Abstract
The association of the U4/U6⋅U5 tri-snRNP with pre-spliceosomes is a poorly understood step in the spliceosome assembly pathway. We have identified two human tri-snRNP proteins (of 65 and 110 kDa) that play an essential role in this process. Characterization by cDNA cloning of the 65 and 110 kDa proteins revealed that they are likely orthologues of the yeast spliceosomal proteins Sad1p and Snu66p, respectively. Immunodepletion of either protein from the HeLa cell nuclear extracts inhibited pre-mRNA splicing due to a block in the formation of mature spliceosomes, but had no effect on the integrity of the U4/U6⋅U5 tri-snRNP. Spliceosome assembly and splicing catalysis could be restored to the respective depleted extract by the addition of recombinant 65 or 110 kDa protein. Our data demonstrate that both proteins are essential for the recruitment of the tri-snRNP to the pre-spliceosome but not for the maintenance of the tri-snRNP stability. Moreover, since both proteins contain an N-terminal RS domain, they could mediate the association of the tri-snRNP with pre-spliceosomes by interaction with members of the SR protein family.
Keywords: RNA splicing/snRNP proteins/spliceosome assembly/SR proteins/U4/U6⋅U5 tri-snRNPs
Introduction
Catalysis of the two transesterification steps of the pre-mRNA splicing reaction takes place in the spliceosome, a large ribonucleoprotein complex that is formed by the ordered interaction of numerous splicing factors and the small nuclear ribonucleoproteins (snRNPs) U1, U2, U5 and U4/U6 with the pre-mRNA (reviewed by Krämer, 1996; Reed and Palandjian, 1997; Staley and Guthrie, 1998; Burge et al., 1999). Splicing complex formation involves the initial interaction of the U1 snRNP with the 5′ splice site and of the U2 snRNP with the branch site, respectively, to form the so-called pre-spliceosome or complex A. Independently of this, the U4/U6 and U5 snRNPs combine to form a U4/U6⋅U5 tri-snRNP. The tri-snRNP, together with an as yet unknown number of non-snRNP splicing factors, associates with the pre-spliceosome to form the mature spliceosome, termed complex B, whereafter the two transesterification steps can occur.
During spliceosome assembly, a complex network of RNA–RNA interactions is formed between the U1, U2, U4/U6 and U5 snRNAs and the pre-mRNA, as well as between a subset of the snRNAs themselves. During pre-spliceosome assembly, the U1 and U2 snRNAs base-pair with the 5′ splice site and the branch site, respectively. Upon integration of the U4/U6⋅U5 tri-snRNP complex, several RNA conformational changes occur. For example, the U4 and U6 snRNAs, which are base-paired within the tri-snRNP complex, dissociate, allowing U6 to base-pair with the 5′ end of U2 snRNA and also with intron sequences at the 5′ splice site. In addition, U5 snRNA interacts with exon nucleotides at the 5′ splice site and, prior to the second step of splicing, additionally with exon nucleotides at the 3′ splice site (reviewed by Nilsen, 1998; Staley and Guthrie, 1998).
In addition to RNA–RNA interactions, protein–protein and protein–RNA interactions are equally important for proper assembly of the spliceosome. In metazoans, members of the SR protein family, a group of essential splicing factors with characteristic arginine–serine C-terminal repeats (RS domain) and one or two N-terminal RNA-binding domains, appear to play an important role at early stages of spliceosome assembly. For example, in addition to the base-pairing interaction between the 5′ end of the U1 snRNA and the 5′ splice site, stable binding of U1 snRNP is mediated by protein–protein contacts involving the RS domains of the SR protein SF2/ASF and the SR-related U1-70K (Kohtz et al., 1994). SR-related proteins contain RS domains but are otherwise structurally and functionally distinct from the SR family proteins (reviewed in Fu, 1995; Blencowe et al., 1999). Moreover, interactions between SR and SR-related proteins are thought to mediate communication between the 5′ and 3′ splice sites during the early stages of spliceosome assembly (for reviews see Fu, 1995; Manley and Tacke, 1996; Stark et al., 1998).
Little is known about the interactions that promote the association of the U4/U6⋅U5 tri-snRNP with pre-spliceosomes. Base-pairing interactions between U5 snRNA and exon–intron junctions have been demonstrated (see above); however, these interactions are too weak to ensure stable binding. It is therefore very likely that protein–RNA and protein–protein interactions will play a dominant role in establishing the initial contacts of the tri-snRNP with the pre-spliceosome. Consistent with this idea, the 5′ splice site and, later in the reaction, the 3′ splice site is contacted by the U5 220 kDa protein (Prp8p in yeast; Teigelkamp et al., 1995; Siatecka et al., 1999), which is likely to stabilize the U5 snRNA interactions with the splice sites. Moreover, indirect evidence has been provided that SR proteins are necessary to recruit the tri-snRNP to the spliceosome (Roscigno and Garcia-Blanco, 1995). However, it is not clear at present which, if any, of the human tri-snRNP proteins is essential for the pre-spliceosome/spliceosome transition.
The purified U4/U6⋅U5 tri-snRNP complex from HeLa cells contains ∼30 distinct proteins, most of which are evolutionarily conserved (reviewed in Will and Lührmann, 1997; see also Gottschalk et al., 1999; Stevens and Abelson, 1999). A set of seven Sm proteins is associated with both the U4 and U5 snRNA, generating two Sm core domains. In addition, one set of the seven Sm-like proteins (LSm 2–8) is bound to U6 snRNA (Achsel et al., 1999; Salgado-Garrido et al., 1999). Of the remaining tri-snRNP proteins, five, with molecular masses of 15.5, 20, 60, 61 and 90 kDa, have been found by biochemical fractionation and/or immunoprecipitation to be associated with the U4/U6 snRNP (Horowitz et al., 1997; Lauber et al., 1997; Teigelkamp et al., 1998; Nottrott et al., 1999; O.Makarova, E.Makarov and R.Lührmann, in preparation). Eight proteins with molecular masses of 15, 40, 52, 100, 102, 116, 200 and 220 kDa are also present in purified 20S U5 snRNPs and thus operationally defined as U5 specific (Bach et al., 1989). Three proteins with molecular masses of 27, 65 and 110 kDa, though abundant in purified tri-snRNPs, are associated less stably or not at all with either of its constituent U4/U6 or U5 snRNP particles when these are isolated. Of these, only the SR-related 27 kDa protein has been characterized by cDNA cloning (Fetzer et al., 1997).
We demonstrate here that the 65 and 110 kDa tri-snRNP proteins are essential for the recruitment of the tri-snRNP to the pre-spliceosome but not for the maintenance of tri-snRNP stability. Characterization by cDNA cloning and sequencing revealed that the 65 and 110 kDa proteins are orthologous to the yeast Sad1p and Snu66p spliceosomal proteins. The 65 kDa protein exhibits structural similarities to members of the family 2 of ubiquitin C-terminal hydrolases (UCHs), while the C-terminal domain of the 110 kDa protein appears to act as a tumour antigen and an autoallergen (Shichijo et al., 1998; Valenta et al., 1998). Both human proteins contain N-terminal RS domains, indicating that they potentially are involved in SR protein-mediated protein–protein interactions between the tri-snRNP and pre-spliceosomes.
Results
Cloning of the cDNAs encoding the human 65 and 110 kDa tri-snRNP proteins
The 65 and 110 kDa proteins were isolated from purified HeLa U4/U6⋅U5 tri-snRNPs and subjected to protein sequencing. The peptide sequences obtained for both proteins were used in database searches of expressed sequence tags (ESTs) and the corresponding cDNAs were identified and cloned as described in Materials and methods. The 65 kDa cDNA contains an open reading frame (ORF) that encodes a protein with 565 amino acids and a predicted mol. wt of 65 400 (see Figure 1). The 110 kDa cDNA encodes a protein with 800 amino acids and a predicted mol. wt of 90 263 (Figure 2). The slower than expected migration behaviour of the 110 kDa protein on SDS–gels (corresponding to an apparent mol. wt of 110 kDa) is assumed to arise from the glutamic acid-rich region comprising amino acids 450–600 (see also Graceffa et al., 1992). A cDNA termed SART-1 encoding the 110 kDa protein was identified previously as a gene encoding antigenic peptides of human squamous cell carcinomas recognized by cytotoxic T lymphocytes (Shichijo et al., 1998). An autoallergen (Hom s 1) against which patients suffering from severe atopic dermatitis often produce IgE autoantibodies appears to comprise a C-terminal domain of the 110 kDa protein (Valenta et al., 1998).
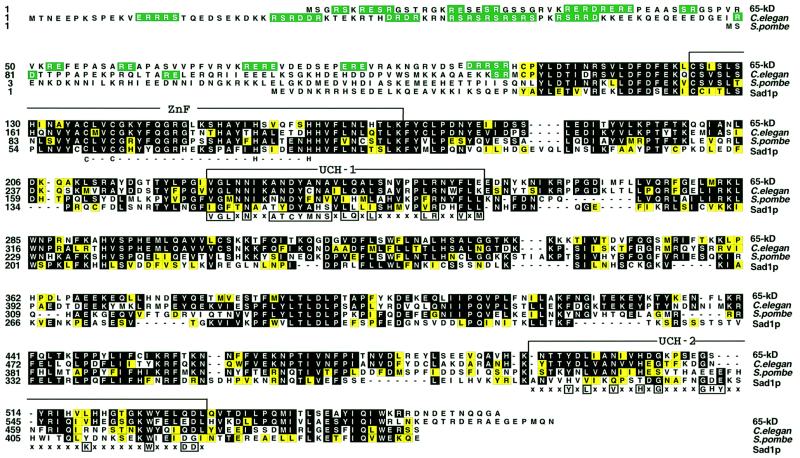
Fig. 1. Sequence alignment of the human 65 kDa protein with homologous proteins. The human 65 kDa protein is shown aligned by the Clustal method with the corresponding proteins from C.elegans, S.pombe and S.cerevisiae (Sad1p). Residues identical to those in the 65 kDa sequence are shown on a black background, and conserved residues (grouped DE, KRH, AFILMPVW and CGNQSTY) are shaded yellow. RS, RD and RE dipeptides in the N-terminal part of the sequence are shown on a green field. The domains are indicated above the sequences, as follows: ZnF, ubiquitin C-terminal hydrolase-like zinc finger; UCH-1 and UCH-2, ubiquitin C-terminal hydrolases family 2. Consensus residues that attribute the sequence to a particular domain are shown below each domain.
Fig. 2. Sequence alignment of the human 110 kDa protein with homologous proteins. The human 110 kDa protein is shown aligned by the Clustal method with the corresponding proteins from D.melanogaster, C.elegans, N.crassa, P.falciparum, S.pombe and S.cerevisae (Snu66p). Residues identical in at least four sequences are shown on a black background, and conserved residues, grouped as in Figure 1, are shaded yellow. RS, RD and RE dipeptides in the N-terminal part of the sequence are in green.
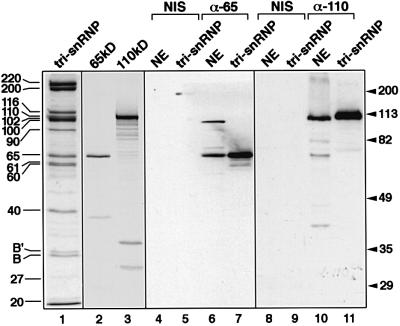
The following criteria were used to verify that the proteins obtained by translation of the respective cDNAs were identical to the human tri-snRNP 65 and 110 kDa proteins. First, the cDNAs contained all the peptides obtained by microsequencing, with 100% matches for all of the amino acid sequences. Secondly, as shown in Figure 3 (lanes 1–3), the transcription/translation products of the cDNAs encoding the 65 and 110 kDa proteins co-migrate with the corresponding proteins isolated from native U4/U6⋅U5 tri-snRNPs. Finally, antisera raised in rabbits against recombinant proteins or peptides react specifically with the corresponding proteins from HeLa cells, both in tri-snRNPs and in nuclear extracts. This is shown for the 65 kDa protein in Figure 3, lanes 4–7. In nuclear extracts, this antibody, which was raised against recombinant 65 kDa, also cross-reacts with a protein that has a molecular mass of ∼100 kDa and is not present in isolated tri-snRNPs. For the 110 kDa protein, antibodies were raised against an N-terminal peptide (residues 117–137), anti-pep 110, and against the C-terminal part of the protein (amino acids 667–800), anti-C110. Both antisera, but not the pre-immune sera, react specifically with the 110 kDa protein (shown for anti-C110 antibodies in Figure 3, lanes 8–11).
Fig. 3. Authenticity of the cDNAs encoding the 65 and 110 kDa proteins. Lane 1: proteins of purified U4/U6⋅U5 tri-snRNPs (indicated on the left) were separated by 10% SDS–PAGE and stained with Coomassie blue. Lanes 2 and 3: the 65 kDa (lane 2) or 110 kDa (lane 3) proteins, prepared by translation in vitro from the corresponding cDNAs in the presence of [35S]methionine, were analysed by 10% SDS–PAGE and made visible by fluorography. Lanes 4–11: western blot of HeLa nuclear extracts (lanes 4, 6, 8 and 10; NE) and purified U4/U6⋅U5 tri-snRNPs (lanes 5, 7, 9 and 11; tri-snRNP) probed with antisera against the 65 kDa (lanes 6 and 7; α-65) or 110 kDa protein (anti-C110) (lanes 10 and 11; α-110). Control experiments with the corresponding pre-immune sera are shown in lanes 4 and 5, and 8 and 9, respectively (NIS). Molecular weight markers are shown on the right.
Human 65 kDa protein contains an RS domain and is related to ubiquitin C-terminal hydrolases
The 65 kDa protein contains an N-terminal domain of ∼100 amino acid residues that is enriched in arginine (∼23%), serine (∼16%) and glutamic acid (∼19%). Several of the arginine and glutamic acid residues form mixed-charged clusters interspersed with RS dipeptides (Figure 1). The biochemical character of this RS domain is thus different from that of the RS domains of classical SR proteins such as SF2/ASF, which contain consecutive RS dipeptides (Ge et al., 1991; Krainer et al., 1991). However, it resembles the RS domains of SR-related proteins such as U1-70K and the tri-snRNP 27 kDa protein which, in addition to RS dipeptides, also contain consecutive RE and/or RD dipeptide repeats.
The PROSITE database search (Bairoch et al., 1996) with the sequence of the 65 kDa protein revealed the presence of three domains characteristic of family 2 of UCHs. These domains are indicated in Figure 1 and consist of a zinc finger (Smart/ZnF_UBP) along with UCH-1 (Pfam/pfam00442) and UCH-2 (Pfam/pfam 00443) domains. In addition database searches also revealed likely orthologues of the 65 kDa protein in various eukaryotic species including Drosophila melanogaster (AAF48897, not shown), Caenorhabditis elegans (T42401), Schizosaccharomyces pombe (T40551), Arabidopsis thaliana (CAB52815, not shown) and Saccharomyces cerevisiae (NP_011195). The S.cerevisiae counterpart of the 65 kDa protein is termed Sad1p and has been shown recently to be essential for pre-mRNA splicing in vivo (Lygerou et al., 1999). Except for the N-terminal domains of the putative orthologues of the 65 kDa protein, which vary in sequence and length among species, the C-terminal parts of the proteins (starting with residue 107 of the human 65 kDa protein) exhibit a high degree of evolutionary conservation. For example, the overall similarity between the human 65 kDa protein and the yeast Sad1 protein is 65%, with regions of homology found throughout the entire sequence.
Human 110 kDa protein is an SR-related protein
The human 110 kDa protein contains an N-terminal domain of ∼120 amino acid residues that include several consecutive RS and RE/RD dipeptide repeats (Figure 2), placing it in the growing group of SR-related proteins. The remaining part of the 110 kDa sequence does not contain known motifs that would suggest additional potential functions of this protein.
A database search with the sequence of the 110 kDa protein revealed likely orthologous proteins in various species such as rat (ABO14722), mouse (ABO4721), A.thaliana (AC01842), D.melanogaster (AAF53138), C.elegans (AAB52287), Neurospora crassa (CAB 91423), Plasmodium falciparum (CAB39050), S.pombe (CAA22848) and S.cerevisiae (SCYOR308, Snu66p). Figure 2 shows selected examples of these. Interestingly, the homologue in S.cerevisiae, Snu66p, has been shown recently to be a component of the yeast U4/U6⋅U5 tri-snRNP (Gottschalk et al., 1999; Stevens and Abelson, 1999) and to be essential for pre-mRNA splicing (Gottschalk et al., 1999). A distinctive feature of the proteins from higher eukaryotes is an N-terminal RS domain (note the extended RS domain in the Drosophila homologue, Figure 2; this domain is absent in the yeast proteins). Regions of homology are found throughout the remaining portion of the putative 110 kDa orthologues. For example, the overall similarity between the human 110 kDa protein and Snu66p from S.cerevisiae is 46%.
Stability of the association of the 65 and 110 kDa proteins with U4/U6⋅U5 tri-snRNPs
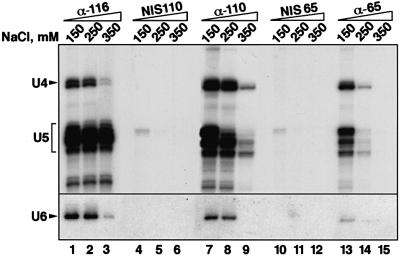
The 65 and 110 kDa proteins are present in almost stoichiometric amounts in isolated U4/U6⋅U5 tri-snRNPs. However, in isolated 13S U4/U6 snRNPs and 20S U5 snRNPs, they are either absent or present in very small quantities (Bach et al., 1989; O.Makarova, E.Makarov and R.Lührmann, in preparation). We therefore investigated the strength of the association between these proteins and U4/U6⋅U5 tri-snRNPs in nuclear extracts by performing immunoprecipitations at various salt concentrations using antibodies against the 65 and 110 kDa proteins. For comparison, we used the 116 kDa protein, which is known to be stably integrated into the U5 snRNP (Fabrizio et al., 1997). As shown in Figure 4, the reference antibody, anti-116 kDa, precipitated the U4/U6⋅U5 tri-snRNP at salt concentrations from 150 to 250 mM with similar efficiency (lanes 1 and 2). At 350 mM NaCl, where the tri-snRNP complex dissociates into U5 and U4/U6 particles, anti-116 kDa antibodies precipitated predominantly the U5 snRNP (lane 3). This is consistent with our earlier observation that the 116 kDa protein is strongly associated with the U5 snRNP (Fabrizio et al., 1997).
Fig. 4. Immunoprecipitation of snRNPs associated with the 65 or the 110 kDa protein. SnRNP particles were precipitated from HeLa nuclear extract, at the salt concentrations indicated above the lanes, by using anti-116 kDa (lanes 1–3), anti-pep 110 kDa (lanes 7–9), anti-65 kDa (lanes 13–15) and the pre-immune serum corresponding to anti-pep 110 kDa (lanes 4–6) or anti-65 kDa (lanes 10–12). Co-precipitated RNAs were extracted, labelled with [32P]pCp, fractionated by 10% urea–PAGE and detected by autoradiography. The positions of snRNAs are indicated on the left. Since U6 snRNA is not labelled efficiently with [32P]pCp (Lund and Dahlberg, 1992), its presence in the various immunoprecipitates was detected by northern blot hybridization (lower panel).
Antibodies against the 110 kDa protein precipitated the tri-snRNP at 150 and 250 mM NaCl with an efficiency similar to that of the anti-116 kDa antibody (compare lanes 7 and 8 with lanes 1 and 2), implying a stable association between this protein and the tri-snRNP. At 350 mM NaCl, where the tri-snRNP is largely dissociated (lane 3), only small quantities of U5 and U4 snRNAs are co-precipitated (lane 9); however, these quantities are roughly equal, which suggests that they represent residual amounts of intact tri-snRNPs. Thus, the 110 kDa protein appears to bind stably only to the tri-snRNP complex as a whole rather than to either of its snRNP constituents.
Analogous experiments with the 65 kDa protein showed that its association with the U4/U6⋅U5 tri-snRNP complex is substantially more sensitive to salt than that of the 110 kDa protein. While the tri-snRNP is still precipitated efficiently at 150 mM NaCl (Figure 4, lane 13), only small quantities of U4/U6 und U5 snRNPs are found in the immunoprecipitate at 250 mM (lane 14), even though at this salt concentration the tri-snRNP is largely intact (as evidenced by the co-precipitation of U4/U6 snRNP with U5 snRNP by the anti-116 kDa antibody at 250 mM NaCl; Figure 4, lane 2). At 350 mM, where the tri-snRNP is dissociated into its subunits, no snRNPs are precipitated by anti-65 kDa antibodies (lane 15). The specificity of the immunoprecipitations with anti-110 kDa and anti-65 kDa is confirmed by the failure of the respective pre-immune sera to precipitate significant quantities of the tri-snRNP at any of the salt concentrations tested (compare lanes 7–9 and 13–15 with lanes 4–6 and 10–12, respectively, in Figure 4). Together, these results suggest that the stable binding of the 65 and 110 kDa proteins requires the presence of both tri-snRNP subunits. A similar conclusion was reached previously for the 27 kDa protein of the tri-snRNP (Fetzer et al., 1997). Thus, of the 29 currently known human tri-snRNP proteins, only three (27, 65 and 110 kDa) may be operationally defined as tri-snRNP-specific, in the sense that they are present in significant quantities in purified tri-snRNPs, but not in individual U4/U6 or U5 snRNPs.
Antibodies specific for human 65 and 110 kDa proteins inhibit pre-mRNA splicing in vitro
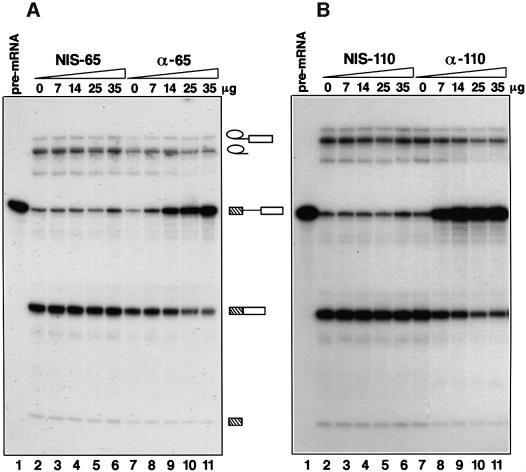
We next investigated the possible roles of the 65 and 110 kDa proteins in pre-mRNA splicing. Initially, we tested whether or not splicing could be inhibited by protein A-purified antibodies against recombinant 65 kDa protein or against the recombinant C-terminal region of the 110 kDa protein. Increasing amounts of either anti-65 kDa (Figure 5A, lanes 7–11) or anti-110 kDa (Figure 5B, lanes 7–11) inhibited the first step of the splicing reaction, as shown by the accumulation of unspliced pre-mRNA. Identical concentrations of IgGs purified from the respective non-immune sera (Figure 5A and B, lanes 2–6) had no effect. These results suggested that the 65 and 110 kDa proteins both play an important role in pre-mRNA splicing.
Fig. 5. The antibodies against the 65 kDa (A) or the 110 kDa protein (B) inhibit splicing in vitro. Splicing reactions containing different amounts of the protein A-purified anti-65 kDa or anti-110 kDa antibody (anti-C110) (lanes 7–11), or corresponding IgGs from pre-immune serum (lanes 2–6), were incubated for 60 min and analysed on a 14% denaturing polyacrylamide gel. The amount of added antibody is stated above each lane. Lane 1 shows the pre-mRNA added to each splicing reaction. Pre-mRNA, splicing intermediates and products are indicated schematically.
The 65 and 110 kDa proteins are essential for pre-mRNA splicing in vitro
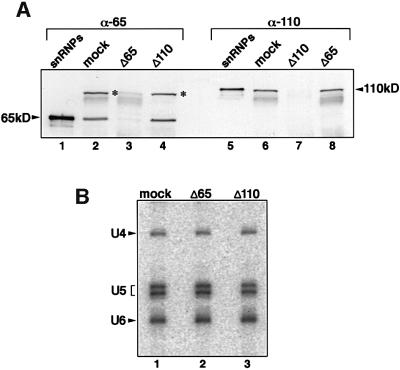
To investigate in more detail the specific roles of these two proteins in pre-mRNA splicing, we used anti-65 kDa and anti-110 kDa antibodies to prepare immunodepleted nuclear extracts. The depletion reactions were carried out at 500 mM NaCl to avoid the removal of U4/U6⋅U5 tri-snRNPs from the extracts. First, the efficiency of the immunodepletion was checked by immunoblotting. As shown in Figure 6A, the anti-65 kDa antibody removed the 65 kDa protein but not the 110 kDa protein from nuclear extracts (compare lanes 3 and 8). At the same time, the 100 kDa non-snRNP protein, with which the anti-65 kDa antibody cross-reacts (marked with asterisks), is also removed from the nuclear extract (Figure 6A, lane 3). Conversely, antibodies against the 110 kDa protein remove this protein quantitatively, while the 65 kDa protein remained in the nuclear extract (compare lanes 7 and 4). Control experiments in which nuclear extracts were incubated with the IgG fraction of the respective pre-immune sera showed no depletion of either the 65 (lane 2) or the 110 kDa (lane 6) protein. Thus, the 65 and 110 kDa proteins can be immunodepleted nearly quantitatively from the splicing extract independently of one another.
Fig. 6. Immunodepletion of the 65 or the 110 kDa protein does not disrupt the U4/U6⋅U5 tri-snRNPs. (A) Western blot of mock-depleted extracts (lanes 2 and 6), 65 kDa-depleted extracts (lanes 3 and 8) and 110 kDa-depleted extracts (lanes 4 and 7) probed with anti-65 kDa or anti-110 kDa antibodies as indicated above each lane. In lanes 1 and 5, total snRNP proteins were blotted as a control. The bands correspond ing to the 65 and 110 kDa proteins are indicated. The bands corres ponding to the uncharacterized protein recognized by the anti-65 kDa antibody are marked with asterisks. (B) Immunoprecipitation of snRNPs with anti-U5 116 kDa antibodies from the mock-depleted (lane 1), 65 kDa-depleted (lane 2) and 110 kDa-depleted (lane 3) extracts. RNAs were isolated and identified by northern blot analysis.
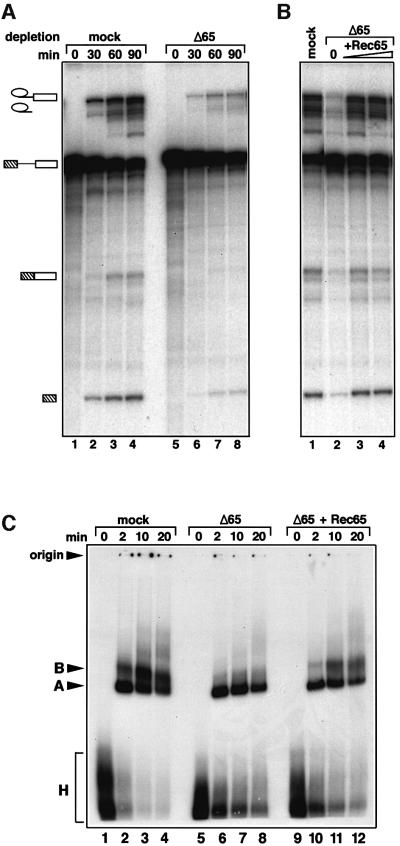
The depleted extracts were used to determine the effect of the absence of the 65 and 110 kDa proteins on the splicing reaction. Figure 7 shows the results of experiments using 65 kDa-depleted nuclear extract. Depletion led to strong inhibition of the splicing reaction, since the amounts of reaction products for both steps were greatly reduced (Figure 7A, lanes 5–8). This is made clear by comparison with a control reaction with ‘mock-depleted’ nuclear extracts (Figure 7A, lanes 1–4); this term is used in the present work to denote nuclear extracts that had been subjected to all stages of depletion, but without the antibody or with IgG from pre-immune serum (for details see Materials and methods). The inference that the inhibition is due specifically to the removal of the 65 kDa protein is demonstrated by the observation that the addition of recombinant 65 kDa protein restores splicing activity to the depleted nuclear extract (Figure 7B; note that treatment of the nuclear extract with anti-65 kDa IgG or IgG from pre-immune serum appears to activate a nuclease activity in the extract, leading to minor degradation of the splicing intermediates and products).
Fig. 7. The 65 kDa protein is required for splicing in vitro. (A) Immunodepletion of the 65 kDa protein from HeLa nuclear extract inhibits splicing in vitro. The time course of splicing reactions was monitored in 65 kDa-depleted (lanes 5–8) and mock-depleted extracts (lanes 1–4). The bands corresponding to pre-mRNA, intermediates and spliced products are indicated on the left. (B) Splicing activity of the depleted extract is restored by addition of the recombinant 65 kDa protein produced in E.coli. Splicing reactions containing the mock-depleted extract (lane 1), or 65 kDa-depleted extracts complemented with 0, 34 and 68 ng of the recombinant protein (lanes 2, 3, and 4), were incubated for 60 min and analysed as described in Materials and methods. (C) Recombinant 65 kDa protein promotes the transition from the A to B complex in the depleted extracts. The spliceosome assembly was analysed by native gel electrophoresis in mock-depleted extracts (lanes 1–4), 65 kDa-depleted extracts (lanes 5–8) and 65 kDa-depleted extracts complemented with 34 ng of recombinant 65 kDa protein (lanes 9–12). The bands corresponding to the H, A and B complexes as well as the gel origin are indicated on the left.
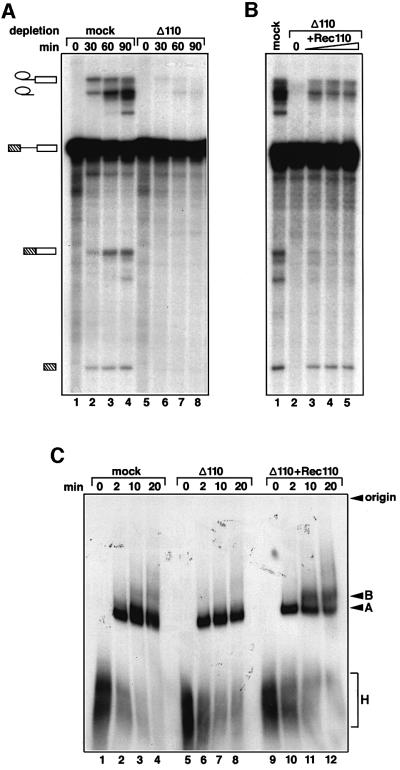
The results for the 110 kDa protein were qualitatively similar (Figure 8). Its removal led to complete inhibition of the splicing reaction, as shown in Figure 8A, lanes 5–8 (depleted extract), compared with lanes 1–4 (mock-depleted extract). Again, the inhibition appears to be caused specifically by the lack of the 110 kDa protein, since the addition of recombinant 110 kDa protein almost completely restored the first step of splicing, as is seen by the formation of the exon lariats and exon 1 (Figure 8B). The second step is also restored, albeit to a lesser extent, as is seen by the presence of the excised intron (Figure 8B). Although, only a low level of mRNA is observed after complementation with recombinant 110 kDa protein, it is also underrepresented in the mock extract (compared with the excised lariat). This may be due to its degradation in both the mock- and the antibody-treated extract.
Fig. 8. The 110 kDa protein is required for splicing in vitro. (A) Immunodepletion of the 110 kDa protein from HeLa nuclear extract inhibits splicing in vitro. The time course of splicing reactions was monitored in 110 kDa-depleted (lanes 5–8) and mock-depleted extracts (lanes 1–4). The bands corresponding to pre-mRNA, inter mediates and spliced products are indicated on the left. (B) Splicing activity of the depleted extract is restored by addition of the recombin ant 110 kDa protein expressed in E.coli. Splicing reactions containing the mock-depleted extract (lane 1), or 110 kDa-depleted extracts complemented with 0, 25, 50 and 100 ng of the recombinant protein (lane 2, 3, 4 and 5), were incubated for 60 min and analysed as described in Materials and methods. (C) The recombinant 110 kDa protein promotes the transition from the A to B complex in the depleted extracts. The spliceosome assembly was analysed by native gel electrophoresis in the mock-depleted extract (lanes 1–4), the 110-kDa-depleted extract (lanes 5–8) and the 110 kDa-depleted extract complemented with 50 ng of the recombinant 110 kDa protein (lanes 9–12). The bands corresponding to the H, A and B complexes as well as the gel origin are indicated on the left.
Taken together, these results demonstrate clearly that the splicing reaction depends upon the presence of both the 65 and the 110 kDa proteins. Neither of these proteins is redundant, i.e. neither one can replace the other. There are several conceivable explanations for this dependence. For example, the 65 and 110 kDa proteins could be involved either in maintaining the stability of the tri-snRNP or in its association with the pre-spliceosome (complex A).
Role of the 65 and 110 kDa proteins in the splicing process
To check for the formation of tri-snRNPs, immunoprecipitations were performed in nuclear extracts depleted of the 65 or 110 kDa protein. Figure 6B shows that in both cases intact U4/U6⋅U5 tri-snRNPs were precipitated from the nuclear extracts with antibodies directed against U5 116 kDa protein (lanes 2 and 3). Thus, the 65 and 110 kDa proteins are not essential for the stability of the tri-snRNPs.
We next examined the influence of the two proteins on the formation of the spliceosome. For this purpose, we analysed splicing complexes by polyacrylamide gel electrophoresis under native conditions. Figures 7C and 8C show that removal of the 65 or the 110 kDa protein blocks the transition from the pre-spliceosome (complex A) to the spliceosome (complex B); in both cases, the main species detected was the pre-spliceosome, with only minimal or no spliceosomes observed with the 65 kDa-depleted or 110 kDa-depleted extract, respectively (Figures 7C and 8C, compare lanes 5–8 with 1–4). After addition of recombinant 65 or 110 kDa protein to the respective immunodepleted splicing extracts, mature spliceosomes (complex B) were formed (Figures 7C and 8C, lanes 9–12) which correlates with the restoration of splicing activity (Figures 7B and 8B). We conclude that the 65 and 110 kDa proteins are both necessary for the association of the tri-snRNP with the pre-spliceosome, but not for the stability of the tri-snRNP itself.
Discussion
Requirement for the 65 and 110 kDa proteins in the binding of the U4/U6⋅U5 tri-snRNP to the pre-spliceosome, but not for the integrity of the tri-snRNP
The final step in the formation of the mature spliceosome is the binding of the U4/U6⋅U5 tri-snRNP to the pre-spliceosome (complex A; see Introduction). The results presented above demonstrate that the 65 and 110 kDa proteins are required for this step. In summary, our key observations are as follows. (i) Antibodies specific for the 65 or the 110 kDa protein inhibited the first step of splicing (Figure 5). (ii) Removal of either the 65 or the 110 kDa protein from the splicing extracts also inhibited the first step of splicing (Figures 7A and 8A). This is a result of a defect in spliceosome assembly, as spliceosome assembly in the absence of either of these proteins is stalled at the stage of complex A formation (Figures 7C and 8C). (iii) The formation of mature spliceosomes (complex B), and thus pre-mRNA splicing, was restored by complementation of the depleted extract with the respective recombinant protein (Figures 7 and 8). (iv) The alternative possibility that immunodepletion of the 65 or the 110 kDa protein might primarily disrupt the tri-snRNPs, and thus hinder the formation of complex B, was excluded by immunoprecipitation experiments that showed that tri-snRNP particles remained stable when either the 65 kDa or the 110 kDa protein was absent (Figure 6B). Moreover, the 65 and 110 kDa proteins have a relatively low affinity for the tri-snRNP particles, as demonstrated by their dissociation at relatively moderate salt concentrations (Figure 4). This low affinity supports the idea that they do not function as structural ‘scaffolding’ in the tri-snRNP. Rather, the experiments described above imply strongly that the function of the 65 and 110 kDa proteins is one of docking. Together, these observations indicate that the 65 and 110 kDa proteins are required for tethering the tri-snRNP particle to the spliceosome, rather than for stabilizing the tri-snRNP particle. To our knowledge, this is the first case where a mammalian tri-snRNP protein has been shown to participate directly in the recruitment of the tri-snRNP to the pre-spliceosome.
In this work, we have demonstrated that the 65 and 110 kDa proteins contain RS domains and thus belong to the expanding group of SR-related proteins. Two other proteins of the tri-snRNP are also known to contain RS domains, namely the 27 and 100 kDa proteins (Fetzer et al., 1997; Teigelkamp et al., 1997). This observation suggests that interactions between SR proteins and one or more of the tri-snRNP SR-related proteins may well contribute to the stable association of the U4/U6⋅U5 tri-snRNP complex with the pre-spliceosome. Consistent with this, indirect evidence has been provided previously that SR proteins are necessary to recruit the tri-snRNP to the spliceosome (Roscigno and Garcia-Blanco, 1995).
Orthology between the human 65 and 110 kDa tri-snRNP proteins and the yeast Sad1p and Snu66p proteins
The human 65 and 110 kDa proteins are evolutionarily conserved and are structurally homologous to proteins Sad1p and Snu66p, respectively, of the yeast S.cerevisiae (Figures 1 and 2). The similarities between the human and yeast counterparts are also apparent when their functions are considered. Sad1p has been shown to be essential for pre-mRNA splicing in vivo and to be required at or prior to the first catalytic step of splicing. This is consistent with our own observation, described herein, that the human 65 kDa protein is required for step 1 of splicing in vitro. Moreover, while Sad1p is not stably associated with the U4/U6⋅U5 tri-snRNP at 150 mM NaCl (Lygerou et al., 1999), significant amounts of it can be co-isolated together with tri-snRNPs from yeast cellular extracts at lower salt concentrations (75 mM NaCl; P.Fabrizio and R.Lührmann, unpublished data). Thus the salt sensitivity of tri-snRNP binding by yeast Sad1p resembles that of the human 65 kDa protein.
Sad1p appears to have a dual role in yeast cells. In addition to its requirement for splicing, it also plays a role in the biosynthesis of U4/U6 snRNPs in yeast (Lygerou et al., 1999). Specifically, evidence was provided that Sad1p is required for the de novo assembly of U4/U6 snRNPs, but not for the stability of the particle or reassembly after each round of splicing (Lygerou et al., 1999). The question of whether the 65 kDa protein also acts in the biosynthetic pathway of U4/U6 snRNPs in HeLa cells remains to be investigated. We can rule out, however, that depletion of the 65 kDa protein from nuclear extracts affects the stability of the U4/U6 snRNP and/or tri-snRNP in vitro (see Figure 6B).
Functional similarities between the yeast protein Snu66p and its human structural homologue, 110 kDa, are even more evident. Not only is Snu66p a component of the yeast U4/U6⋅U5 tri-snRNP (Gottschalk et al., 1999; Stevens and Abelson, 1999), but it is also essential for the first step of pre-mRNA splicing in vitro (Gottschalk et al., 1999). Moreover, and most significantly, using a fusion protein of Snu66p with GST, we have shown in pull-down experiments that Snu66p, at low salt concentrations, co-precipitates from yeast cellular extracts both tri-snRNPs and U2 snRNPs, most probably in the form of a [U2⋅U4/U6⋅U5] tetra-snRNP (Gottschalk et al., 1999). These data are consistent with the idea that Snu66p is involved in promoting the interaction between the tri-snRNP and the U2 snRNP in yeast, as we believe its human counterpart, 110 kDa, does in the human splicing apparatus. Thus, in view of the similar functions of the human 65 and 110 kDa proteins and yeast Sad1p and Snu66p, respectively, we conclude that the two protein pairs are orthologues.
A ubiquitin C-terminal hydrolase-related protein in the spliceosome
The human 65 kDa protein and its evolutionarily conserved orthologues exhibit significant homology with proteins known as ubiquitin C-terminal hydrolases (UCHs). The structural similarities include an N-terminal zinc finger domain and UCH-1- and UCH-2-like domains of UCH family 2 proteins (Figure 1). It will be interesting to investigate whether the 65 kDa protein plays a part in reactions involving ubiquitin. While, to our knowledge, no ubiquitinylated spliceosomal protein has yet been reported, the U2 snRNP protein SF3a120 has been shown to contain a C-terminal ubiquitin-like domain (Krämer et al., 1995). We note, however, that a functionally critical cysteine residue in the catalytic centre of UCHs is replaced in the 65 kDa protein and its orthologues by an aspartate (Asp234 in the 65 kDa protein, Figure 1). Moreover, one of the two histidine residues which are functionally important for UCHs is absent from the UCH-2-like domain of the 65 kDa protein (Figure 1). It is therefore questionable whether the 65 kDa protein exhibits hydrolase activity. It could however, function as a competitor of UCHs. Alternatively, the UCH-like domains in the 65 kDa protein may function as protein–protein interaction motifs. Clearly, more work is required to settle these interesting questions.
Materials and methods
Isolation of the 65 and 110 kDa proteins
Nuclear extracts were prepared from HeLa cells by the method of Dignam (1983). Total snRNPs and the 25S U4/U6⋅U5 tri-snRNPs were isolated as described previously (Laggerbauer et al., 1996). Proteins were extracted from tri-snRNPs and separated by SDS–PAGE. One-dimensional 12% SDS–PAGE clearly revealed at least three proteins with mol. wts in the 60 kDa range, of which the 60 and 61 kDa proteins have already been identified (Lauber et al., 1997; O.Makarova, E.Makarov and R.Lührmann, in preparation). The broad band exhibiting a mol. wt of 63–65 kDa was excised and the protein digested with trypsin or cyanogen bromide.
To separate the (at least three) proteins in the 100 kDa range, tri-snRNP proteins were fractionated in two dimensions, first by non-equilibrium pH-gradient gel electrophoresis (pH 3–11) and then by 9% SDS–PAGE (Teigelkamp et al., 1997). The spot corresponding to the 110 kDa protein was excised and the protein digested with trypsin. Peptides from the 65 and 110 kDa proteins were sequenced as described by Lauber et al. (1996).
Identification of cDNAs encoding the 65 and 110 kDa proteins
Fragments of the protein migrating in the range of 63–65 kDa were sequenced, yielding 12 oligopeptides in all: ERDREREPEAASSRGS PVRVK (residues 31–51 in the final sequence, Figure 1), REFEPA SAREAPASVVPFVR (52–71), SHAYIHSVQFSH (150–161), QQIAN LDK (200–207), AYXGTXYLPGXVGL (214–227), FLLVQRFGEL (271–280), LWNPRNFK (284–291), AHVSPHEMLQAVVLXSK (292–308), VESTF (384–386), TIVTDVFQGSMRIFTK (343–358), RFQLTK (440–445) and NNFFVEK (460–466). Database searches at NCBI using these peptide sequences revealed that all of them match the sequence of a single EST, AA176284 from the human Stratagene neuroepithelium cDNA library. The EST AA176284 was obtained commercially and re-sequenced. It contained an ORF that appeared to correspond to the full-length 65 kDa protein. The cDNA sequence for the human 65 kDa protein has been submitted to DDBJ/EMBL/GenBank under accession No. AF353989.
The microsequencing of the 110 kDa protein revealed two peptides: DGYKPDVKIEYVDETGRK (residues 702–719 in the final sequence, Figure 2) and EAFRQLSHRFHGK (724–736). Both of these were found in a number of human ESTs, as well as in two human proteins deposited in DDBJ/EMBL/GenBank: SART-1 (accession No. AB006198) and Hom s 1 (accession No. Y14314). The nucleotide sequence of Hom s 1 cDNA is identical to that of the SART-1 cDNA, except that the latter contains an additional stretch of 164 bases at its 5′ terminus of which 126 encode the N-terminal amino acids. To obtain independent evidence that SART-1 contains an ORF encoding the full-length 110 kDa protein, we isolated and sequenced the corresponding cDNA from a HeLa cell cDNA library. Two fragments were amplified by PCR from a HeLa Marathon cDNA library (Clontech): (i) a 587 bp fragment containing the 5′-untranslated region (UTR) and the N-terminus of the protein; and (ii) a 1081 bp fragment encoding a central part of the protein. The third fragment containing the C-terminus of the protein and the 3′-UTR and poly(A) tail was taken from the human EST R00027 that was obtained commercially. These three fragments were ligated to one another by using unique restriction sites, and were subcloned into the pBluescript KS(+) vector under the control of the T7 promoter. Several clones were sequenced to confirm the identity of the cDNA. The cDNA sequence for the human 110 kDa protein has been submitted to DDBJ/EMBL/GenBank under accession No. AF353625.
Production of the recombinant 65 and 110 kDa proteins
The full-length coding region of the 65 kDa protein was amplified by PCR from the cDNA and subcloned into the pQE-70 expression vector (Qiagen), resulting in a plasmid that was used to express the 65 kDa protein with a C-terminal histidine tag in Escherichia coli. The full-length coding region of the 110 kDa protein was cloned into the pET-28a expression vector (Novagen), resulting in a plasmid with which the N-terminally histidine-tagged 110 kDa protein was expressed in E.coli. The 65 and 110 kDa proteins were purified on an Ni-NTA column (Qiagen) under native conditions (20 mM Tris–HCl, pH 8.0; 0.3 M NaCl, 0.05% NP-40), dialysed against D-buffer (Dignam et al., 1983) and stored at –70°C.
Antibodies
Antibodies were raised in a rabbit against the full-length recombinant 65 kDa protein. Two antibodies, anti-pep 110 and anti-C110, were raised in rabbits against the 110 kDa protein. For anti-pep 110, the antigen was the conserved N-terminal peptide (residues 117–137) conjugated to ovalbumin. The anti-pep 110 antibodies were affinity purified by using a SulfoLink column (Pierce) with the immobilized peptide. For anti-C110, the antigen was the GST fusion of the C-terminal part of the protein (residues 667–800). The anti-65 kDa and anti-C110 antibodies were purified on protein A–Sepharose (Pharmacia). Anti-116 kDa antibodies were used as antiserum (Fabrizio et al., 1997). For western blot analysis, proteins were separated on a 10% SDS–polyacrylamide gel, transferred to a membrane, and subsequently immunostained with antibodies at a 1:2000 dilution and developed with the ECL detection kit (Amersham).
Immunoprecipitation and immunodepletion
Procedures were based upon those described previously (Lamond and Sproat, 1994; Hermann et al., 1995). A 60 µl aliquot of protein A–Sepharose beads (bed volume) was incubated with 100 µl of either protein A-purified antibodies or affinity-purified antibodies in a total volume of 400 µl of phosphate-buffered saline (PBS), pH 8.0, containing 0.5 mg/ml of bovine serum albumin (BSA), 50 µg/ml total yeast tRNA and 0.01% NP-40; incubation was for 2 h at 4°C with head-over-tail rotation. Subsequently, beads were washed four times with 1 ml of PBS and then equilibrated with the buffer in which immunodepletion or immunoprecipitation was to be carried out. For immunoprecipitation, 20 µl of beads with coupled antibodies were incubated with 60 µl of HeLa nuclear extracts in a total volume of 400 µl of IP buffer (20 mM HEPES, pH 7.9, 1.5 mM MgCl2, 0.05% NP-40) containing different concentrations of NaCl (150–500 mM) for 1–2 h at 4°C. Subsequently, the beads were washed five times with 1 ml of IP buffer at the corresponding salt concentration. Co-precipitated RNAs were released from beads by phenol–chloroform extraction, precipitated with ethanol, 3′ end-labelled with [32P]pCp (3000 Ci/mmol; Amersham) by the method of England and Uhlenbeck (1978), separated on 10% polyacrylamide–7 M urea gels and detected by autoradiography. Alternatively, snRNAs were detected by northern hybridization using probes generated from plasmids encoding U4, U5 or U6 snRNAs (Pikielny et al., 1989) by using a random primer labelling kit (Prime-It II, Stratagene).
For immunodepletion of HeLa nuclear extracts, 200 µl of the extract (stored in C-buffer; Dignam et al., 1983) was first brought to 500 mM NaCl and then incubated with 60 µl of beads with coupled antibodies for 2 h at 4°C with head-over-tail rotation. The nuclear extract was then separated from the beads and dialysed for 4 h against D-buffer in a Slide-A-Lyzer 3.5K (Pierce). Control (‘mock-depleted’) extracts were treated in a manner identical to that used for the depleted extracts, except that protein A–beads pre-blocked with BSA and tRNA or beads coupled to pre-immune serum were used; these gave very similar results.
Splicing in vitro
Capped, internally labelled MINX pre-mRNA derived from the adenovirus major late transcription unit (Zillmann et al., 1988) was produced by transcription in vitro in the presence of [32P]UTP (3000 Ci/mmol; Amersham) and purified on a polyacrylamide gel. The splicing reactions contained 40% (v/v) HeLa nuclear extract, 50 mM KCl, 3.25 mM MgCl2, 2 mM ATP, 20 mM creatine phosphate and 3 nM MINX pre-mRNA, and were incubated at 30°C. At defined time intervals, aliquots of 10–20 µl were transferred to the wells of a microtitre plate and processed as described by Eperon and Krainer (1994). Subsequently, RNA was analysed on a gel containing 14% polyacrylamide and 7 M urea and detected by autoradiography.
For complementation of depleted extracts, 0–2 µl of recombinant proteins (as detailed in the respective figure legends) were added to the splicing reaction in a total volume of 50 µl before addition of pre-mRNA.
For inhibition of splicing with antibodies, antiserum and corresponding pre-immune serum were purified on protein A–Sepharose, dialysed against D-buffer, concentrated in Centricon filter units (Amicon) and added to the splicing reaction before addition of pre-mRNA.
For analysis of spliceosomal complexes, 10 µl aliquots of the splicing reaction were mixed with 2 µl of heparin (5 mg/ml) and 2 µl of 87% glycerol at defined time intervals, and placed on ice. Splicing complexes were separated on a composite gel containing 3.5% polyacrylamide and 0.5% agarose, and detected by autoradiography as described by Behrens et al. (1993).
Acknowledgments
Acknowledgements
We thank P.Kempkes, I.Öchsner and A.Badouin for excellent technical assistance, and J.Lauber for his help in the initial phase of the project. We are also grateful to C.L.Will for critical comments on the manuscript. This work was supported by the Gottfried Wilhelm Leibniz Program and grants from the Deutsche Forschungsgemeinschaft (SFB 397) and Fonds der Chemischen Industrie (to R.L.).
References
- Achsel T., Brahms,H., Kastner,B., Bachi,A., Wilm,M. and Lührmann,R. (1999) A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J., 18, 5789–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach M., Winkelmann,G. and Lührmann,R. (1989) 20S small nuclear ribonucleoprotein U5 shows a surprisingly complex protein composition. Proc. Natl Acad. Sci. USA, 86, 6038–6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A., Bucher,P. and Hofmann,K. (1996) The PROSITE database, its status in 1995. Nucleic Acids Res., 24, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens S.E., Galisson,F., Legrain,P. and Lührmann,R. (1993) Evidence that the 60 kDa protein of 17S U2 small nuclear ribonucleoprotein is immunologically and functionally related to the yeast PRP9 splicing factor and is required for the efficient formation of prespliceosomes. Proc. Natl Acad. Sci. USA, 90, 8229–8233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe B.J., Bowman,J.A., McCracken,S. and Rosonina,E. (1999) SR-related proteins and the processing of messenger RNA precursors. Biochem. Cell Biol., 77, 277–291. [PubMed] [Google Scholar]
- Burge C.B., Tuschl,T. and Sharp,P.A. (1999) Splicing of precursors to mRNAs by the spliceosomes. In Gesteland,R.F. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–560.
- Dignam J.D., Lebovitz,R.M. and Roeder,R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England T.E. and Uhlenbeck,O.C. (1978) 3′-Terminal labelling of RNA with T4 RNA ligase. Nature, 275, 560–561. [DOI] [PubMed] [Google Scholar]
- Eperon I.C. and Krainer,A.R. (1994) Splicing of mRNA precursors in mammalian cells. In Higgins,S.J. and Hames,B.D. (eds), RNA Processing. Oxford University Press, New York, USA, pp. 57–101.
- Fabrizio P., Laggerbauer,B., Lauber,J., Lane,W.S. and Lührmann,R. (1997) An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J., 16, 4092–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetzer S., Lauber,J., Will,C.L. and Lührmann,R. (1997) The U4/U6⋅U5 tri-snRNP-specific 27K protein is a novel SR protein that can be phosphorylated by the snRNP-associated protein kinase. RNA, 3, 344–355. [PMC free article] [PubMed] [Google Scholar]
- Fu X.D. (1995) The superfamily of arginine/serine-rich splicing factors. RNA, 1, 663–680. [PMC free article] [PubMed] [Google Scholar]
- Ge H., Zuo,P. and Manley,J.L. (1991) Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell, 66, 373–382. [DOI] [PubMed] [Google Scholar]
- Gottschalk A., Neubauer,G., Banroques,J., Mann,M., Lührmann,R. and Fabrizio,P. (1999) Identification by mass spectrometry and functional analysis of novel proteins of the yeast U4/U6⋅U5 tri-snRNP. EMBO J., 18, 4535–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graceffa P., Jancso,A. and Mabuchi,K. (1992) Modification of acidic residues normalizes sodium dodecyl sulfate–polyacrylamide gel electrophoresis of caldesmon and other proteins that migrate anomalously. Arch. Biochem. Biophys., 297, 46–51. [DOI] [PubMed] [Google Scholar]
- Hermann H., Fabrizio,P., Raker,V.A., Foulaki,K., Hornig,H., Brahms,H. and Lührmann,R. (1995) snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein–protein interactions. EMBO J., 14, 2076–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz D.S., Kobayashi,R. and Krainer,A.R. (1997) A new cyclophilin and the human homologues of yeast Prp3 and Prp4 form a complex associated with U4/U6 snRNPs. RNA, 3, 1374–1387. [PMC free article] [PubMed] [Google Scholar]
- Kohtz J.D., Jamison,S.F., Will,C.L., Zuo,P., Lührmann,R., Garcia-Blanco,M.A. and Manley,J.L. (1994) Protein–protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature, 368, 119–124. [DOI] [PubMed] [Google Scholar]
- Krainer A.R., Mayeda,A., Kozak,D. and Binns,G. (1991) Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K and Drosophila splicing regulators. Cell, 66, 383–394. [DOI] [PubMed] [Google Scholar]
- Krämer A. (1996) The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem., 65, 367–409. [DOI] [PubMed] [Google Scholar]
- Krämer A., Mulhauser,F., Wersig,C., Groning,K. and Bilbe,G. (1995) Mammalian splicing factor SF3a120 represents a new member of the SURP family of proteins and is homologous to the essential splicing factor PRP21p of Saccharomyces cerevisiae. RNA, 1, 260–272. [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B., Lauber,J. and Lührmann,R. (1996) Identification of an RNA-dependent ATPase activity in mammalian U5 snRNPs. Nucleic Acids Res., 24, 868–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond A.I. and Sproat,B.S. (1994) Isolation and characterization of ribonucleoprotein complexes. In Higgins,S.J. and Hames,B.D. (eds), RNA Processing. Oxford University Press, New York, pp. 103–140.
- Lauber J., Fabrizio,P., Teigelkamp,S., Lane,W.S., Hartmann,E. and Lührmann,R. (1996) The HeLa 200 kDa U5 snRNP-specific protein and its homologue in Saccharomyces cerevisiae are members of the DEXH-box protein family of putative RNA helicases. EMBO J., 15, 4001–4015. [PMC free article] [PubMed] [Google Scholar]
- Lauber J., Plessel,G., Prehn,S., Will,C.L., Fabrizio,P., Gröning,K., Lane,W.S. and Lührmann,R. (1997) The human U4/U6 snRNP contains 60 and 90 kDa proteins that are structurally homologous to the yeast splicing factors Prp4p and Prp3p. RNA, 3, 926–941. [PMC free article] [PubMed] [Google Scholar]
- Lund E. and Dahlberg,J.E. (1992) Cyclic 2′,3′-phosphates and nontemplated nucleotides at the 3′ end of spliceosomal U6 small nuclear RNAs. Science, 255, 327–430. [DOI] [PubMed] [Google Scholar]
- Lygerou Z., Christophides,G. and Seraphin,B. (1999) A novel genetic screen for snRNP assembly factors in yeast identifies a conserved protein, Sad1p, also required for pre-mRNA splicing. Mol. Cell. Biol., 19, 2008–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley J.L. and Tacke,R. (1996) SR proteins and splicing control. Genes Dev., 10, 1569–1579. [DOI] [PubMed] [Google Scholar]
- Nilsen T.W. (1998) RNA–RNA interactions in nuclear pre-mRNA splicing. In Simons,R.W. and Grunberg-Manago,M. (eds), RNA Structure and Function. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 279–307.
- Nottrott S., Hartmuth,K., Fabrizio,P., Urlaub,H., Vidovic,I., Ficner,R. and Lührmann,R. (1999) Functional interaction of a novel 15.5 kDa U4/U6⋅U5 tri-snRNP protein with the 5′ stem–loop of U4 snRNA. EMBO J., 18, 6119–6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikielny C.W., Bindereif,A. and Green,M.R. (1989) In vitro reconstitution of snRNPs: a reconstituted U4/U6 snRNP participates in splicing complex formation. Genes Dev., 3, 479–487. [DOI] [PubMed] [Google Scholar]
- Reed R. and Palandjian,L. (1997) Spliceosome assembly. In Krainer,A.R. (ed.), Eukaryotic mRNA Processing. Oxford University Press, New York, pp. 103–129.
- Roscigno R.F. and Garcia-Blanco,M.A. (1995) SR proteins escort the U4/U6⋅U5 tri-snRNP to the spliceosome. RNA, 1, 692–706. [PMC free article] [PubMed] [Google Scholar]
- Salgado-Garrido J., Bragado-Nilsson,E., Kandels-Lewis,S. and Seraphin,B. (1999) Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J., 18, 3451–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shichijo S. et al. (1998) A gene encoding antigenic peptides of human squamous cell carcinoma recognized by cytotoxic T lymphocytes. J. Exp. Med., 187, 277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siatecka M., Reyes,J.L. and Konarska,M.M. (1999) Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes Dev., 13, 1983–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley J.P. and Guthrie,C. (1998) Mechanical devices of the spliceosome: motors, clocks, springs and things. Cell, 92, 315–326. [DOI] [PubMed] [Google Scholar]
- Stark J.M., Bazett-Jones,D.P., Herfort,M. and Roth,M.B. (1998) SR proteins are sufficient for exon bridging across an intron. Proc. Natl Acad. Sci. USA, 95, 2163–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens S.W. and Abelson,J. (1999) Purification of the yeast U4/U6⋅U5 small nuclear ribonucleoprotein particle and identification of its proteins. Proc. Natl Acad. Sci. USA, 96, 7226–7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teigelkamp S., Newman,A.J. and Beggs,J.D. (1995) Extensive interactions of PRP8 protein with the 5′ and 3′ splice sites during splicing suggest a role in stabilization of exon alignment by U5 snRNA. EMBO J., 14, 2602–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teigelkamp S., Mundt,C., Achsel,T., Will,C.L. and Lührmann,R. (1997) The human U5 snRNP-specific 100 kDa protein is an RS domain-containing, putative RNA helicase with significant homology to the yeast splicing factor Prp28p. RNA, 3, 1313–1326. [PMC free article] [PubMed] [Google Scholar]
- Teigelkamp S., Achsel,T., Mundt,C., Gothel,S.F., Cronshagen,U., Lane,W.S., Marahiel,M. and Lührmann,R. (1998) The 20 kDa protein of human U4/U6⋅U5 tri-snRNPs is a novel cyclophilin that forms a complex with the U4/U6-specific 60 kDa and 90 kDa proteins. RNA, 4, 127–141. [PMC free article] [PubMed] [Google Scholar]
- Valenta R. et al. (1998) Molecular characterization of an autoallergen, Hom s 1, identified by serum IgE from atopic dermatitis patients. J. Invest. Dermatol., 111, 1178–1183. [DOI] [PubMed] [Google Scholar]
- Will C.L. and Lührmann,R. (1997) Protein functions in pre-mRNA splicing. Curr. Opin. Cell Biol., 9, 320–328. [DOI] [PubMed] [Google Scholar]
- Zillmann M., Zapp,M.L. and Berget,S.M. (1988) Gel electrophoretic isolation of splicing complexes containing U1 small nuclear ribonucleoprotein particles. Mol. Cell. Biol., 8, 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]