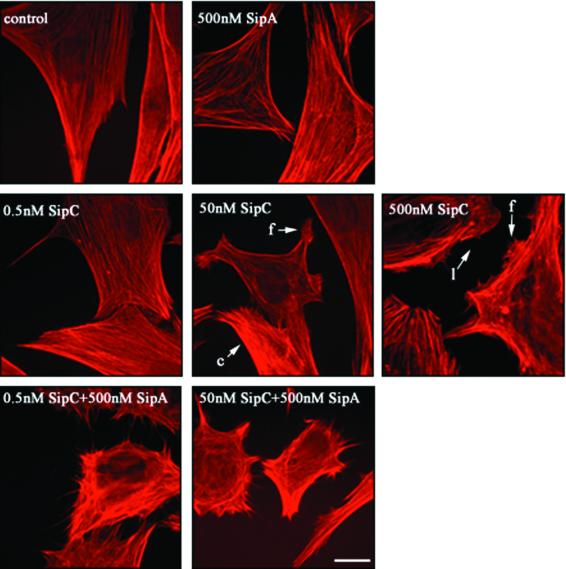
To investigate whether SipA can also enhance SipC-induced cytoskeletal rearrangements in vivo, we used a permeabilized cell assay employed previously to study the effects of Shigella secreted proteins on host cell cytoskeletal architecture (Tran Van Nhieu et al., 1999). This enabled defined concentrations of SipA and SipC, alone or in combination, to be introduced into the cytoplasm of cultured cells. Semi-permeabilization was selected in preference to microinjection or co-transfection since it is less invasive and protein concentrations can be more accurately quantified. After permeabilization, semi-confluent Swiss 3T3 cells were stained with Texas Red-conjugated phalloidin to visualize F-actin directly. They exhibited no significant cytoskeletal rearrangements (Figure 6, control); filopodial and lamellipodial structures were rarely observed (<5% total cells), and were restricted to interfaces of cell–cell contact or where cells were sub-confluent (data not shown). Incubation with 500 nM purified SipA induced no cytoskeletal rearrangements (Figure 6). However, when cells were treated with 500 nM SipC, many cells (>85% total) showed zones of submembranous actin condensation, and lamellipodia- and filopodia-like structures formed at the cell periphery (Figure 6). These data are in agreement with our previous observations that microinjection of 3 µM purified SipC or 300 nM SipC-derived polypeptides induced actin condensation in cultured HeLa cells (Hayward and Koronakis, 1999).
Fig. 6. Cooperation of SipA and SipC induces actin rearrangements in permeabilized Swiss 3T3 cells. Semi-permeabilized cultured Swiss 3T3 cells fixed following 5 min incubation in the presence of permeabilization buffer alone (control), purified SipA, SipC or mixtures of both proteins at the concentrations indicated. Cells were stained with Texas Red-conjugated phalloidin to visualize F-actin and viewed by fluorescence microscopy. Arrows indicate representative actin condensation (c), filopodial (f) and lamellipodial (l) structures. For each typical frame, >200 cells were observed from three independent experiments. Scale bar, 0.5 µm.
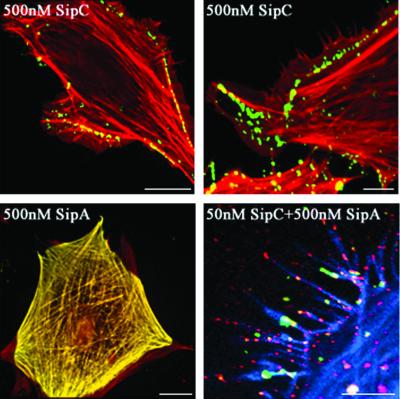
To assess the threshold SipC concentration required to induce cytoskeletal rearrangements, permeabilized cells were treated with lower concentrations. With 50 nM SipC, only ∼30% of cells exhibited zones of actin condensation or filopodial protrusions (Figure 6), and when the concentration was reduced further to 0.5 nM SipC, cytoskeletal architecture appeared indistinguishable from that of control cells (Figure 6, compare 0.5 nM SipC with control). To determine whether SipA influences the ability of SipC to induce cytoskeletal rearrangements, cells were incubated with 0.5 nM SipC in combination with 500 nM SipA. Dramatic cytoskeletal rearrangements were evident (∼50% total cells), including extensive peripheral actin condensation and filopodial extensions (Figure 6). Similar effects were observed when 50 nM SipC and 500 nM SipA were combined, and many cells (∼30%) ‘rounded-up’, a phenotype consistent with gross morphological change (Figure 6). To confirm that these observed cytoskeletal rearrangements were indeed caused by the Sip proteins, staining with anti-SipA or anti-SipC polyclonal antisera and phalloidin was used to visualize intracellular Sip proteins and F-actin simultaneously. Double staining of SipC and F-actin in permeabilized cells treated with 500 or 50 nM SipC alone revealed that SipC localized exclusively to the cell periphery beneath foci where lamellipodial or filopodial protrusions were generated (Figure 7, upper panels). SipC and F-actin staining were apparently coincident at many peripheral sites, consistent with its in vitro actin-binding activity (Hayward and Koronakis, 1999) and its observed membrane affinity in vivo (Scherer et al., 2000) and in vitro (Hayward and Koronakis, 1999). When SipA and F-actin were similarly double stained in permeabilized cells treated with 500 nM SipA alone, SipA was seen coincident with F-actin throughout the cell body, but cytoskeletal architecture remained unchanged (Figure 7, lower left). To confirm that a combination of SipC and SipA induced the dramatic cytoskeletal rearrangements observed in Figure 6, permeabilized cells treated with 50 nM SipC and 500 nM SipA were triple stained to visualize SipC, SipA and F-actin simultaneously; 0.5 nM SipC was below the threshold for reproducible detection by immunostaining. Both SipC and SipA localized to the filopodial structures at the cell periphery (Figure 7, lower right). These data indicate that SipA also potentiates the ability of SipC to induce cytoskeletal rearrangements in cultured mammalian cells.
Fig. 7. Localization of SipA and SipC in semi-permeabilized Swiss 3T3 cells. Semi-permeabilized cultured Swiss 3T3 cells fixed following 5 min incubation with SipA and/or SipC at the concentrations indicated. Upper panels: merged immunofluorescence images following staining for SipC with anti-SipC IgG/AlexaFluor 488-conjugated anti-rabbit IgG (green), and cellular F-actin with Texas Red-conjugated phalloidin (red). Coincident staining appears yellow. Scale bars, 5 µm (left) and 1 µm (right). Similar SipC staining was observed in cells treated with 50 nM SipC and correlated with actin rearrangements. Lower left panel: merged immunofluorescence image following staining for SipA with anti-SipA IgG/AlexaFluor 488-conjugated anti-rabbit IgG (green), and cellular F-actin with Texas Red-conjugated phalloidin (red). Coincident staining appears yellow. Scale bar, 5 µm. Lower right panel: merged image following staining for SipA with anti-SipA IgG/AlexaFluor 488-conjugated anti-rabbit IgG (green), SipC with anti-SipC IgG/Texas Red-conjugated anti-sheep IgG (red) and cellular F-actin with AlexaFluor 350-conjugated phalloidin (blue). Scale bar, 0.3 µm.
Discussion
In eukaryotic cells, remodelling of the cellular actin filament network is determined by ABPs that direct the nucleation of actin polymerization or the destabilization of F-actin to promote rapid, spatial control of filament assembly or disassembly, respectively. Most cellular ABPs bundle, cross-link or stabilize F-actin to generate higher order supramolecular structures that sustain membrane deformations such as those required for lamellipodial or filopodial protrusions (Ayscough, 1998). Invasive bacterial pathogens achieve comparable manipulations by subverting cytoskeletal function. Most of their effectors act indirectly by stimulating or inhibiting a host cell signalling pathway involved in peripheral actin organization, or by modulating the activities of a host ABP (Chen et al., 1996; Persson et al., 1997; Tran Van Nhieu et al., 1997). It has recently emerged that, in contrast, the pathogenic bacterium Salmonella encodes two invasion proteins, SipC and SipA, which interact directly with cellular actin and modulate its dynamics (Hayward and Koronakis, 1999; Zhou et al., 1999a).
Our data show that Salmonella SipA substantially potentiates the nucleation of actin polymerization and F-actin bundling, which are mediated by SipC in vitro. Comparable effects were observed in vivo, SipA enhancing the ability of SipC to induce cytoskeletal rearrangements in semi-permeabilized cultured mammalian cells. To our knowledge, this is the first demonstration of specific cooperation between bacterial effector proteins that subvert target cell function. Such cooperation is also consistent with the phenotype of Salmonella sipA mutants that, although able to enter cultured mammalian cells, induce diffuse rather than localized actin polymerization and cytoskeletal rearrangements. This results in a decreased invasion efficiency in comparison with wild-type strains over short infection times (Zhou et al., 1999a). While the semi-permeabilization assay provided confirmation of the detailed in vitro experiments, it should be emphasized that during bacterial entry such dramatic cytoskeletal rearrangements do not occur throughout the entire cell. It is therefore likely that high local concentrations of these effectors direct cytoskeletal rearrangements at sites of bacterial contact.
Comparison of our findings with those describing eukaryotic ABPs suggests possible mechanisms by which SipA could stimulate SipC-mediated actin nucleation. SipA-induced reduction of the critical actin concentration could indirectly decrease the threshold SipC concentration required to induce nucleation, while also favouring rapid filament elongation. This could result from an interaction between SipA and actin monomers or dimers before nucleation (Mitra et al., 2000), or from SipA binding and stabilization of SipC-nucleated actin filaments. Alternatively, an actin-dependent or -independent SipA–SipC interaction may stimulate the formation of actin nuclei. In vitro studies have demonstrated that Listeria ActA and cellular WASP/Scar proteins bind and significantly enhance the nucleating activity of the Arp2/3 complex (reviewed by Svitkina and Borisy, 1999). Since N-WASP activity is further enhanced by binding of activated Cdc42 and phosphatidylinositol (4,5) bisphosphate (Rohatgi et al., 1999), and Scar assembles an actin-associated multi-kinase signalling scaffold (Westphal et al., 2000), WASP/Scar proteins are proposed to provide a direct connection between signal transduction pathways and the stimulation of actin polymerization (Rohatgi et al., 1999; Prehoda et al., 2000). It is tempting to speculate that a direct interaction between membrane-integral SipC and translocated SipA may determine localized efficient nucleation of actin polymerization beneath the membrane (Finlay et al., 1991). Nevertheless, since WASP/Scar and ActA bind G- rather than F-actin (Machesky and Insall, 1998; Miki and Takenawa, 1998; Zalevsky et al., 2000), and SipA has no primary sequence similarity to these proteins, any mechanistic similarities are at present speculative.
How could SipA and SipC cooperate in the early stages of bacterial internalization during infection? A possible scenario would be that SipC, potentially in association with co-secreted SipB, could insert into the target cell plasma membrane (Hayward and Koronakis, 1999; Scherer et al., 2000) from where SipA translocation could stimulate SipC-mediated nucleation of actin polymerization and enhance the subsequent bundling of actin filaments, which are stabilized beneath the invading bacteria. This would allow rapid, localized, cell-independent reorganization of the cytoskeleton at this very early stage in bacterial endocytosis. Other effector proteins, such as the inositol phosphatase SopB and the Rac-1/Cdc42-activating SopE (Hardt et al., 1998; Norris et al., 1998), subvert host cell signal transduction pathways, amplify cytoskeletal rearrangements and recruit host ABPs to the sites of bacterial entry. Further in vitro and in vivo analysis of effector protein interplay will be needed to establish a comprehensive view of how Salmonella spatially and temporally subverts host cell function during the complex internalization process.
Materials and methods
Expression and purification of recombinant SipA and SipC proteins
Plasmid pHSipA containing the entire sipA was generated by PCR amplification from the S.typhimurium SJW1103 chromosome (Yamaguchi et al., 1984). PCR products, engineered to contain NdeI and BamHI sites at the start codon ATG and 3′ of the stop codon, respectively, were ligated into the corresponding sites of T7 expression vector pET15b (Novagen), allowing an in-frame N-terminal fusion of His6. The resulting recombinant plasmid, pHSipA, directed high level expression of SipA in laboratory E.coli BL21 (DE3). Transformed E.coli BL21 (DE3) cells (Studier and Moffat, 1986) were cultured in 2 l of aerated tryptone yeast (TY) medium with additional 50 µg/ml ampicillin at 37°C to A600 = 1.0 and induced with 0.1 mM isopropyl-β-d-thio galactopyranoside (IPTG) for 2 h. Cells were pelleted (10 000 g, 10 min), resuspended in 1/20 culture volume and lysed through a French Press (82 800 kPa, Aminco). Full-length His-tagged SipA inclusion bodies were pelleted, washed with 1 M NaCl, 2% (v/v) Triton X-100 then distilled water, solubilized in binding buffer [6 M guanidine hydrochloride (GuHCl), 20 mM Tris–HCl pH 8, 150 mM NaCl, 5 mM imidazole], and bound to and eluted from nickel nitrilotri acetic acid–agarose (Qiagen) under denaturing conditions according to the manufacturer’s instructions. Eluted SipA was concentrated by precipitation with 80% (v/v) ethanol before resuspension in 6 M GuHCl, 20 mM 2-mercaptoethanol. SipA was refolded by rapid dilution of the concentrated stock 1:100 with F-buffer [20 mM Tris–HCl pH 8, 100 mM KCl, 1 mM MgCl2, 0.1 mM ATP, 0.1 mM CaCl2, 0.5 mM dithiothreitol (DTT), 0.01% (v/v) NaN3], dialysed against F-buffer (5 × 1 l) and subsequently filtered through a 0.2 µm syringe filter before use. SipC and SipC-C were purified and concentrated to 600 µM as described previously (Hayward and Koronakis, 1999). Purified protein concentration was determined with a Coomassie Blue-based staining solution (Pierce).
Actin binding assays
F-actin was pre-assembled from purified rabbit skeletal muscle actin (2 µM) in G-buffer [5 mM Tris–HCl pH 8, 0.1 mM ATP, 0.2 mM CaCl2, 0.02% (v/v) NaN3] by the addition of 0.02 vols 50× initiation buffer (100 mM MgCl2, 50 mM ATP, 2.5 M KCl) (Cytoskeleton Inc.) and incubated for 30 min at 25°C. For co-sedimentation, mixtures of the full-length SipA protein and F-actin in F-buffer were centrifuged (100 000 g, 25 min, 4°C) and the supernatants and pellets analysed by Coomassie Blue-stained SDS–PAGE. To determine the stoichiometry of the SipA–F-actin binding, a constant concentration of F-actin was incubated with a range of SipA concentrations and the resulting pellets and supernatants were analysed as above. Densitometric quantitation of pelleted SipA was made from the Coomassie Blue-stained SDS–PAGE gel. For the low-speed sedimentation assay, a 1:1 mixture (2 µM) of SipA to F-actin was centrifuged (12 000 g, 15 min, 4°C) and pellets and supernatants analysed as above.
Actin bundling assay (light scattering)
F-actin bundling was monitored by light scattering (25°C, excitation 520 nm, emission 520 nm, slit widths 2.5 nm) in an LS50B fluorescence spectrophotometer (Perkin Elmer). SipA and/or SipC was added to preassembled F-actin (2.5 µM) in different combinations in F-buffer and monitored for 30 min at 25°C.
Actin polymerization assay
Pyrene-labelled G-actin and unlabelled G-actin were mixed 1:10 in fresh G-buffer (2.5 µM), equilibrated on ice and centrifuged to remove F-actin. Initiation buffer (50×, 0.02 vols) was used to trigger actin polymerization, and pyrene fluorescence was monitored by spectrophotometry (excitation 365 nm, emission 385 nm, slit widths 5 nm) using FLWinLab software (Perkin Elmer). Purified SipA and/or SipC-C (or control buffers) was added before or during polymerization. His tag alone has no effect on the polymerization of actin (Welch et al., 1998).
Actin depolymerization assay
Pyrene-labelled F-actin was pre-assembled by polymerization of purified pyrene-labelled and unlabelled G-actin mixed 1:10 in fresh G-buffer (24 µM) using 0.02 vols 50× initiation buffer (45 min, 25°C), diluted to 5 µM in F-buffer and equilibrated for 1 h at room temperature. F-actin aliquots, vortexed reproducibly, were mixed with equal concentrations of SipA or phalloidin. F-actin was diluted to 0.1 µM in F-buffer and the mixture transferred to a fluorimeter cuvette. Fluorescence intensity (excitation 365 nm, emission 385 nm, slit widths, 5 nm) was measured over incubation time. To account for fluorescence quenching induced by SipA or phalloidin binding, the percentage of F-actin remaining was determined from the initial fluorescence of each mixture at t = 0.
Electron microscopy
Proteins were adsorbed on to freshly glow-discharged, carbon-coated copper grids for 1–2 min and washed in water before negative staining with 2% (w/v) uranyl acetate or 2% (w/v) phosphotungstic acid pH 7.0 (Harris, 1997). Grids were examined using a Philips CM100 transmission electron microscope operated at 80 kV.
Cell permeabilization
SipC (600 µM) in 4 M GuHCl was diluted to 5 µM in 1 ml of 1 mM MES pH 6, and dialysed extensively against 5 × 5 l of the same buffer. This stock was diluted appropriately in 50 mM HEPES pH 7.3, 100 mM KCl, 3 mM MgCl2, 0.1 mM DTT, 0.2% (w/v) bovine serum albumin (BSA) (permeabilization buffer). SipA (250 µM) in 6 M GuHCl, 20 mM 2-mercaptoethanol was similarly refolded by dilution to 2.5 µM in, and dialysed against, permeabilization buffer without BSA. Swiss 3T3 cells were grown (37°C, 10% CO2) in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% (v/v) fetal calf serum, l-glutamine and penicillin/streptomycin (Sigma). Cells were plated on to 13 mm diameter glass coverslips (BDH). After 12 h, semi-confluent cells were washed with warm phosphate-buffered saline (PBS) and incubated in serum-free DMEM for a further 36–48 h. After washing with permeabilization buffer, coverslips were inverted into 100 µl droplets of permeabilization buffer with additional 1 mM ATP, 100 µM GTP, 100 µM UTP and 0.0002% (w/v) saponin. Following incubation (37°C, 5 min), permeabilized cells were washed three times with the same buffer lacking saponin, and incubated in 100 µl of buffer (without saponin) containing SipC and/or SipA as appropriate (37°C, 5 min). Cells were fixed after treatment in 3.7% formaldehyde [15 min, room temperature (RT)], and processed for fluorescence microscopy. For each representative frame, >200 cells were observed from three independent experiments.
Fluorescence microscopy
Fixed samples were permeabilized with PBS, 0.2% Triton X-100 (PBST) for 10 min at RT, then blocked in PBST containing 3% (w/v) BSA (1 h, RT). For single staining, blocked samples were incubated (1 h, RT) with 1:250 dilution in PBST/BSA of anti-SipC or anti-SipA IgG purified from polyclonal antisera raised in rabbits. For double staining, samples were sequentially incubated with 1:200 dilution of anti-SipC IgG purified from polyclonal antisera raised in sheep (1 h, RT), then 1:250 dilution of rabbit anti-SipA IgG (1 h, RT), both in PBST/BSA. AlexaFluor 488 anti-rabbit IgG (Molecular Probes) and Texas Red-conjugated anti-sheep IgG (Vector Laboratories) diluted in PBST/BSA at recommended concentrations were used to detect SipA and/or SipC, and AlexaFluor 350 or Texas Red-conjugated phalloidin (Molecular Probes) to visualize the actin cytoskeleton. Coverslips were mounted on to slides using ProLong Antifade reagent (Molecular Probes) and visualized under a fluorescence microscope (Leica DM IRBE). Images were captured using a CCD digital camera (Hamamatsu) and processed using OpenLab software and Photoshop 5.5 (Adobe).
Acknowledgments
Acknowledgements
We thank Colin Hughes for critical discussion of this manuscript. R.D.H. thanks Collette Tourlamain for the gift of Swiss 3T3 cells and discussions. This work was supported by a Wellcome Trust project grant to V.K.
References
- Ayscough K.R. (1998) In vivo functions of actin-binding proteins. Curr. Opin. Cell Biol., 10, 102–111. [DOI] [PubMed] [Google Scholar]
- Chen L.-M., Hobbie,S. and Galán,J.E. (1996) Requirement of CDC42 for Salmonella-induced cytoskeletal and nuclear responses. Science, 274, 2115–2118. [DOI] [PubMed] [Google Scholar]
- Cooper J.A. (1987) Effects of cytochalasin and phalloidin on actin. J. Cell Biol., 105, 1473–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay B.B. and Cossart,P. (1997) Exploitation of mammalian host cell functions by bacterial pathogens. Science, 276, 718–725. [DOI] [PubMed] [Google Scholar]
- Finlay B.B., Ruschkowski,S. and Dedhar,S. (1991) Cytoskeletal rearrangements accompanying Salmonella entry into epithelial cells. J. Cell Sci., 99, 283–296. [DOI] [PubMed] [Google Scholar]
- Galán J.E. (1999) Interaction of Salmonella with host cells through the centisome 63 type III secretion system. Curr. Opin. Microbiol., 2, 46–50. [DOI] [PubMed] [Google Scholar]
- Garcia-Del Portillo F., Pucciarelli,M.G., Jefferies,W.A. and Finlay,B.B. (1994) Salmonella typhimurium induces selective aggregation and internalization of host cell surface proteins during invasion of epithelial cells. J. Cell Sci., 107, 2005–2010. [DOI] [PubMed] [Google Scholar]
- Goode B.L., Wong,J.J., Butty,A.-C., Peter,M., McCormack,A.L., Yates,J.R., Drubin,D.G. and Barnes,G. (1999) Coronin promotes the rapid assembly and cross-linking of actin filaments and may link the actin and microtubule cytoskeletons in yeast. J. Cell Biol., 144, 83–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardt W.-D., Chen,L.-M., Schuebel,K.E., Bustelo,X.R. and Galán,J.E. (1998) Salmonella typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell, 93, 815–826. [DOI] [PubMed] [Google Scholar]
- Harris J.R. (1997) Negative Staining and Cryoelectron Microscopy. BIOS Scientific Publishers, Oxford, UK, in association with the Royal Microscopical Society.
- Hayward R.D. and Koronakis,V. (1999) Direct nucleation and bundling of actin by the SipC protein of invasive Salmonella. EMBO J., 18, 4926–4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward R.D., McGhie,E.J. and Koronakis,V. (2000) Membrane fusion activity of purified SipB, a Salmonella surface protein essential for mammalian cell invasion. Mol. Microbiol., 37, 727–739. [DOI] [PubMed] [Google Scholar]
- Kaniga K., Tucker,S., Trollinger,D. and Galán,J.E. (1995) Homologs of the Shigella IpaB and IpaC invasins are required for Salmonella typhimurium entry into cultured epithelial cells. J. Bacteriol., 177, 3965–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky L.M. and Insall,R.H. (1998) Scar1 and the related Wiskott–Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol., 8, 1347–1356. [DOI] [PubMed] [Google Scholar]
- Machesky L.M., Mullins,R.D., Higgs,H.N., Kaiser,D.A., Blanchion,L., May,R.C., Hall,M.E. and Pollard,T.D. (1999) Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl Acad. Sci. USA, 96, 3739–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H. and Takenawa,T. (1998) Direct binding of the verprolin-homology domain in N-WASP to actin is essential for cytoskeletal reorganization. Biochem. Biophys. Res. Commun., 243, 73–78. [DOI] [PubMed] [Google Scholar]
- Mitra K., Zhou,D. and Galán,J.E. (2000) Biophysical characterization of SipA, an actin-binding protein from Salmonella enterica. FEBS Lett., 482, 81–84. [DOI] [PubMed] [Google Scholar]
- Mullins R.D., Heuser,J.A. and Pollard,T.D. (1998) The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl Acad. Sci. USA, 95, 6181–6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris F.A., Wilson,M.P., Wallis,T.S., Galyov,E.E. and Majerus,P.W. (1998) SopB, a protein required for virulence of S. dublin, is an inositol phosphate phosphatase. Proc. Natl Acad. Sci. USA, 95, 14057–14059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson C., Carballeira,N., Wolf-Watz,H. and Fällman,M. (1997) The PTPase YopH inhibits uptake of Yersinia, tyrosine phosphorylation of p130Cas and FAK, and the associated accumulation of these proteins in peripheral focal adhesions. EMBO J., 16, 2307–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prehoda K.E., Scott,J.A., Mullins,R.D. and Lim,W.A. (2000) Integration of multiple signals through cooperative regulation of the N-WASP–Arp2/3 complex. Science, 290, 801–806. [DOI] [PubMed] [Google Scholar]
- Rohatgi R., Ma,L., Miki,H., Lopez,M., Kirchhausen,T., Takenawa,T. and Kirschner,M.W. (1999) The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell, 97, 221–231. [DOI] [PubMed] [Google Scholar]
- Scherer C.A., Cooper,E. and Miller,S.I. (2000) The Salmonella type III secretion translocation protein SspC is inserted into the epithelial cell plasma membrane upon infection. Mol. Microbiol., 37, 1133–1145. [DOI] [PubMed] [Google Scholar]
- Studier W. and Moffat,B.A. (1986) Use of bacteriophage T7 RNA polymerase to direct selective high level expression of cloned genes. J. Mol. Biol., 189, 113–130. [DOI] [PubMed] [Google Scholar]
- Svitkina T.M. and Borisy,G.G. (1999) Progress in protrusion: the tell-tale scar. Trends Biochem. Sci., 24, 432–436. [DOI] [PubMed] [Google Scholar]
- Tran Van Nhieu G. and Sansonetti,P.J. (1999) Mechanism of Shigella entry into epithelial cells. Curr. Opin. Microbiol., 2, 51–55. [DOI] [PubMed] [Google Scholar]
- Tran Van Nhieu G., Ben Ze’ev,A. and Sansonetti,P.J. (1997) Modulation of bacterial entry into epithelial cells by interaction between vinculin and Shigella IpaA invasin. EMBO J., 16, 2717–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran Van Nhieu G., Caron,E., Hall,A. and Sansonetti,P.J. (1999) IpaC induces actin polymerization and filopodia formation during Shigella entry into epithelial cells. EMBO J., 18, 3249–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M.D., Rosenblatt,J., Skoble,J., Portnoy,D.A. and Mitchison, T.J. (1998) Interaction of human Arp2/3 complex with the Listeria monocytogenes ActA protein in actin filament nucleation. Science, 281, 105–108. [DOI] [PubMed] [Google Scholar]
- Westphal R.S., Soderling,S.H., Alto,N.M., Langeberg,L.K. and Scott, J.D. (2000) Scar/WAVE-1, a Wiskott–Aldrich syndrome protein, assembles an actin-associated multi-kinase scaffold. EMBO J., 19, 4589–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Fujita,H., Sugata,K., Taira,T. and Iino,T. (1984) Genetic analysis of H2, the structural gene for phase-2 flagellin in Salmonella. J. Gen. Microbiol., 130, 255–265. [DOI] [PubMed] [Google Scholar]
- Zalevsky J., Grigorova,I. and Mullins,R.D. (2001) Activation of the Arp2/3 complex by the Listeria ActA protein: ActA binds two actin monomers and three subunits of the Arp2/3 complex. J. Biol. Chem., 276, 3468–3475. [DOI] [PubMed] [Google Scholar]
- Zhou D., Mooseker,M.S. and Galán,J.E. (1999a) Role of the Salmonella actin-binding protein SipA in bacterial internalization. Science, 283, 2092–2095. [DOI] [PubMed] [Google Scholar]
- Zhou D., Mooseker,M.S. and Galán,J.E. (1999b) An invasion-associated Salmonella protein modulates the actin-binding activity of plastin. Proc. Natl Acad. Sci. USA, 96, 10176–10181. [DOI] [PMC free article] [PubMed] [Google Scholar]