Abstract
Streptococcus pneumoniae peptidoglycan GlcNAc deacetylase (SpPgdA) protects the Gram-positive bacterial cell wall from host lysozymes by deacetylating peptidoglycan GlcNAc residues. Deletion of the pgda gene has been shown to result in hypersensitivity to lysozyme and reduction of infectivity in a mouse model. SpPgdA is a member of the family 4 carbohydrate esterases, for which little structural information exists, and no catalytic mechanism has yet been defined. Here we describe the native crystal structure and product complexes of SpPgdA biochemical characterization and mutagenesis. The structural data show that SpPgdA is an elongated three-domain protein in the crystal. The structure, in combination with mutagenesis, shows that SpPgdA is a metalloenzyme using a His-His-Asp zinc-binding triad with a nearby aspartic acid and histidine acting as the catalytic base and acid, respectively, somewhat similar to other zinc deacetylases such as LpxC. The enzyme is able to accept GlcNAc3 as a substrate (Km = 3.8 mM, kcat = 0.55 s-1), with the N-acetyl of the middle sugar being removed by the enzyme. The data described here show that SpPgdA and the other family 4 carbohydrate esterases are metalloenzymes and present a step toward identification of mechanism-based inhibitors for this important class of enzymes.
Keywords: crystal structure, metalloenzyme
Peptidoglycan, the peptide-linked heteropolymer of GlcNAc and N-acetylmuramic acid, is one of the main protective barriers in the bacterial cell wall. The mammalian immune system uses a range of hydrolytic enzymes (peptidases/glycosidases) to fragment and destroy the peptidoglycan layer, allowing activation of specific immune responses through recognition of peptidoglycan components by Toll-like receptors and Nod proteins (reviewed in ref. 1). However, as a defense mechanism against the mammalian hydrolases, bacteria have developed strategies for modifying peptidoglycan so that it is no longer recognized by these enzymes (1–3). One example of this is the de-N-acetylation of the GlcNAc residues. Recent work has identified the protein responsible for this deacetylation step in the pathogen Streptococcus pneumoniae (4). S. pneumoniae GlcNAc peptidoglycan deacetylase (SpPgdA) was found to de-N-acetylate GlcNAc sugars in pneumococcal peptidoglycan, with overall deacetylation levels up to 80% (4). Furthermore, a Δpgda strain was hypersensitive to exogenous lysozyme, resulting in complete lysis (4). Subsequently, it was shown that this also extends to a mouse model of S. pneumoniae infection where the Dpgda strain showed significantly reduced virulence (5). Thus, the SpPgdA enzyme may represent an attractive drug target, and structural and biochemical characterization is urgently required to aid exploitation as such.
SpPgdA is a member of the family 4 carbohydrate esterases (CE-4) as defined in the CAZy database (http://afmb.cnrs-mrs.fr/CAZy), which has been reviewed recently (6). NodB, the first enzyme of this family to be described in detail, deacetylates a GlcNAc residue in the synthesis of Nod factors, bacterial signaling molecules that regulate the symbiotic relationship with leguminous plants (7). Subsequently, it was discovered that chitin deacetylases also contain a NodB homology domain (8) as well as other enzymes that deacetylate carbohydrate polymers, such as the xylan esterases and peptidoglycan deacetylases (6). These enzymes all share a number of sequence motifs, including several conserved aspartic acid and histidine residues. There are several reports showing that the activity of these enzymes can be increased by addition of divalent cations (6, 9); other studies, however, report full activity without metal supplementation (e.g., ref. 10). Substrate affinity and specificity have been studied in detail for the chitin deacetylases, showing that these enzymes require longer oligosaccharides for optimal activity and produce specific patterns of deacetylation (10). However, the catalytic mechanism of these enzymes is as yet unknown, and no mutagenesis studies probing the role of the conserved residues have been described.
Recently, the first structure of a CE-4 family member, the Bacillus subtilis peptidoglycan deacetylase (BsPdaA), has been reported (11). The BsPdaA enzyme deacetylates peptidoglycan N-acetyl muramic acid residues, leading to the formation of d-lactam structures (12). The BsPdaA NodB homology domain appears to adopt an (α/β)8 fold, with a groove running over the surface of the protein harboring the majority of the conserved residues (11). Two of the conserved histidines come together in the bottom of the active site, yet soaking studies were inconclusive as to the possible role of these histidines as metal-binding residues. A complex with GlcNAc was also reported, although it was not clear whether this represented productive substrate binding (11). Thus, further structural and biochemical studies are required to define the mechanism of action of the CE-4 family.
Here, x-ray crystallography, enzymology, mutagenesis, and mass spectrometry data are used to define the structure, mechanism, and substrate binding of SpPgdA, showing that the enzyme has an unusual multidomain structure and that SpPgdA and the CE-4 family are metalloenzymes, which use acid/base catalysis, with a conserved aspartic acid and histidine.
Materials and Methods
Expression, Structure Solution, and Docking. SpPgdA was cloned and overexpressed in Escherichia coli and analyzed by x-ray crystallography and docking approaches, using standard methods. Further details of these procedures are given supporting information, which is published on the PNAS web site.
Enzymology. Standard reactions consisted of 100 nM peptidoclycan deacetylase, 5 μM CoCl2, 50 mM Bis·Tris (pH 7.0), and 2 mM GlcNAc3 (Sigma) in a total volume of 50 ml, incubated for 60 min at 37°C. Fifty microliters of 0.4 M borate buffer (pH 9.0) was then added and free amines labeled with 20 μl of 2 mg/ml fluorescamine in dimethylformamide (DMF) for 10 min at room temperature. The labeling reaction was terminated by the addition of 150 μl of DMF/H2O (1:1). Fluorescence was quantified by using an FLX 800 Microplate Fluorescence Reader (Bio-Tek, Burlington, VT), with excitation and emission wavelengths of 360 and 460 nm, respectively. The production of free amine was quantified with a glucosamine standard. The fluorescence intensity data were analyzed by using nonlinear regression analysis with grafit (13), with the default equations for first-order reaction rates and Michaelis–Menten steady-state kinetics. All measurements were performed in triplicate.
Crystallization, Phasing, and Refinement. The sitting-drop vapor diffusion method was used to produce crystals by mixing 1 μl of protein solution with an equal volume of mother liquor [35% (vol/vol) polyethylene glycol (PEG)200/5% PEG 3000 (wt/vol)/0.1 M Mes, pH 6]. Hexagonal crystals grew within 4 days. Crystals were transferred to mother liquor containing 10 mM ZnCl2 and were frozen in a nitrogen gas stream cooled to 100 K. A two-wavelength zinc multiwavelength anomalous dispersion experiment was carried out at beamline BM14 at the European Synchrotron Radiation Facility in Grenoble, France. Data were processed with the hkl suite (14). Phasing, phase extension, and solvent flattening were performed with the shelx suite (15), exploiting the anomalous signal of three zinc sites. The resulting 1.75-Å electron density map was partially autotraced with warpntrace (16), followed by refinement with cns (17) and refmac (18), interspersed with model building with o (19) and coot (20). Ligands were included when unambiguously defined by unbiased |Fo|-|Fc|, φcalc maps. Ligand topologies and coordinates were generated with prodrg (21). All figures were made with pymol.†
Mass Spectrometry. Five-nanomolar GlcNAc3 was digested with SpPgdA for 5 h at 37°C. A 10% (500 pmol) aliquot of the reaction mixture was re-N-acetylated with 100 μl of saturated NaHCO3 and three 2.5-μl aliquots of deuterated acetic anhydride (Sigma-Aldrich), incubated at 0°C for 10 min twice and finally at room temperature for >1 h. The sample was desalted, and boric acid and acetic acid were removed via passage through a prewashed column of 0.25 ml of AG50W-X12 resin (Bio-Rad) over 0.25 ml of AG3-X4 resin (Bio-Rad) and elution by 4 × 0.25 μl of water. The samples were then freeze-dried. Electrospray MS were recorded on a Micromass (Manchester, U.K.) Q-ToF2 instrument, in positive ion mode. Freeze-dried samples were dissolved in 100 μl of 50% acetonitrile/2% formic acid and were introduced into the mass spectrometers by using nanospray tips. The capillary and cone voltages were 0.7–1.2 kV and 25–35 V, respectively. Product ion electrospray–tandem MS were recorded by using collision voltages of 35–40 V and collision gas of argon at 3 × 10-3 1 Torr (1 torr = 133 Pa). All data were collected and processed with masslynx software (Waters).
Results
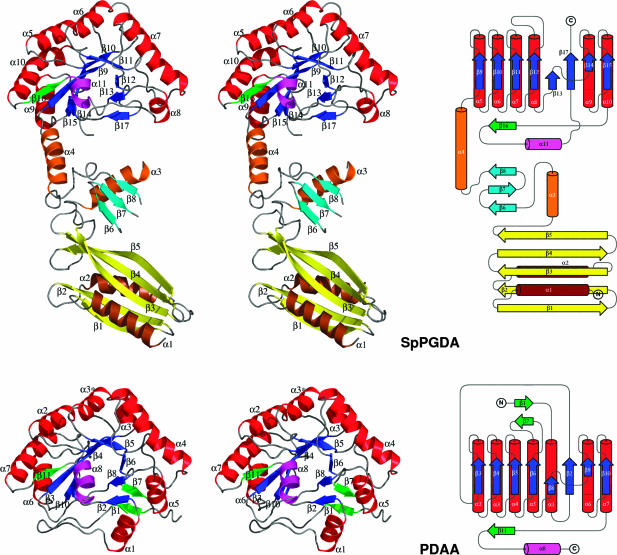
SpPgdA Has a Unique Three-Domain Structure. SpPgdA was cloned, overexpressed, and purified from E. coli, crystallized from PEG 200 solutions followed by a zinc–multiwavelength anomalous dispersion structure solution. The SpPgdA structure reveals three separate domains, an N-terminal domain (residues 46–160), a middle domain (residues 161–268), and the C-terminal catalytic domain (residues 269–463), which adopts a fold reminiscent of an (β/α)8 topology (Fig. 1). The catalytic domain contains the NodB homology domain, present in all CE-4 esterases (6). Strikingly, however, structural comparison with the only currently known structure of a CE-4 family member, B. subtilis PdaA (11), shows that, although the overall fold of the catalytic core is the same (rms deviation = 2.0 Å on 193 equivalenced Cα atoms), there are significant topological differences (Fig. 1). The N/C termini are on opposite ends of the barrel; furthermore, the first strand of the barrel in PdaA (β2) is in a topologically equivalent position to the last strand of the barrel in SpPgdA (β17).
Fig. 1.
SpPgdA structure. Stereo image of the SpPgdA (SpPGDA) and BsPdaA [PDAA, from Protein Data Bank entry 1W1A (11)] structures, alongside topological representations as constructed with topdraw (27). In the SpPgdA N-terminal domain, helices are colored brown and strands, yellow. In the SpPgdA middle domain, helices are colored orange and strands, cyan. In the catalytic domains, helices are colored red and strands, blue, except for the helices (magenta) and strands (green) that do not fit the canonical (β/α)8 fold. Secondary structure elements are named as indicated in the sequence alignment in Fig. 2.
Whereas the PdaA structure consists only of a catalytic core, SpPgdA contains two additional domains (Fig. 1). The middle domain (orange/cyan in Fig. 1) is a small α/β fold, with a three-stranded antiparallel β-sheet (β6–8) packed against an α-helix (α3), connected to the catalytic core through a long helix (α4). This domain contains two large loops (187–210 and 224–232) for which no defined electron density was present, and the average B factor in this domain is relatively high (44 Å2, compared with 26 Å2 for the rest of the protein). The N-terminal section of the SpPgdA structure (residues 46–160) contains an α/β-fold, consisting of a mixed five-stranded β sheet, flanked on both faces by an α helix. A range of fold recognition servers (ce, dali, dejavu, vast, and stamp) were searched with this domain, yielding no significant hits. A detailed structural analysis of this domain will be reported elsewhere. Although N-terminal extensions are found in other CE-4 members, the N-terminal domains of SpPgdA appear to be unique to this enzyme.
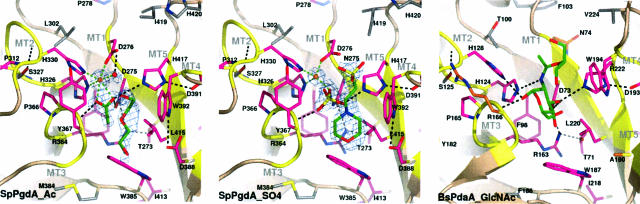
The Active Site Shows a Conserved Zinc-Binding Triad. Analysis of the cleft running over the surface of the protein revealed the presence of a zinc ion complexed to two histidines (His-326 and His-330) and an aspartic acid (Asp-276; Fig. 2). This metal-binding triad is conserved throughout the CE-4 family (see supporting information). Interestingly, in what was presumed to be a native uncomplexed, SpPgdA structure, a well ordered acetate (7-σ peak in difference map, average B factor 26 Å2) molecule is seen to interact with the zinc (Fig. 2). Acetate is one of the products of the deacetylation reaction and is most likely a contaminant of the PEG crystallization solutions. The acetate molecule makes several further interactions in the active site. One of the oxygens interacts with the conserved Asp-275, which is tethered by a buried and conserved Arg-364 (Fig. 2). Similarly, the same acetate oxygen interacts with the conserved His-417, which is tethered by the buried and conserved Asp-319. The other acetate oxygen accepts a hydrogen bond from the backbone nitrogen of Tyr-367. The terminal acetate methyl occupies a small hydrophobic pocket generated by the conserved Leu-415 and Trp-385. In addition to the acetate, an ordered PEG200 molecule is seen to interact with the solvent exposed conserved Trp-392. PEG200 is present as 30% of the crystallization mother liquor. Together with His-326, His-330, Asp-276, and a tightly bound water molecule the two acetate oxygens coordinate the zinc in a distorted octahedral fashion (the average ligand-Zn-ligand angle for neighboring ligands is 90 ± 14°).
Fig. 2.
Details of the SpPgdA active site. Close-up of the active sites of: the native SpPgdA structure in complex with the acetate product and PEG200 (SpPGDA_Ac), the SpPgdA D275N mutant in complex with sulfate and Mes (SpPGDA_SO4), and the previously determined complex of B. subtilis PdaA in complex with GlcNAc and a glycerol molecule (PDAA_GlcNAc). The five CE-4 sequence motifs (MT1–5) are shown in yellow. Side chains lining the active site cleft are shown as sticks. Residues conserved in all CE-4 esterases are magenta. Water molecules (spheres) and ligands (green sticks) are also shown. Unbiased Fo - Fc, φcalc maps are shown at 2.25 σ (Mes in SpPGDA_SO4) and 12 σ (Zn in SpPGDA_Ac/SO4). Hydrogen bonds are shown as dashed lines in black and zinc–ligand interactions, as green dashed lines.
By combination of the SpPgdA structure (Fig. 2) and a multiple sequence alignment of representatives of the CE-4 family (see supporting information), it becomes apparent that five distinct sequence motifs assemble the active site (marked MT1–MT5 in Fig. 2). All of these motifs lie at the C-terminal ends of the β strands making up the (β/α)8 barrel. Motif 1 (TFDD) contributes the zinc-binding aspartic acid Asp-276 and the acetate-binding Asp-275. Motif 2 constitutes a zinc-binding motif, H(S/T)xxHP, where both histidines (His-326 and His-330) bind the zinc, whereas the conserved serine/threonine residue accepts a hydrogen bond from His-330, stabilizing the conformation of the loop (Fig. 2). Structure and sequence searches did not reveal other examples of this zinc-binding motif outside the CE-4 family. Motif 3 forms one of the sides of the active site groove. It contains the backbone hydrogen bond donor for one of the acetate oxygens (Tyr-367), a conserved proline (Pro-366) that wedges the two zinc-binding histidines and the buried arginine (Arg-364) which coordinates the conserved Asp-275 (Fig. 2). Motif 4 forms the other side of the active site groove, dominated by a solvent-exposed tryptophan (Trp-392) and also containing a buried aspartic acid (Asp-391), which tethers His-417. Motif 5 contains a leucine (Leu-415), which, together with the conserved Trp-385, forms the hydrophobic pocket for the acetate methyl group, and His-417, which hydrogen bonds the acetate.
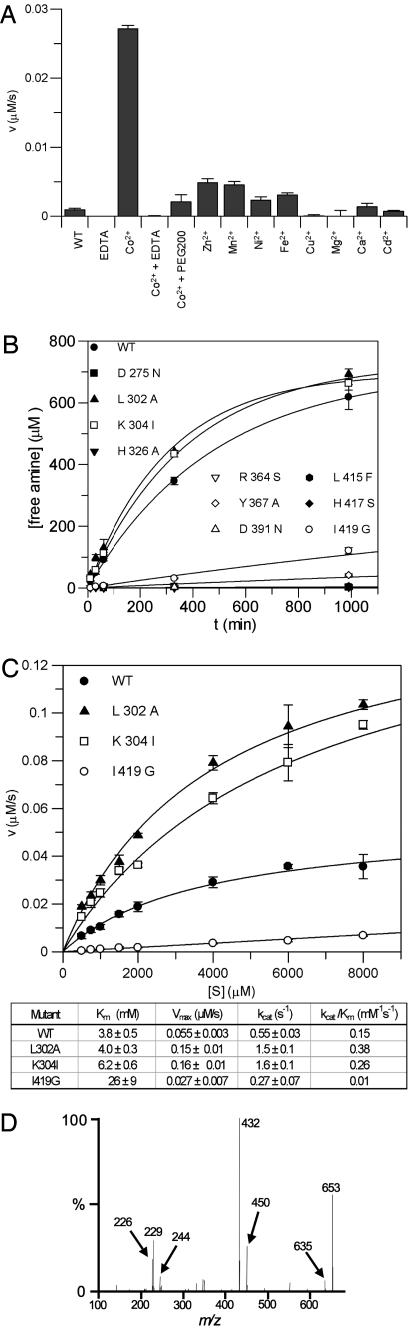
Activity of SpPgdA Is Metal-Dependent. Activity for CE-4 esterases has traditionally been reported by using one of three possible approaches: a substrate with radioactive acetyl groups (e.g., ref. 9), the direct spectroscopic detection of the acetate product (e.g., ref. 10), or detection of acetate by using a commercially available coupled assay involving three additional enzymatic steps. None of these is particularly suitable for rapid screening of different substrates/inhibitors/reaction conditions. We therefore developed an assay based on fluorogenic labeling with fluorescamine, which labels the free amines generated from the GlcNAc3 substrate by the enzyme reaction. This allowed miniaturization of the assay to 50 μl volumes suitable for a 96-well format (see supporting information) (Fig. 3A).
Fig. 3.
SpPgdA enzymology. (A) Metal-dependent activity. The reaction was run for 60 min with 2 mM GlcNAc3 and 0.1 μM SpPgdA. WT, native protein after purification. All metals were added as chloride salts at a concentration of 5μM. EDTA was added at 100μM and PEG200, at 30%. Bars represent triplicate experiments with standard deviation. (B) Time-dependent deacetylation of GlcNAc3 by wild-type (WT) and mutant SpPgdA, followed by labeling of the resulting free amine with fluorescamine. (C) Steady-state kinetics by using GlcNAc3 as a substrate. Initial velocities were measured after 60 min at different substrate concentrations. (D) Positive ion electrospray tandem MS of deutero-re-N-acetylated GlcNAc-GlcN-GlcNAc. The ions at m/z 226, 229, 432, and 653 represent the Y series [M+Na]+ ions of GlcNAc, GlcNAcD, GlcNAcGlcNAcD, and GlcNAcGlcNAcDGlcNAc, respectively. The ions at m/z 244 and 450 represent the C series [M+Na]+ ions of GlcNAc and GlcNAcGlcNAcD, respectively. The ion at m/z 635 represents the [M+Na-H2O]+ ion of GlcNAcGlcNAcDGlcNAc.
The structure of the active site suggests that SpPgdA and, by sequence conservation, other CE-4 esterases are metalloenzymes. Indeed, several reports have described an increase in activity when supplementing purified CE-4 enzymes with divalent cations, in particular Co2+ (6). The recombinant SpPgdA enzyme, exposed to buffers containing EDTA during the gel filtration step, shows barely detectable enzymatic activity (Fig. 3A), although under standard laboratory conditions, activity increases over the course of several weeks (data not shown). Supplementing the recombinant enzyme with EDTA inactivates the enzyme. Addition of 5 μM of a range of divalent cations dramatically increases the enzyme activity of the enzyme, with the largest effects for Co2+ (30-fold) and Zn2+ (5.5-fold) (Fig. 3A). With further addition of EDTA to the protein–cobalt complex, activity is abrogated. Thus, SpPgdA and, most likely, the CE-4 family as a whole are metalloenzymes. Although the highest activity is seen with cobalt, bioavailability may well limit these enzymes to be zinc-dependent.
SpPgdA Deacetylates GlcNAc3 on the Middle Sugar. SpPgdA has been shown to deacetylate GlcNAc residues in native peptidoglycan (4). Here, GlcNAc3 is used as a pseudosubstrate, with production of free amine fitting a first-order reaction rate (Fig. 3B, k = 0.0021 ± 0.0002 s-1). Initial velocity measurements with increasing substrate concentrations fitted Michaelis–Menten kinetics, showing Km to be 3.8 ± 0.5 mM with a kcat of 0.03-1 to 0.03 s-1 (Fig. 3C). Although SpPgdA is a peptidoglycan deacetylase, these values are surprisingly similar to those reported for deacetylation of GlcNAc3 by the CE-4 enzyme Colletotrichum lindemuthianum chitin deacetylase (Km = 4.3 mM, kcat = 6 s-1) (10).
We next determined which of the three possible GlcNAc3 N-acetyl groups is initially removed by SpPgdA. GlcNAc3 was partially deacetylated with SpPgdA and then re-N-acetylated with deuterated acetic anhydride, generating a 3 Da mass increase for enzymatically deacetylated GlcNAc. Analysis by electrospray mass spectra revealed an ion at m/z 653.4 (supporting information) that was shown to correlate with the sodiated molecule [GlcNAcGlcNAcDGlcNAc+Na]+ after collision-induced dissociation (Fig. 5D, GlcNAcD = deuterated GlcNAc). The absence of an ion of m/z 447 indicates there is no loss of a 206-Da residue associated with deacetylation of a terminal GlcNAc. Furthermore, the absence of an ion of m/z 424 shows there no nondeuterated GlcNAc2 was present. This, together with detailed interpretation of the other product ions (Fig. 3D), shows that the central GlcNAc residue, but not the terminal residues, had been deacetylated by the enzyme. In addition, an ion at m/z 656.4 shows that a subsequent deacetylation step is possible, as confirmed by collision-induced dissociation (supporting information). Thus, when defining the deacetylated sugar as the 0 subsite [following previously proposed nomenclature (10)], the data show that GlcNAc3 initially binds in the -1, 0, +1 subsites of the enzyme.
Deacetylation Involves a Conserved Aspartic Acid and Histidine. In an attempt to trap a substrate complex, the conserved Asp-275, which is seen to coordinate the acetate in the SpPgdA structure (Fig. 2), was mutated to asparagine, and the D275N mutant protein was purified. Significantly, this rather conservative substitution completely inactivates the enzyme (Fig. 3 B and C). The mutant crystallized under similar conditions as the wild type, and the structure was solved (see supporting information). A well ordered oxyanion (10 σ peak in difference map, average B factor 16 Å2), presumed to be sulfate, is seen to interact with zinc and active-site residues (Asn-275, Tyr-367, and His-417) in a similar way as observed for acetate (Fig. 3). The mutant residue, Asn-275, donates a hydrogen bond to one of the oxyanion oxygens. In a position similar to that of PEG200 in the acetate complex, an ordered Mes buffer molecule is seen to bind in the sulfate complex (Fig. 3). It is noteworthy that the exchange of acetate for sulfate, together with a movement of the metal-coordinating water, results in a decrease of the coordination number of the zinc from 6 to the preferred 4, with the three protein ligands and one sulfate oxygen producing a tetrahedral coordination sphere (average angle, 109 ± 11°).
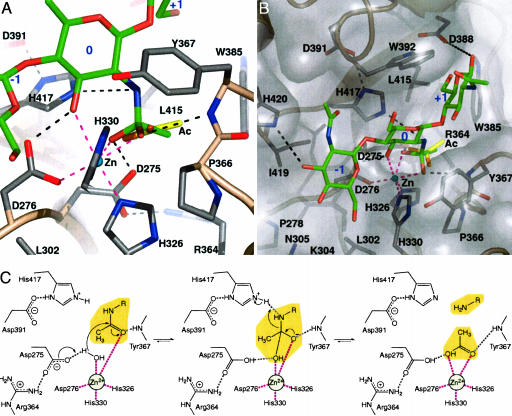
To be able to postulate a mechanism and guide subsequent mutagenesis studies, computer docking was used to generate a putative SpPgdA complex. Current knowledge on other zinc-dependent de-N-acetylases shows that a conserved carboxylic acid residue near the zinc acts as a base to activate a water molecule, which subsequently acts as a nucleophile to attack the acetate, generating an oxyanion tetrahedral intermediate (recently reviewed in ref. 22). In docking experiments with gold, a rigid protein receptor was used with a GlcNAc3 trimer carrying the proposed oxyanion intermediate on the middle sugar, producing gold scores in the 60–75 range. A representative complex is shown in Fig. 4 A and B. The middle sugar occupies a position near the zinc (the 0 subsite), interacting directly with the metal and one of the metal ligands (Asp-276) through O3. Furthermore, the 0 subsite sugar shows extensive stacking interactions with Trp-392 (which in the crystal structures is seen to stack with PEG200/Mes, Figs. 2 and 4). The terminal sugars occupy a groove running over the face of the protein (the -1 and +1 subsites; Fig. 4B). Because the two O3 oxygens are pointing up, this binding mode would be accessible to peptidoglycan, where the -1 and +1 sugars would both be N-acetylmuramic acid, with the O3 lactoyl-peptide groups pointing out of the active site. The tetrahedral intermediate is positioned on the zinc, with the methyl group and the two oxygens occupying similar positions to the equivalent atoms in the acetate complex (maximum shift, 0.8 Å; Fig. 4). One of the two oxygens of the docked intermediate tightly interacts with Asp-275, which could represent the catalytic base. The other oxygen interacts directly with the zinc ion and the backbone nitrogen of Tyr-367; this could represent the classic “oxyanion hole.” The nitrogen of the tetrahedral intermediate approaches the His-417 Nε2 atom to within 3.7 Å (Fig. 4A). Interestingly, the protonation algorithm predicts His-417 to be in the histidinium form. This can be understood by the interaction with the buried and conserved Asp-391 that interacts with the His-417 Nδ1 atom, probably raising the imidazole pKa such that the Nε2 atom is also protonated at physiological pH. This leads to the hypothesis that His-417 could be the acid protonating the nitrogen in the reaction intermediate, generating a good leaving group. This hypothesis is supported by the analysis of two additional mutants. H417S and the conservative D319N mutants were both completely inactive (Fig. 4D), suggesting that both the presence of the histidine itself and the tuning of its pKa are essential for the reaction.
Fig. 4.
Docked GlcNAc3 complex and reaction mechanism. (A) Sp-PgdA is shown as a ribbon, with conserved side chains. The acetate molecule as observed in the SpPgdA–acetate complex (Fig. 2) is shown as yellow sticks. GlcNAc3 is shown as green sticks with subsites labeled in blue. Hydrogen bonds are shown as dashed lines in black, and zinc–ligand interactions are shown as magenta dashed lines. (B) As A, but viewed from the top. (C) Proposed catalytic mechanism.
SpPgdA Acid/Base Reaction Mechanism. From structural, mutagenesis, and in silico data, a reaction mechanism can be proposed as shown in Fig. 4C. SpPgdA would catalyze the removal of an N-acetyl group by binding a water molecule on its tightly bound zinc. The catalytic base, Asp-275, abstracts a proton from this water molecule, creating a nucleophile to attack the carbonyl carbon in the substrate to produce a tetrahedral oxyanion intermediate. The charge on this intermediate is stabilized by the metal and the Tyr-367 backbone nitrogen. His-417 then protonates this intermediate on the nitrogen, generating a free amine and also the acetate product on the zinc, as observed in the crystal structure.
Additional mutants were generated to study the proposed reaction mechanism and GlcNAc3-binding mode. Mutation of His-326 (involved in zinc binding; Fig. 4A) and Arg-364 (positioning of the catalytic base) resulted in inactive protein (Fig. 3), showing these are also essential for the reaction, in line with their roles proposed in the reaction mechanism (Fig. 4C). Other mutants were made to study the roles of residues further away from the active site. Lys-304 is an isoleucine in yeast chitin deacetylase 2, and the K304I mutant increases catalytic efficiency (Fig. 4C). Leu-302 is a nonconserved residue interacting with the zinc-binding histidines (Fig. 4A), and the L302A mutant also shows increased catalytic efficiency (Fig. 4C). Mutation of residues Tyr-367, Leu-415, and Ile-419, lining the cleft and showing hydrophobic interactions with one or more of the sugars in the docked complex, results in either a 10-fold increase in Km (for I419G) or no detectable activity (Y367A/L415F) (Fig. 4).
Discussion
Enzymatic and structural data presented here show that SpPgdA is a metalloenzyme binding zinc in the crystal structure through a previously undescribed zinc-binding motif, which is conserved in all CE-4 esterases. Structural analyses and mutagenesis show that two “charge relay” pairs, a conserved aspartic acid (Asp-275, the catalytic base) tethered by a conserved arginine (Arg-364), and a conserved histidine (His-417, the catalytic acid) tethered by a conserved aspartic acid (Asp-319), perform acid/base catalysis exploiting a zinc-bound water molecule as the nucleophile. These data firmly put SpPgdA, and by similarity other CE-4 esterases, in the domain of the zinc-dependent deacetylases, whose reaction mechanisms have recently been reviewed in detail (23). The structures of several of these enzymes, such as the bacterial LpxC, involved in lipid A biosynthesis (24, 25) and Mycobacterium tuberculosis MshB, involved in mycothiol biosynthesis (16), have recently been solved. Although SpPgdA does not share any sequence or structural homology with these enzymes, the reaction mechanism proposed here is similar to those proposed for these other zinc-dependent deacetylases, although details concerning the identity of the catalytic acid and oxyanion hole differ (23).
So far, only one other CE-4 structure has been reported, that of BsPdaA (11). Strikingly, the equivalent of SpPgdA Asp-276, one of the zinc-binding ligands, is an asparagine in PdaA, and the backbone around this residue adopts a dramatically different conformation, with the side chain buried in the core of the protein (Fig. 2). A BsPdaA cadmium complex has also been reported, with the Cd2+ and SpPgdA Zn2+ ions occupying equivalent positions (distance = 0.8 Å after alignment on selected active site residues). Interestingly, BsPdaA de-N-acetylates only N-acetylmuramic acid residues in peptidoglycan, generating muramic δ-lactam residues (12). It is tempting to speculate that in this, perhaps unusual, member of the CE-4 family, it is the carboxylate on the O-lactoyl group of the substrate itself that interacts with the zinc. Additional experiments are required to investigate this hypothesis.
The demonstration that SpPgdA is a metalloenzyme, similar to other zinc-dependent deacetylases, offers exciting possibilities for inhibitor design (23). Hydroxamates and sulfonamides are just some examples of classes of inhibitors of these enzymes. Significant progress has already been made in the design and structural analysis of LpxC inhibitors (e.g., ref. 26). The structural data and enzyme assay described here offer invaluable tools for the evaluation of similar inhibitors.
Supplementary Material
Acknowledgments
We thank the European Synchrotron Radiation Facility (Grenoble, France) for time at beamline BM14 and Deutsches Elektronen Synchrotron European Molecular Biology Laboratory staff, Dmitri Svergun, and Peter Konarev for small-angle x-ray scattering time and help. We thank Doug Lamont and the Post-Genomics and Molecular Interactions Centre, University of Dundee (supported by Wellcome Trust Grant 060269), for access to mass spectrometers and Mike Ferguson for discussions of the data. D.M.F.v.A. is supported by a Wellcome Trust Senior Research Fellowship and the European Molecular Biology Organization Young Investigator Programme. D.E.B. is supported by a Biotechnology and Biological Sciences Research Council studentship.
Author contributions: D.M.F.v.A. designed research; D.E.B., A.W.S., and J.I.M. performed research; D.E.B., A.W.S., D.M.F.v.A., and J.I.M. analyzed data; and A.W.S., J.I.M., and D.M.F.v.A. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: GlcNAcD, deuterated GlcNAc; SpPgdA, Streptococcus pneumoniae GlcNAc peptidoglycan deacetylase; CE-4, family 4 carbohydrate esterases; BsPdaA, Bacillus subtilis peptidoglycan deacetylase; PEG, polyethylene glycol.
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 2c1g (WT Zn-acetate) and 2c1i (D275N Zn-sulphate)].
Footnotes
DeLano, W. L. (2004) Abstr. Pap. Am. Chem. Soc. 228, 030-CHED.
References
- 1.Boneca, I. G. (2005) Curr. Opin. Microbiol. 8, 46-53. [DOI] [PubMed] [Google Scholar]
- 2.Severin, A., Tabei, K. & Tomasz, A. (2004) Microb. Drug Resist.-Mechan. Epidemiol. Dis. 10, 77-82. [DOI] [PubMed] [Google Scholar]
- 3.Bera, A., Herbert, S., Jakob, A., Vollmer, W. & Gotz, F. (2005) Mol. Microbiol. 55, 778-787. [DOI] [PubMed] [Google Scholar]
- 4.Vollmer, W. & Tomasz, A. (2000) J. Biol. Chem. 275, 20496-20501. [DOI] [PubMed] [Google Scholar]
- 5.Vollmer, W. & Tomasz, A. (2002) Infect. Immun. 70, 7176-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caufrier, F., Martinou, A., Dupont, C. & Bouriotis, V. (2003) Carbohydr. Res. 338, 687-692. [DOI] [PubMed] [Google Scholar]
- 7.Long, S. R. (1989) Cell 56, 203-214. [DOI] [PubMed] [Google Scholar]
- 8.Kafetzopoulos, D., Thireos, G., Vournakis, J. N. & Bouriotis, V. (1993) Proc. Natl. Acad. Sci. USA 90, 8005-8008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinou, A., Koutsioulis, D. & Bouriotis, V. (2002) Protein Expr. Purif. 24, 111-116. [DOI] [PubMed] [Google Scholar]
- 10.Hekmat, O., Tokuyasu, K. & Withers, S. G. (2003) Biochem. J. 374, 369-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blair, D. E. & van Aalten, D. M. F. (2004) FEBS Lett. 570, 13-19. [DOI] [PubMed] [Google Scholar]
- 12.Fukushima, T., Kitajima, T. & Sekiguchi, J. (2005) J. Bacteriol. 187, 1287-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leatherbarrow, R. J. (2001) grafit (Erithacus Software, Horley, U.K.), Ver. 5.
- 14.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 15.Sheldrick, G. M. & Schneider, T. R. (1997) Methods Enzymol. 277, 319-343. [PubMed] [Google Scholar]
- 16.Perrakis, A., Morris, R. & Lamzin, V. S. (1999) Nat. Struct. Biol. 6, 458-463. [DOI] [PubMed] [Google Scholar]
- 17.Brunger, A. T., Adams, P. D., Clore, G. M., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 18.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240-255. [DOI] [PubMed] [Google Scholar]
- 19.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 20.Emsley, P. & Cowtan, K. (2004) Acta Crystallogr. D 60, 2126-2132. [DOI] [PubMed] [Google Scholar]
- 21.Schuettelkopf, A. W. & van Aalten, D. M. F. (2004) Acta Crystallogr. D 60, 1355-1363. [DOI] [PubMed] [Google Scholar]
- 22.Whittington, D. A., Rusche, K. M., Shin, H., Fierke, C. A. & Christianson, D. W. (2003) Proc. Natl. Acad. Sci. USA 100, 8146-8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernick, M. & Fierke, C. A. (2005) Arch. Biochem. Biophys. 433, 71-84. [DOI] [PubMed] [Google Scholar]
- 24.Coggins, B. E., Li, X. C., McClerren, A. L., Hindsgaul, O., Raetz, C. R. H. & Zhou, P. (2003) Nat. Struct. Biol. 10, 645-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCarthy, A. A., Peterson, N. A., Knijff, R. & Baker, E. N. (2004) J. Mol. Biol. 335, 1131-1141. [DOI] [PubMed] [Google Scholar]
- 26.Jackman, J. E., Fierke, C. A., Tumey, L. N., Pirrung, M., Uchiyama, T., Tahir, S. H., Hindsgaul, O. & Raetz, C. R. H. (2000) J. Biol. Chem. 275, 11002-11009. [DOI] [PubMed] [Google Scholar]
- 27.Bond, C. S. (2003) Bioinformatics 19, 311-312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.