Abstract
The nuclear envelope proteins LAP2, emerin and MAN1 share a conserved ∼40-residue ‘LEM’ motif. Loss of emerin causes Emery–Dreifuss muscular dystrophy. We have solved the solution NMR structure of the constant region of human LAP2 (residues 1–168). Human LAP21–168 has two structurally independent, non-interacting domains located at residues 1–50 (‘LAP2-N’) and residues 111–152 (LEM-domain), connected by an ∼60-residue flexible linker. The two domains are structurally homologous, comprising a helical turn followed by two helices connected by an 11–12-residue loop. This motif is shared by subdomains of T4 endonuclease VII and transcription factor rho, despite negligible (≤15%) sequence identity. NMR chemical shift mapping demonstrated that the LEM-domain binds BAF (barrier-to-autointegration factor), whereas LAP2-N binds DNA. Both binding surfaces comprise helix 1, the N-terminus of helix 2 and the inter-helical loop. Binding selectivity is determined by the nature of the surface residues in these binding sites, which are predominantly positively charged for LAP2-N and hydrophobic for the LEM-domain. Thus, LEM and LEM-like motifs form a common structure that evolution has customized for binding to BAF or DNA.
Keywords: LAP2/LEM motif/NMR/structure
Introduction
Lamin-associated polypeptide 2 (LAP2) comprises a family of alternatively spliced proteins that are associated with the inner nuclear membrane (Foisner and Grace, 1993; Dechat et al., 2000a). All isoforms of LAP2 share a constant N-terminal region, encoded by exons 1–3 (Berger et al., 1996), which spans residues 1–187 in the case of human LAP2 (Harris et al., 1994). Within this constant region resides the so-called LEM motif (Lin et al., 2000), which is also conserved in a growing number of other nuclear membrane proteins, including emerin and MAN1. Collectively, these proteins are termed LEM (LAP2-emerin-MAN1) proteins (Lin et al., 2000). LEM proteins are found in multicellular eukaryotes but not in single cell eukaryotes or plants (Cohen et al., 2001). The cellular functions of LEM proteins are not understood. LAP2, the best characterized LEM protein, binds to chromatin and to B-type lamins (nuclear intermediate filament proteins) in vitro (Foisner and Gerace, 1993; Furukawa et al., 1997, 1998). LAP2 isoforms are abundant in the nucleus, both at the nuclear envelope (e.g. LAP2β; Foisner and Gerace, 1993) and within the nuclear interior in association with A-type lamins (e.g. LAP2α; Dechat et al., 2000b). Interest in the molecular functions of LAP2 and other LEM proteins has intensified since the unexpected discovery of the null phenotype for emerin in humans; loss of emerin causes the X-linked recessive form of Emery–Dreifuss muscular dystrophy (Bione et al., 1994; Manilal et al., 1996; Nagano et al., 1996). This disease affects skeletal muscle and tendons, and causes potentially life-threatening cardiac conduction system defects; its mechanism is not understood (reviewed by Wilson et al., 2001). To further understand emerin and other LEM proteins, we have investigated the structure of the constant region of human LAP2.
The constant region of LAP2 has two binding activities. Residues 1–88 of rat LAP2 are sufficient to bind chromatin in vitro (Furukawa et al., 1997); within this region Worman and colleagues found a ‘LEM-like’ motif of unknown functional significance (Lin et al., 2000). The constant domain also interacts with barrier-to-autointegration factor (BAF) (Furukawa, 1999), a protein first identified for its role in retroviral DNA integration (Chen and Engelman, 1998; Lee and Craigie, 1998). Through deletion analysis, Furukawa (1999) narrowed the BAF-binding region to residues 67–137 of LAP2, which includes most of the LEM-domain and is distinct from the ‘chromatin-binding’ region. Alanine substitution mutagenesis of Xenopus LAP2 mapped most of the mutants defective for BAF interaction to the LEM-domain (Shumaker et al., 2001). BAF is an 89-residue protein that is highly conserved in multicellular eukaryotes with 60% sequence identity between the human and Caenorhabditis elegans homologs (Cai et al., 1998). BAF dimers bind to double-stranded DNA non-specifically and thereby bridge DNA molecules to form a large, discrete nucleoprotein complex (Zheng et al., 2000). This DNA-bridging property of BAF is proposed to block the autointegration of retroviral DNA by compacting it into a rigid structure. Most BAF is located inside the nucleus (Furukawa, 1999). BAF’s ability to interact simultaneously with both LAP2 and DNA in vitro (Shumaker et al., 2001), and other results (Yang et al., 1997; Gant et al., 1999), are consistent with a model in which LEM proteins and BAF mediate chromatin attachment to the nuclear envelope during nuclear assembly or interphase, or both.
In this paper we report the solution structure of the con stant region of human LAP2 (residues 1–168) using multidimensional NMR. We show that it comprises two small (∼40–50 residue) independent helical domains that are structurally very similar. Using chemical shift mapping, we demonstrate that the LEM-domain (here termed LAP2-C on account of its location in the C-terminal half of the constant region) interacts with BAF, while theanalogous `LEM-like' domain at the N-terminus (LAP2-N) unexpectedly binds DNA. In addition, we identify the binding surface for LAP2-C on BAF, and show that its shape and composition is complementary to the BAF binding surface on LAP2-C. The functional implications of these findings for LAP2 and emerin are discussed.
Results and discussion
Structure determination
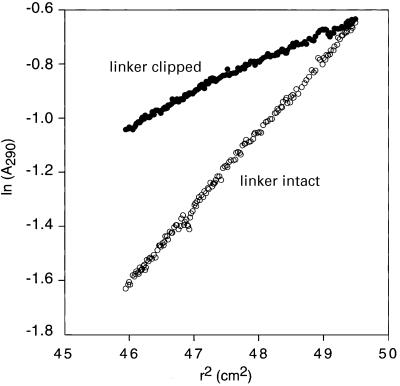
The structure of the constant region of human LAP2 (residues 1–168; LAP21–168) was solved by heteronuclear double and triple resonance NMR spectroscopy (Clore and Gronenborn, 1991; Bax and Grzesiek, 1993). The 1H-15N correlation spectrum of a longer construct comprising residues 1–187 is the same as that of LAP1–168, and the presence or absence of residues 169–187 does not affect the binding properties of the constant region of LAP2 (data not shown). The NMR data indicate that LAP21–168 comprises two globular domains, referred to hereafter as LAP2-N (residues 1–50) and LAP2-C (residues 111–153), which are connected by a highly flexible 60-residue linker. The two domains essentially tumble in solution independently from each other, as manifested by different alignment tensors in a liquid crystalline medium of Pf1 phage. The 1H-15N correlation spectrum of LAP21–168 is unaffected by thrombin cleavage between Arg86 and Ser87, and there are no NOEs observed between the two domains. These results are independently supported by data from analytical ultracentrifugation (Figure 1). Intact LAP21–168, including a His-tag at the N-terminus, has a mass of 20 700 ± 700 Da, showing that it exists as a monomer in solution. The average molecular weight of thrombin-cleaved LAP21–168 is 9040 ± 200 Da, consistent with the presence of two non-interacting, independently folded domains.
Fig. 1. Analytical ultracentrifugation of LAP21–168. Sedimentation equilibrium profiles, plotted in terms of ln(A290) versus the square of the radius (r2), for intact LAP21–168 (open circles) and thombin-cleaved LAP21–168 (filled circles). In the case of thrombin-cleaved LAP21–168 the N-terminal His-tag and the polypeptide chain between Arg86 and Ser87 are completely clipped as judged by SDS–PAGE. The sample comprises 10 mg/ml LAP21–168 in 50 mM potassium phosphate pH 7.2 and the data were collected at 20°C and 16 000 r.p.m. The molecular weight for intact LAP21–168 is 20 700 ± 700, indicating that LAP21–168 exists as a single monomeric species in solution. The average molecular weight of thrombin-cleaved LAP21–168 is 9040 ± 200. These data indicate that LAP21–168 includes two independently folded domains that do not associate with each other in solution, consistent with the NMR data.
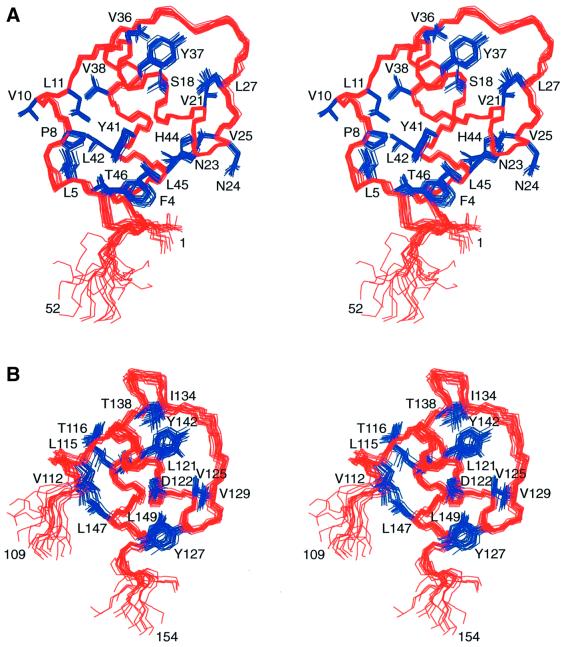
The structures of LAP2-N and LAP2-C were solved on the basis of 769 and 715 experimental restraints, respectively. The experimental restraints included a large number of residual dipolar couplings, measured in a liquid crystalline medium of phage Pf1, which provide long-range orientational information. The structural statistics are summarized in Table I, and Figure 2 shows best-fit superpositions of the final ensemble of 20 simulated annealing structures of LAP2-N and LAP2-C.
Table I. Structural statisticsa.
| LAP2-N |
LAP2-C |
|||||||
|---|---|---|---|---|---|---|---|---|
| <SAN> | (SAN)r | <SAC> | (SAC)r | |||||
| A. Experimental restraints | ||||||||
| R.m.s.ds from experimental restraints | ||||||||
| distance restraints (Å) (395/401)b | 0.023 ± 0.004 | 0.024 | 0.024 ± 0.008 | 0.012 | ||||
| torsion angle restraints (°) (151/121)b | 0.22 ± 0.05 | 0.49 | 0.36 ± 0.11 | 0.13 | ||||
| 13Cα/13Cβ shift restraints (p.p.m.) (105/91) | 1.13 ± 0.10 | 0.92 | 0.82 ± 0.10 | 0.81 | ||||
| Dipolar coupling R-factors (%)c | ||||||||
| 1DNH (39/34) | 5.7 ± 0.3 | 6.4 | 4.7 ± 0.5 | 2.9 | ||||
| 1DCH (36/29) | 6.1 ± 0.2 | 6.4 | 3.4 ± 0.8 | 1.9 | ||||
| 1DNC′ (24/19) | 27.3 ± 1.0 | 28.2 | 26.4 ± 2.6 | 26.2 | ||||
| 2DHNC′ (24/20) |
31.0 ± 0.7 |
30.1 |
28.2 ± 2.4 |
25.3 |
||||
| B. Measures of structure quality |
|
|
|
|
||||
| R.m.s.ds from idealized covalent geometry | ||||||||
| bonds (Å) | 0.003 ± 0 | 0.003 | 0.002 ± 0 | 0.003 | ||||
| angles (°) | 0.50 ± 0.10 | 0.48 | 0.28 ± 0.01 | 0.54 | ||||
| improper torsions (°) | 0.51 ± 0.03 | 0.49 | 0.5 ± 0.13 | 0.39 | ||||
| % residues in most favorable region of Ramachandran plotd | 87.9 ± 2.7 | 84.1 | 91.7 ± 2.5 | 89.0 | ||||
| Number of bad contacts per 100 residuesd |
6.6 ± 1.8 |
4.1 |
7.0 ± 2.5 |
9.5 |
||||
| C. Coordinate precision (Å)e |
|
|
|
|
||||
| Backbone (N, Cα, C′,O) | 0.43 ± 0.09 | 0.64 ± 0.10 | ||||||
| All heavy atoms | 1.05 ± 0.13 | 1.15 ± 0.10 | ||||||
aThe notation of the NMR structures is as follows: <SAN> and <SAC> are the final 20 simulated annealing structures for LAP2-N (residues 1–60) and LAP2-C (residues 101–159), respectively; SAN and SAC are the mean structures obtained by averaging the coordinates of the individual SA structures best fitted to each other (with respect to the backbone atoms of residues 2–50 for LAP2-N and 111–152 for LAP2-C); (SAN)r and (SAC)r are the corresponding restrained regularized mean structures. The number of terms for the various restraints is given in parentheses, the first number corresponding to LAP2-N, the second to LAP2-C. None of the structures exhibited interproton distance violations >0.5 Å or torsion angle violations >5°. Note that residue 1 and residues 51–60 in LAP2-N and residues 101–110 and 153–159 in LAP2-C are disordered in solution.
bThe distance restraints comprise the following (LAP2-N/LAP2-C): 361/363 NOE-derived interproton distance restraints within the protein subdivided into 4/19 intraresidue and 184/184 sequential (|i – j| = 1), 119/126 medium-range (1 < |i – j| ≤ 5) and 58/34 long-range (|i – j| > 5) interresidue restraints; and 34/38 distance restraints for 17/19 backbone hydrogen bonds located in helices and added during the final stages of refinement according to standard criteria. The torsion angle restraints for LAP2-N and LAP2-C comprised 59/47 φ, 59/47 ψ, 23/19 χ1 and 10/8 χ2.
cThe dipolar coupling R-factor is defined as the ratio of the r.m.s.d. between observed and calculated values to the expected r.m.s.d. if the vectors were randomly oriented. The latter is given by {2Da2[4 + 3η2]/5}1/2 where Da is the magnitude of the alignment tensor and η the rhombicity (Clore and Garrett, 1999). Note that the alignment tensors for LAP2-N and LAP2-C are different.
dThe % residues in the most favorable region of the Ramachandran plot and the number of bad contacts per 100 residues (which provides a measure of the quality of the non-bonded contacts) were determined using the program PROCHECK (Laskowski et al., 1993). There are no φ/ψ angles in the disallowed region of the Ramachandran plot and the number of bad contacts per 100 residues lies inside the range observed for high resolution (≤2 Å) crystal structures. The dihedral angle G-factors for φ/ψ, χ1/χ2, χ1 and χ3/χ4 are 0.19 ± 0.03, 0.22 ± 0.04, 0.36 ± 0.15 and 0.29 ± 0.12, respectively, for LAP2-N, and 0.18 ± 0.08, 0.30 ± 0.13, –0.36 ± 0.13 and –0.02 ± 0.17, respectively, for LAP2-C. The values refer to the ordered regions of LAP2-N and LAP2-C, which comprise residues 2–50 and 111–152, respectively.
eThe precision of the coordinates is defined as the average atomic r.m.s.d. between the 20 individual simulated annealing structures and the corresponding mean coordinates. The values refer to residues 2–50 of LAP2-N and 111–152 of LAP2-C.
Fig. 2. Best-fit superpositions of the final 20 simulated annealing structures of (A) the LAP2-N domain and (B) the LAP2-C domain. The backbone is shown in red and ordered side chains in blue.
Description of the structure
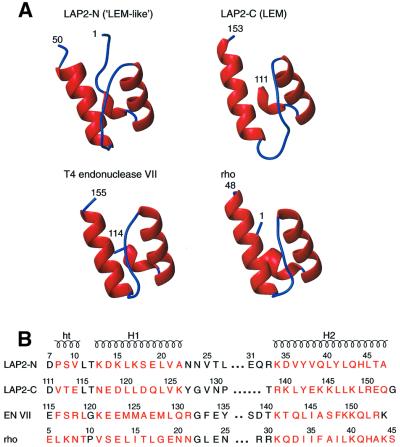
The two domains, LAP2-N (LEM-like) and LAP2-C (LEM), are structurally very similar (Figures 2 and 3). Both have a three-residue helical turn at their N-termini (residues 8–10 and 112–114, respectively) and two helices (residues 13–22 and 34–45 for LAP2-N, and residues 117–126 and 139–152 for LAP2-C). The two helices, which are connected by a long loop (11 residues for LAP2-N and 12 for LAP2-C) are oriented at an angle of ∼45°. The Cα atomic root mean square deviation (r.m.s.d.) between LAP2-N and LAP2-C is 1.4 Å for 33 residues (residues 7–25 and 33–46 of LAP2-N corresponding to residues 111–129 and 33–46, respectively, of LAP2-C). This Cα r.m.s.d. is consistent with the ∼25% sequence identity between the two domains for this structural alignment (Figure 3). Indeed, there are only two regions of structural difference between LAP2-N and LAP2-C. The first is at the N-terminus: for LAP2-N, the six residues preceding the helical turn are well defined, whereas the polypeptide chain preceding the helical turn in LAP2-C is not defined by the present NMR data (Figures 2 and 3A). The second difference involves the loop connecting the two helices (Figure 3A).
Fig. 3. (A) Ribbon diagrams of the LAP2-N domain (‘LEM-like’), the LAP2-C domain (LEM-domain), the C-terminal domain of T4 endonuclease VII and the N-terminal domain of the transcription termination factor rho. (B) Structure-based sequence alignment of the four domains. Residues in helices or helical turns are colored red. The coordinates of T4 endonuclease VII and rho were taken from Allison et al. (1998; PDB accession code 1A62) and Raaijmakers et al. (1999; PDB accession code 1EN7), respectively.
The N-terminus in LAP2-N is anchored to the body of the domain by packing interactions between Phe4 and hydrophobic residues (Tyr41 and Leu45) of helix 2, between Pro8 (at the beginning of the helical turn) and Tyr41 (helix 2), and between Leu11 (at the end of the helical turn) and Lys15 and Leu16 (helix 1). The packing of helices 1 and 2 involves Leu16, Leu20, Tyr37, Leu40 and Tyr41. The conformation of the loop is stabilized by a number of hydrophobic and electrostatic interactions. The hydrophobic interactions comprise Val25, Leu20 (helix 1) and His44 (helix 2); Leu27, Leu20 (helix 1) and Leu40 (helix 2); Pro28 and Leu40 (helix 2); and Glu31 and Val36 (helix 2). There are also two electrostatic interactions involving the backbone carbonyl of Glu31 and the hydroxyl of Tyr37 (helix 2), and the backbone carbonyl of Arg33 and the amino group of Lys13 (helix 1).
As noted above, LAP2-C is shorter than LAP2-N. The orientation of the helical turn at the N-terminus of LAP2-C is stabilized by electrostatic (between the hydroxyl group of Thr113 and the carboxylate of Glu143) and hydrophobic (between Val112, Leu146 and Leu147) interactions with helix 2. The packing of helices 1 and 2 involves Asn117 and Arg139, Leu120 and Glu143, Leu124 and Tyr142, and Tyr127 and Leu149. The loop is stabilized by hydrophobic contacts with helix 2 (between Val129 and Leu149, Pro131 and Tyr142, and Ile134 and Tyr142) and helix 1 (between Ile134 and Leu120).
A search of the DALI structural database (Holm and Sander, 1993) indicates the existence of structurally homologous domains in a number of DNA and RNA binding proteins. The two closest matches with Cα atomic r.m.s.ds ranging from 1.3 to 1.5 Å (for between 33 and 37 atoms) are the C-terminal domain of T4 endonuclease VII (Raaijmakers et al., 1999) and the N-terminal domain of the transcription termination factor rho (Allison et al., 1998). The percentage sequence identities for the structure-based sequence alignments shown in Figure 3B are ∼10% between LAP2-C and either endonuclease VII or rho, ∼15% between LAP2-N and either endonuclease VII or rho, and ∼20% between endonuclease VII and rho. Although T4 endonuclease VII binds DNA and rho binds RNA, the exact function of the subdomains that are structurally homologous to LAP2-N and LAP2-C are currently unknown.
Interaction of LAP21–168 with DNA, BAF and the BAF–DNA nucleoprotein complex
The interactions of LAP21–168 with DNA, BAF and the BAF–DNA nucleoprotein complex were analyzed by chemical shift mapping using 1H-15N correlation spectroscopy with either LAP21–168 or BAF uniformly labeled with 15N. These results are summarized in Figures 4 and 5.
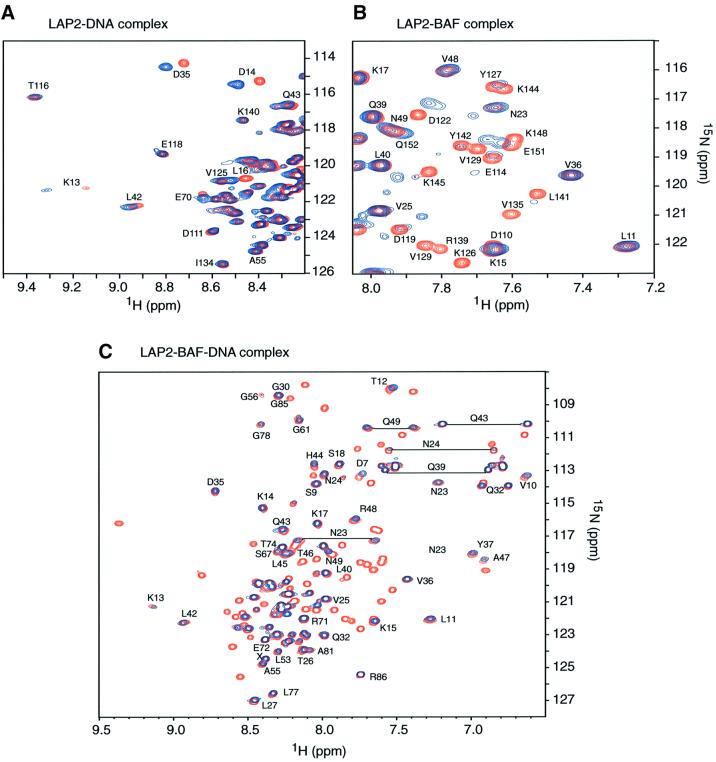
Fig. 4. 1H-15N correlation spectra illustrating the interactions between LAP21–168, DNA and BAF. (A) Comparison of LAP2 in the absence (red) and presence of 1.5 equivalents (blue) of a duplex DNA dodecamer. Only cross-peaks of the LAP2-N domain shift upon DNA binding, while those of the LAP2-C domain remain unchanged, indicating that duplex DNA interacts exclusively with the LAP2-N (LEM-like) domain. (B) Comparison of the spectrum of LAP21–168 (selected region) in the absence (red) and presence (blue) of one equivalent (in terms of monomer) of BAF. Only the cross-peaks in the LAP2-C domain change significantly upon binding to BAF, while the cross-peaks of the LAP2-N domain remain unchanged. Thus, BAF only interacts with the LAP2-C domain (LEM-domain). (C) Comparison of LAP21–168 in the absence (red) and presence (blue) of the BAF–DNA nucleoprotein complex. All resolved peaks in the complex are labeled and arise from the LAP2-N domain. All peaks originating from the LAP2-C domain disappear upon binding the BAF–DNA nucleoprotein complex. These data indicate that only the LAP2-C domain interacts with the BAF–DNA nucleoprotein complex.
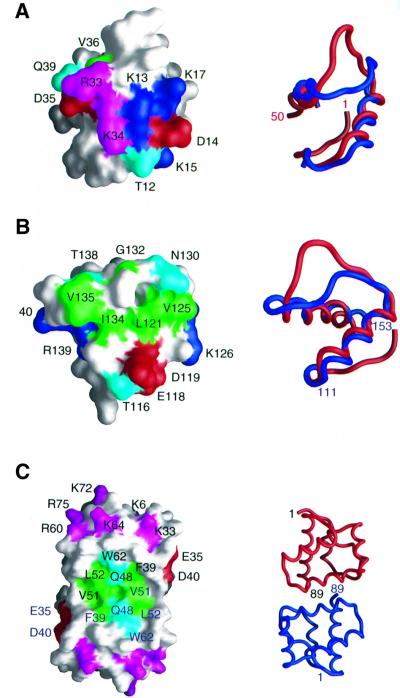
Fig. 5. Interaction surfaces on (A) LAP2-N for DNA, (B) LAP2-C for BAF and (C) BAF for LAP2-C derived from chemical shift mapping. The left side of the figure shows the domains as molecular surface representations with residues that either shift or broaden significantly upon binding colored in green (hydrophobic), cyan (hydrophilic), blue (positively charged) and red (negatively charged). For LAP2-N (A), Arg33 and Lys34 are displayed in magenta since they are clearly located in the DNA binding site but their backbone amide resonances were too broad to be observed in the 1H-15N correlation spectra either in the free state or bound to DNA. For BAF (C), residues known to interact with DNA from single site mutational analysis are also displayed in magenta (Umland et al., 2000). To the right of the figure are ribbon diagrams in the same orientations as the molecular surfaces; best-fits of LAP2-N (red) and LAP2-C (blue) are displayed in (A) and (B); and the two subunits of BAF are shown in red and blue in (C). The views shown in (A) and (B) differ by an ∼90° rotation. The structures in (A) and (B) are displayed on the same scale, while that in (C) is on a smaller scale. Coordinates for BAF are from Cai et al. (1998; PDB accession code 1QCK).
Upon titration of a double-stranded DNA 12mer or 21mer to 15N-labeled LAP21–168, only cross-peaks from the LAP2-N domain are shifted, while the spectrum of the LAP2-C domain remains unchanged (Figure 4A). Thus, only the LAP2-N domain interacts with free DNA. The exchange between free and DNA-complexed LAP2 is fast on the chemical shift scale; the maximal chemical shift difference observed is 120 Hz at a 1H frequency of 750 MHz, indicating that the lifetime of the complex is less than ∼1.5 ms. Significant 1H-15N cross-peak shifts (>0.05 p.p.m. in 1H and/or >0.2 p.p.m. in 15N) are seen for residues in helix 1 (Thr12, Lys13, Asp14, Lys15, Leu16 and Lys17) and at the N-terminal end of helix 2 (Asp35, Tyr36 and Gln39). The location of these residues on the surface of LAP2-N defines the proposed DNA-binding site and is depicted in Figure 5A. The surface between these two locations is bridged by Arg33 and Lys34 (Figure 5A). These resonances were broadened in both free and DNA-complexed LAP2-N such that their 1H-15N correlation peaks could not be observed. However, it seems likely that these two positively charged residues, which are located in the loop connecting helices 1 and 2, are also part of the DNA-binding site.
When unlabeled BAF is titrated into 15N-labeled LAP21–168, only 1H-15N cross-peaks arising from residues in the LAP2-C domain are either shifted or disappear. Exchange is on the slow side of intermediate; the maximal shift difference at a 1H frequency of 750 MHz is ∼190 Hz, indicative of a lifetime of ∼1.5–2 ms. Cross-peaks exhibiting significant changes (>0.1 p.p.m. in 1H and/or >0.4 p.p.m. in 15N) involve helix 1 (Thr116, Glu118, Asp119, Leu121, Val125 and Lys126), the loop connecting helices 1 and 2 (Asn130, Gly132, Ile134, Val135 and Thr138) and the N-terminal end of helix 2 (Arg139 and Lys140) (Figure 5B). These residues are located on a convex protrusion on the surface of LAP2-C, which is characterized by a central hydrophobic region (Leu121, Val125, Ile134 and Val135) surrounded by a ring of hydrophilic or charged residues (Figure 5B).
When unlabeled LAP21–168 is added to 15N-labeled BAF, many cross-peaks in the 1H-15N correlation spectrum are shifted but only the cross-peaks of Glu35, Phe39, Asp40, Gly47, Gln48, Leu50, Val51 and Trp62 (backbone and sidechain) disappear. These residues are located in a concave cleft bridging the two subunits of BAF, and again comprise a central hydrophobic patch surrounded by hydrophilic residues (Figure 5C). Also shown in Figure 5C are BAF residues involved in DNA binding as deduced from single site mutational analysis, specifically Lys6, Lys33, Arg60, Lys64, Lys72 and Arg75 (Umland et al., 2000). These positively charged residues form a contiguous surface located at either end of the BAF dimer, which does not overlap with the LAP2-C binding surface.
Thus, the two proposed interaction surfaces on LAP2-C and BAF are complementary to one another and resemble many protein–protein interaction surfaces (Wang et al., 2000). The interaction between LAP2-C and BAF is predominantly hydrophobic in nature, and this is supported by the observation that the complex, as judged by NMR, cannot be disrupted by high salt (up to 0.5 M NaCl; higher salt concentrations were not tested; data not shown).
We also carried out a titration experiment involving 15N-labeled LAP21–168 and the unlabeled BAF–DNA nucleoprotein complex (Figure 4C), a discrete entity consisting of six BAF dimers plus an estimated six molecules of DNA, with a molecular mass in excess of 150 kDa (Zheng et al., 2000). Upon adding the BAF–DNA nucleoprotein complex, the 1H-15N cross-peaks of the LAP2-C domain completely disappear, while those of the LAP2-N domain remain unchanged. Thus, both free BAF and the BAF–DNA nucleoprotein complex interact exclusively with the LAP2-C domain.
Concluding remarks
We have solved the solution structure of the constant region of LAP2. We show that it consists of two structurally independent, non-interacting domains: residues 1–50 (here termed LAP2-N), which correspond to the LEM-like motif predicted by hydrophobic cluster analysis (Lin et al., 2000), and residues 111–152 (here termed LAP2-C), which correspond to the LEM motif. These two structural domains are connected by a long (∼60 residue), highly flexible linker. Similar structural motifs were found at the C-terminal domain of T4 endonuclease VII and the N-terminal domain of transcription termination factor rho, respectively, despite having no significant sequence identity. Our findings strongly suggest that the LEM and LEM-like motifs comprise a structural module that is well suited for interacting with either protein or DNA (or possibly RNA), depending upon the nature of the surface residues.
The interaction surfaces for DNA and BAF are located exclusively in the LAP2-N and LAP2-C domains, respectively. These interaction surfaces involve similar regions of the two domains, specifically helix 1, the loop that connects helices 1 and 2, and the N-terminal residues of helix 2. The distinct selectivities of LAP2-N (which binds DNA) and LAP2-C (which binds BAF) are determined by their surface residues in these locations: predominantly positively charged in the case of LAP2-N, and mainly hydrophobic for LAP2-C. In addition, we show that the convex interaction surface on LAP2-C is complementary both in shape and composition to the concave interaction surface on BAF. Our results suggest that LEM and ‘LEM-like’ motifs, originally defined by sequence homology and hydrophobic cluster analysis (Lin et al., 2000), form a conserved structural module that has been customized during evolution for binding to either BAF or DNA (and possibly additional ligands) through changes in its surface residues. We have designated the BAF-binding and DNA-binding modules as LEM-B and LEM-D, respectively.
We discovered that the constant region of LAP2 interacts directly with DNA, through its LEM-D domain. The LEM-D domain (residues 1–50) appears to be biologically relevant. It is located within residues 1–88 of rat LAP2, which interact with chromatin in vitro (Furukawa et al., 1997). Furthermore, in a mutational analysis of Xenopus LAP2, alanine substitutions in LEM-D (equivalent to human residues Arg33/Lys34/Asp35; mutant m4) completely blocked the activity of the LAP2 constant region in nuclear assembly extracts (Shumaker et al., 2001). This same mutant protein exhibited normal binding to BAF and the BAF–DNA nucleoprotein complex, suggesting that direct binding between LAP2 and DNA is relevant to its function during nuclear assembly. Our present data indicate that the two domains are structurally independent of each other. This does not mean, however, that they are functionally independent. We propose that DNA binding by LEM-D may be important in vivo, because it could stabilize the attachment of LAP2 to BAF–DNA complexes on chromosomal DNA. Interestingly, emerin and MAN1 both lack the LEM-D domain, and their interactions with BAF bound to chromosomal DNA might therefore be weaker than LAP2.
We propose that the constant region of LAP2 consists of two small functional ‘beads’ on a flexible ‘string’. A flexible, modular arrangement for membrane-anchored LAP2 proteins is logical, given that LAP2 may interlink the lamin filaments and chromatin, both of which are unusually long, large and dynamic structures. Flexible modular function is also consistent with the existence of numerous splicing isoforms of LAP2, many of which differ by the loss of one or a few exons (Berger et al., 1996; Gant et al., 1999). It would be interesting to determine the structure of the predicted lamin-binding ‘module(s)’ in the variable regions of LAP2 (Furukawa et al., 1998; Dechat et al., 2000b), and to look for new structural modules, to test the idea that LAP2 consists entirely of structural ‘beads on a string’.
Our work has further implications for the structure of emerin, which is linked to Emery–Dreifuss muscular dystrophy. In addition to sharing the LEM-B domain, LAP2β and emerin polypeptides also have moderate to high sequence similarity outside the LEM-B domain. A mutational analysis of human emerin has identified conserved residues that are critical for binding to either BAF or lamin A, but not both (K.K.Lee, R.S.Lee, T.Haraguchi, T.Koujin, Y.Hiraoka and K.L.Wilson, submitted), consistent with a flexible, modular structure for emerin. The structure determination of the LEM-B domain in LAP2 now allows one to predict precisely the structure of the LEM-B domain in emerin. Further work is expected to shed light on the structure of this interesting and medically relevant family of nuclear envelope proteins.
Materials and methods
Protein expression and purification
The constant region of human LAP2 (residues 1–168; LAP21–168) was cloned in the pET15b vector (Novagen) as a fusion protein with a His6-tag at its N-terminus, and expressed in Escherichia coli BL21 (DE3) cells grown in minimal medium using 15NH4Cl and/or 13C6-glucose as the sole nitrogen and carbon sources. Cells were grown at 37°C to an OD600 of 1.0 and induced with isopropyl-β-d-thiogalactopyranoside for 3 h at 37°C. Cells were suspended in 50 mM HEPES pH 7.5 containing 200 mM NaCl and 40 µg of lysozyme per ml of suspension, and further lysed through a French press. LAP2, which was found mostly in the soluble fraction, was bound to a Ni affinity column, thoroughly washed with washing buffer (50 mM HEPES, 1 M NaCl pH 7.5) and eluted with an imidazole gradient (25 mM to 0.7 M within 150 ml elution volume). Fractions containing LAP2 were pooled and further purified by gel filtration on a Superdex75 column (Pharmacia) using 50 mM HEPES, 200 mM NaCl pH 7.5 as the running buffer. Samples for NMR contained ∼1 mM protein in 50 mM phosphate buffer pH 7.2.
LAP21–168 contains a long flexible linker spanning residues 51–110, which is easily cleaved by various proteases. Thrombin specifically cleaves the peptide bond between Arg86 and Ser87, as determined by mass spectrometry and N-terminal amino acid sequence analysis. To assess the significance of the linker, two constructs were made: one in which the thrombin site (LVPRGSH) between the His-tag and LAP2 was replaced with an enterokinase site (DDDDDK), and the other in which Arg86 was replaced by Gln. In the first construct, enterokinase cleaved the His-tag, leaving the linker intact; in the second construct, the Gln86–Ser87 peptide bond was insensitive to thrombin. In both cases, the linker region was protected from cleavage by thrombin. 1H-15N correlation spectra of linker-intact and linker-clipped LAP21–168 were the same, indicating that the linker has no structural significance. In addition, BAF bound equally well to linker-intact and linker-clipped LAP21–168, as judged by the disappearance or shifting of cross-peaks in the 1H-15N HSQC spectra of 15N-labeled LAP21–168 or BAF upon mixing with unlabeled BAF or LAP21–168, respectively.
BAF was expressed, purified and 15N-labeled as described previously (Cai et al., 1998), except that gel filtration was substituted for the reverse phase chromatography step (Zheng et al., 2000). The DNA 12mer and 21mer used in the titration studies were purchased from Midlands Certified Reagent Co. and purified by anion exchange chromatography. The two sequences were as follows: 5′d(ATCTCTAGCAGT). 5′d(ACTGCTAGAGAT) and 5′d(GTGTGGAAAATCTCTAGCAGT). 5′d(ACTGCTAGA GATTTTCCACAC). The BAF–DNA nucleoprotein complex was prepared as described previously (Zheng et al., 2000).
Analytical ultracentrifugation experiments were conducted using a Beckman Optima XL-A analytical ultracentrifuge. The data were analyzed in terms of a single ideal solute to obtain the buoyant molecular mass, M(1–νρ) using the Optima XL-A data analysis software (Beckman).
NMR spectroscopy
Spectra were recorded at 27°C on Bruker DMX500, DRX600 and DMX750 spectrometers. Spectra were processed using the program NMRPipe (Delaglio et al., 1995), and analyzed using the programs PIPP, CAPP and STAPP (Garrett et al., 1991). Spectra were collected on both linker intact LAP21-168 and the Arg86–Ser87 peptide bond-cleaved LAP21–168. Sequential assignment of 1H, 15N and 13C resonances was achieved by means of through-bond heteronuclear scalar correlations along the protein backbone and side chains (Clore and Gronenborn, 1991; Bax and Grzesiek, 1993) using 3D HNCO, CBCACONH, HNCACB, (H)C(CO)NH TOCSY, H(CCO)NH-TOCSY and CCH-COSY experiments. Interproton distance restraints were derived from 3D 15N- and 13C-separated NOE experiments. Stereospecific assignments of valine and leucine methyl groups were obtained from a 1H-13C HSQC spectrum recorded on 10% 13C-labeled LAP21–168 (Neri et al., 1989). Side chain rotamers were derived from 3JNCγ(aromatic, methyl and methylene) and 3JC′Cγ(aromatic, methyl and methylene) scalar couplings measured by quantitative J correlation spectroscopy (Bax et al., 1994), in combination with data from a short mixing time (40 ms) 3D 13C-separated NOE spectrum recorded in H2O.
Residual 1DNH, 1DCαH, 1DNC′ and 2DHNC′ dipolar couplings (Tjandra and Bax, 1997) were measured in a liquid crystalline medium of phage Pf1 (15 mg/ml) (Clore et al., 1998a; Hansen et al., 1999). The magnitudes of the axial (DaNH) and rhombic (η) components of the alignment tensor DNH were obtained by examining the distribution of the normalized residual dipolar couplings (Clore et al., 1998b). The magnitudes of the alignment tensor for the LAP2-N (DaNH = –7.8 Hz, η = 0.38) and LAP2-C (DaNH = 10.7 Hz and η = 0.18) domains were completely different, indicating that they are non-interacting and align independently of each other.
Structure calculations
Approximate interproton distance restraints were grouped into four distance ranges: 1.8–2.7 Å (1.8–2.9 Å for NOEs involving NH protons), 1.8–3.3 Å (1.8–3.5 Å for NOEs involving NH protons), 1.8–5.0 Å and 1.8–6.0 Å, corresponding to strong, medium, weak and very weak NOEs, respectively. In addition, 0.5 Å was added to the upper limit of inter proton distance restraints involving methyl groups. Distances involving ambiguous NOEs, non-stereospecifically assigned methylene protons, methyl groups, and the Hδ and Hε protons of Tyr and Phe were represented as a (Σr–6)–1/6 sum (Nilges, 1993). Backbone torsion angles were derived from backbone 1H, 15N and 13C chemical shifts using the program TALOS (Cornilescu et al., 1999). Side chain torsion angle restraints were derived from heteronuclear coupling and NOE data as described previously (Omichinski et al., 1997).
Structures were calculated by simulated annealing (Nilges et al., 1988) using the NIH version (J.Kuszewski, C.D.Schwieters and G.M.Clore, available by anonymous ftp on portal.niddk.nih.gov in /pub/clore/xplor_nih) of XPLOR (Brünger, 1993), which has been highly modified to incorporate numerous features relevant to NMR (Clore and Gronenborn, 1998), as well as new and highly efficient algorithms for torsion angle dynamics and minimization (Schwieters and Clore, 2001a). All simulated annealing (Nilges et al., 1988) and minimization calculations were carried out in torsion angle space; the torsion angle dynamics algorithm employed a sixth-order predictor-corrector integrator with automatic time-step selection, which varied during the course of the calculation (Schwieters and Clore, 2001a). The simulated annealing protocol employed was essentially that described by Omichinski et al. (1997) with the difference that torsion angle dynamics rather than Cartesian coordinate dynamics were employed, and that the target function included a few additional terms. Bond lengths and angles were constrained to idealized covalent geometry. The target function for simulated annealing consisted of: harmonic terms for covalent geometry (i.e. improper torsion angles used to define chirality and planarity, and bond lengths and angles associated with the closed ring systems of proline; note that the other bonds and angles are held fixed by constraints); square-well potentials for the interproton distance, torsion angle and hydrogen bonding restraints; harmonic potentials for the 13Cα and 13Cβ secondary chemical shift, and residual dipolar coupling restraints (Clore et al., 1998c); and three terms for the non-bonded contacts. The latter comprise a quartic van der Waals repulsion term (Nilges et al., 1988), the DELPHIC torsion angle database potential term of mean force (Kuszewski and Clore, 2000) and the radius of gyration restraint (Kuszewski et al., 1999). No hydrogen bonding, electrostatic or 6–12 Lennard–Jones empirical potential energy terms were present in the target function used for simulated annealing or restrained regularization.
Structure figures were generated using the programs VMD-XPLOR (Schwieters and Clore, 2001b), MOLMOL (Koradi et al., 1996) and GRASP (Nicholls et al., 1991). The coordinates have been deposited in the RCSB Protein Data Bank (accession code IGJJ).
Acknowledgments
Acknowledgements
We thank Dan Garrett, Charles Schwieters and Frank Delaglio for software support; and James Chou and Marcus Zweckstetter for useful discussions. This work was supported by the AIDS Targeted Antiviral Program of the Office of the Director of the National Institutes of Health (to G.M.C. and R.C.) and by a grant from the National Institutes of Health (ROI GM48646, to K.L.W.).
References
- Allison T.J., Wood,T.C., Briercheck,D.M., Rastinejad,F., Richardson,J.P. and Rule,G.S. (1998) Crystal structure of the RNA-binding domain from transcription termination factor rho. Nature Struct. Biol., 5, 352–356. [DOI] [PubMed] [Google Scholar]
- Bax A. and Grzesiek,S. (1993) Methodological advances in protein NMR. Acc. Chem. Res., 26, 131–138. [Google Scholar]
- Bax A., Vuister,G.W., Grzesiek,S., Delaglio,F., Wang,A.C., Tschudin,R. and Zhu,G. (1994) Measurement of homo- and heteronuclear J couplings from quantitative J correlation. Methods Enzymol., 239, 79–106. [DOI] [PubMed] [Google Scholar]
- Berger R. et al. (1996) The characterization and localization of the mouse thymopoietin/lamina-associated polypeptide 2 gene and its alternatively spliced products. Genome Res., 6, 361–370. [DOI] [PubMed] [Google Scholar]
- Bione S., Maestrini,E., Rivella,S., Mancini,M., Regis,S., Romeo,G. and Toniolo,D. (1994) Identification of a novel X-linked gene responsible for Emery–Dreifuss muscular dystrophy. Nature Genet., 8, 323–327. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. (1993) XPLOR: A System for X-ray Crystallography and NMR. Yale University Press, New Haven, CT.
- Cai M., Huang,Y., Zheng,R., Wei,S.-Q., Ghirlando,R., Lee,M.S., Craigie,R., Gronenborn,A.M. and Clore,G.M. (1998) Solution structure of the cellular factor BAF responsible for protecting retroviral DNA from autointegration. Nature Struct. Biol., 5, 903–909. [DOI] [PubMed] [Google Scholar]
- Chen H. and Engelman,A. (1998) The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc. Natl Acad. Sci. USA, 95, 15270–15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore G.M. and Garrett,D.S. (1999) R-factor, free R and complete cross-validation for dipolar coupling refinement of NMR structures. J. Am. Chem. Soc., 121, 9008–9012. [Google Scholar]
- Clore G.M. and Gronenborn,A.M. (1991) Structures of larger proteins in solution: three- and four-dimensional heteronuclear NMR spectroscopy. Science, 252, 1390–1399. [DOI] [PubMed] [Google Scholar]
- Clore G.M. and Gronenborn,A.M. (1998) New methods of structure refinement for macromolecular structure determination by NMR. Proc. Natl Acad. Sci. USA, 95, 5891–5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore G.M., Starich,M.R. and Gronenborn,A.M. (1998a) Measurement of residual dipolar couplings of macromolecules aligned in the nematic phase of a colloidal suspension of rod-shaped viruses. J. Am. Chem. Soc., 120, 10571–10572. [Google Scholar]
- Clore G.M., Gronenborn,A.M. and Bax,A. (1998b) A robust method for determining the magnitude of the fully asymmetric alignment tensor of oriented macromolecules in the absence of structural information. J. Magn. Reson., 133, 216–221. [DOI] [PubMed] [Google Scholar]
- Clore G.M., Gronenborn,A.M. and Tjandra,N. (1998c) Direct refinement against residual dipolar couplings in the presence of rhombicity of unknown magnitude. J. Magn. Reson., 131, 159–162. [DOI] [PubMed] [Google Scholar]
- Cohen M., Lee,K.K., Wilson,K.L. and Gruenbaum,Y. (2001) Transcriptional repression, apoptosis, human disease and the functional evolution of the nuclear lamina. Trends Biochem. Sci., 26, 41–47. [DOI] [PubMed] [Google Scholar]
- Cornilescu G., Delaglio,F. and Bax,A. (1999) Protein backbone angle restraints from searching a database for protein chemical shift and sequence homology. J. Biomol. NMR, 13, 289–302. [DOI] [PubMed] [Google Scholar]
- Dechat T., Vlcek,S. and Foisner,R. (2000a) Lamina-associated polypeptide 2 isoforms and related proteins in cell cycle-dependent nuclear structure dynamics. J. Struct. Biol., 129, 335–345. [DOI] [PubMed] [Google Scholar]
- Dechat T., Korbei,B., Vaughan,O.A., Vlcek,S., Hutchinson,C.J. and Foisner,R. (2000b) Lamina-associated polypeptide 2α binds intranuclear A-type lamins. J. Cell Sci., 113, 3473–3483. [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek,S., Vuister,G.W., Zhu,G., Pfeifer,P. and Bax,A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX PIPES. J. Biomol. NMR, 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Foisner R. and Gerace,L. (1993) Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes and binding is modulated by mitotic phosphorylation. Cell, 73, 1267–1279. [DOI] [PubMed] [Google Scholar]
- Furukawa K. (1999) LAP2 binding protein 1 (L2BP1/BAF) is a candidate mediator of LAP2–chromatin interaction. J. Cell Sci., 112, 2485–2492. [DOI] [PubMed] [Google Scholar]
- Furukawa K., Glass,C. and Kondo,T. (1997) Characterization of the chromatin-binding activity of lamina-associated polypeptide (LAP) 2. Biochem. Biophys. Res. Commun., 238, 240–246. [DOI] [PubMed] [Google Scholar]
- Furukawa K., Fritze,C.E. and Gerace,L. (1998) The major nuclear envelope targeting protein LAP2 coincides with its lamin binding region but is distinct from its chromatin interaction domain. J. Biol. Chem., 273, 4213–4219. [DOI] [PubMed] [Google Scholar]
- Gant T.M., Harris,C.A. and Wilson,K.L. (1999) Roles of LAP2 proteins in nuclear assembly and DNA replication: truncated LAP2β proteins alter lamina assembly, envelope formation, nuclear size and DNA replication efficiency in Xenopus laevis extracts. J. Cell Biol., 144, 1083–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett D.S., Powers,R., Gronenborn,A.M. and Clore,G.M. (1991) A common sense approach to peak picking in two-, three- and four-dimensional spectra using automatic computer analysis of contour diagrams. J. Magn. Reson., 94, 214–220. [DOI] [PubMed] [Google Scholar]
- Hansen M.R., Mueller,L. and Pardi,A. (1999) Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nature Struct. Biol., 5, 1065–1074. [DOI] [PubMed] [Google Scholar]
- Harris C.A., Andryuk,P.J., Cline,S., Chan,H.K., Natarajan,A., Siekierka,J.J. and Goldstein,G. (1994) Three distinct human thymopoietins are derived from alternatively spliced mRNAs. Proc. Natl Acad. Sci. USA, 91, 6283–6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Koradi R., Billeter,M. and Wüthrich,K. (1996) MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph., 14, 51–55. [DOI] [PubMed] [Google Scholar]
- Kuszewski J. and Clore,G.M. (2000) Source of and solutions to problems in the refinement of protein NMR structures against torsion angle potentials of mean force. J. Magn. Reson., 146, 249–254. [DOI] [PubMed] [Google Scholar]
- Kuszewski J., Gronenborn,A.M. and Clore,G.M. (1999) Improving the packing and accuracy of NMR structures with a pseudopotential for the radius of gyration. J. Am. Chem. Soc., 121, 2337–2338. [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lee M.S. and Craigie,R. (1998) A previously unidentified host protein protects retroviral DNA from autointegration. Proc. Natl Acad. Sci. USA, 95, 1528–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F., Blake,D.L., Callebaut,I., Skerjanc,I.S., Holmer,L., McBurney,M.W., Paulin-Levasseur,M. and Worman,H.J. (2000) An inner nuclear membrane protein that shares the LEM domain with lamina associated polypeptide 2 and emerin. J. Biol. Chem., 275, 4840–4847. [DOI] [PubMed] [Google Scholar]
- Manilal S., Man,N.T., Sewry,C.A. and Morris,G.E. (1996) The Emery–Dreifuss muscular dystrophy protein, emerin, is a nuclear membrane protein. Hum. Mol. Genet., 5, 801–808. [DOI] [PubMed] [Google Scholar]
- Nagano A., Koga,R., Ogawa,M., Kurano,Y., Kawada,J., Okada,R., Hayashi,Y.K., Tsukahara,T. and Arahata,K. (1996) Emerin deficiency at the nuclear membrane in patients with Emery–Dreyfuss muscular dystrophy. Nature Genet., 12, 254–259. [DOI] [PubMed] [Google Scholar]
- Neri D., Szyperski,T., Otting,G., Senn,H. and Wüthrich,K. (1989) Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry, 28, 7510–7516. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from interfacial and thermodynamic properties of hydrocarbons. Proteins, 17, 297–309. [DOI] [PubMed] [Google Scholar]
- Nilges M. (1993) A calculational strategy for the structure determination of symmetric dimers by 1H NMR. Proteins, 17, 297–309. [DOI] [PubMed] [Google Scholar]
- Nilges M., Gronenborn,A.M., Brünger,A.T. and Clore,G.M. (1988) Determination of three-dimensional structures of proteins by simulated annealing with interproton distance restraints: application to crambin, potato carboxypeptidase inhibitor and barley serine proteinase inhibitor 2. Protein Eng., 2, 27–38. [DOI] [PubMed] [Google Scholar]
- Omichinski J.G., Pedone,P.V., Felsenfeld,G., Gronenborn,A.M. and Clore,G.M. (1997) The solution structure of a specific GAGA factor–DNA complex reveals a modular binding mode. Nature Struct. Biol., 4, 122–132. [DOI] [PubMed] [Google Scholar]
- Raaijmakers H., Vix,O., Törö,I., Golz,S., Kemper,B. and Suck,D. (1999) X-ray structure of T4 endonuclease VII: a DNA junction resolvase with a novel fold and unusual domain-swapped dimer architecture. EMBO J., 18, 1447–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieters C.D. and Clore,G.M. (2001a) Internal coordinates for molecular dynamics and minimization in structure determination and refinement. J. Magn. Reson., in press. [DOI] [PubMed] [Google Scholar]
- Schwieters C.D. and Clore,G.M. (2001b) The VMD-XPLOR visualization package for NMR structure refinement. J. Magn. Reson., 149, 239–244. [DOI] [PubMed] [Google Scholar]
- Shumaker D.K., Lee,K.L., Tanhehco,Y.C., Craigie,R. and Wilson,K.L. (2001) LAP2 binds to BAF–DNA complexes: requirement for the LEM domain and modulation by variable regions. EMBO J., 20, 1754–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjandra N. and Bax,A. (1997) Direct measurement of distances and angles in biomolecules by NMR in dilute liquid crystalline medium. Science, 278, 1111–1114. [DOI] [PubMed] [Google Scholar]
- Umland T.C., Wei,S.-Q., Craigie,R. and Davies,D.R. (2000) Structural basis of DNA bridging by barrier-to-autointegration factor. Biochemistry, 39, 9130–9138. [DOI] [PubMed] [Google Scholar]
- Wang G., Louis,J.M., Sondej,M., Seok,Y.-J., Peterkofsky,A. and Clore,G.M. (2000) Solution structure of the phosphoryl transfer complex between the signal transducing proteins HPr and IIAGlucose of the Escherichia coli phosphoenolpyruvate:sugar phosphotransferase system. EMBO J., 19, 5635–5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K.L., Zastrow,M.S. and Lee,K.K. (2001) Lamins and disease: insights into nuclear infrastructure. Cell, 104, 647–650. [PubMed] [Google Scholar]
- Yang L., Guan,T. and Gerace,L. (1997) Lamin-binding fragment of LAP2 inhibits increase in nuclear volume during the cell cycle and progression into S phase. J. Cell Biol., 139, 1077–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng R., Ghirlando,R., Lee,M.S., Mizuuchi,K., Krause,M. and Craigie,R. (2000) Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex. Proc. Natl Acad. Sci. USA, 97, 8997–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]