Abstract
Recent studies show that heterochromatin-associated protein-1 (HP1) recognizes a ‘histone code’ involving methylated Lys9 (methyl-K9) in histone H3. Using in situ immunofluorescence, we demonstrate that methyl-K9 H3 and HP1 co-localize to the heterochromatic regions of Drosophila polytene chromosomes. NMR spectra show that methyl-K9 binding of HP1 occurs via its chromo (chromosome organization modifier) domain. This interaction requires methyl-K9 to reside within the proper context of H3 sequence. NMR studies indicate that the methylated H3 tail binds in a groove of HP1 consisting of conserved residues. Using fluorescence anisotropy and isothermal titration calorimetry, we determined that this interaction occurs with a KD of ∼100 µM, with the binding enthalpically driven. A V26M mutation in HP1, which disrupts its gene silencing function, severely destabilizes the H3-binding interface, and abolishes methyl-K9 H3 tail binding. Finally, we note that sequence diversity in chromo domains may lead to diverse functions in eukaryotic gene regulation. For example, the chromo domain of the yeast histone acetyltransferase Esa1 does not interact with methyl- K9 H3, but instead shows preference for unmodified H3 tail.
Keywords: chromo domain/Esa1/heterochromatin-associated protein 1 (HP1)/histone tail/lysine methylation
Introduction
It is now recognized that heritable changes in gene expression can occur without changes in gene DNA sequence. Elegant studies in Drosophila have provided insights into an epigenetic phenomenon known as position effect variegation (PEV) (Spofford, 1976; Eisseberg, 1989; Weiler and Wakimoto, 1995). Through rearrangement or transposition from its normal euchromatic position to one in the vicinity of heterochromatin, the ‘on/off’ state of a particular gene can be altered, resulting in PEV or mosaic silencing. Heterochromatin regions classically are observed as chromocenters of polytene chromosomes found in salivary glands of Drosophila (Figure 1C). Importantly, a group of dominant suppressors of PEV has been identified that are referred to collectively as Su(var) genes. For example, heterochromatin protein 1 (HP1) was first identified in Drosophila as Su(var)2-5 (James and Elgin, 1986; Eissenberg et al., 1990). HP1 proteins have been reported in organisms as diverse as fission yeast (Swi6p) and mammals (HP1α, β and γ) (Eissenberg and Elgin, 2000), with a conserved role in epigenetic control of heterochromatin assembly that has recently been characterized (Bannister et al., 2001; Nakayama et al., 2001). In the absence of HP1, multiple telomere–telomere fusions occur, resulting in a striking spectrum of abnormal chromosomal configurations (Fanti et al., 1998).
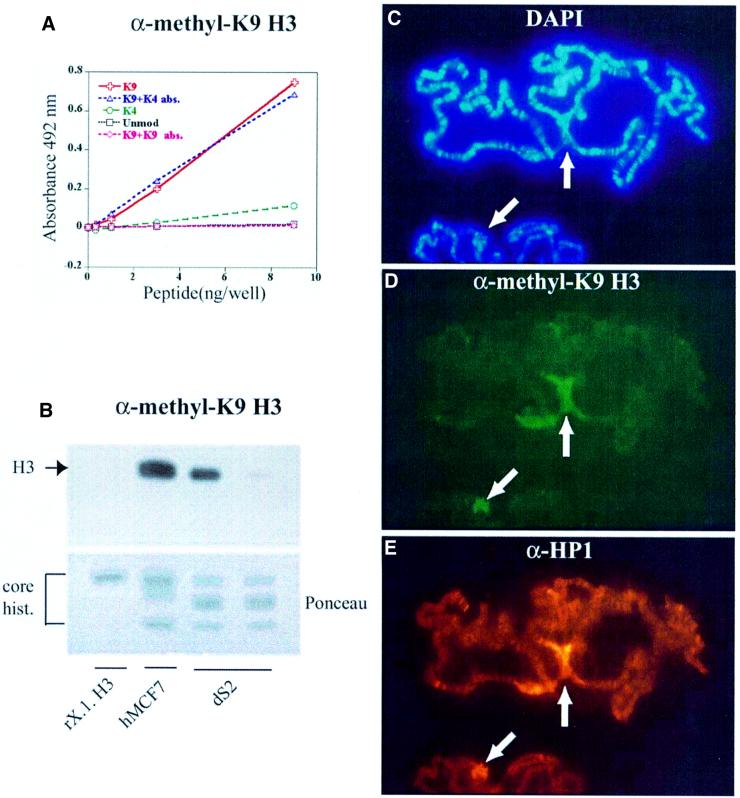
Fig. 1. A specific antibody for methyl-K9 H3, α-methyl-K9 H3, co-localizes with HP1 in polytene chromosomes. (A) ELISA analysis of α-methyl-K9 H3 showing specificity of the antiserum for methyl-K9 H3 peptide. The α-methyl-K9 H3 antibody was produced by using a methyl-lysine-containing H3 sequence, TARKSTGGC, coupled to keyhole limpet hemocyanin (KLH). (B) Western analysis (12% SDS–PAGE) of recombinant Xenopus H3 (rX.l. H3) (gift from K.Luger), human core histones (hMCF7) and Drosophila core histones (dS2) using α-methyl-K9 H3. The right lane of the two dS2 lanes corresponds to the experiment where antibodies were pre-absorbed with methyl-K9 H3 peptide. (C–E) Third instar polytene chromosomes were analyzed by indirect immunofluoresence using the general DNA stain DAPI (C), α-methyl-K9 H3 (D) and anti-HP1 antibodies, α-HP1 (E). The white arrows point to chromocenters.
Chromatin structure and function can be influenced by distinct patterns of post-translational modification within histone tails (Turner, 2000; Wolffe and Guschin, 2000). The ‘histone code’ hypothesis suggests that multiple histone modifications, acting in a combinatorial or sequential fashion on one or multiple histone tails, specify unique functions along the eukaryotic genome (Strahl and Allis, 2000). In some cases, subunits of transcriptional regulators contain one or multiple bromodomains that recognize histone tails acetylated on lysines normally required for active transcription (Dhalluin et al., 1999; Hudson et al., 2000; Jacobson et al., 2000; Owen et al., 2000). Methylation of specific lysines on histone H3 has also been reported (Ohe and Iwai, 1981; Strahl et al., 1999). In this regard, human SUV39H1 and Schizo saccharomyces pombe Clr4, two SET domain-containing homologs of Drosophila Su(var)3-9, have been identified to be histone methyltransferases that specifically methylate Lys9 of histone H3 (Rea et al., 2000; Nakayama et al., 2001; for reviews see Jenuwein, 2001; Rice and Allis, 2001). Very recently, a series of in vitro pull-down assays showed that methylated Lys9 on histone H3 (methyl-K9 H3) might be a molecular handle for the chromo domain of HP1 (Bannister et al., 2001; Lachner et al., 2001). In particular, Bannister et al. showed that the HP1 association with methylated mononucleosomes could be completely disrupted by the addition of excess methyl-K9 H3 peptide, suggesting that HP1 recognizes methyl-K9 H3 in the context of mononucleosomes. Pull-down assays have also indicated that the HP1 chromo domain interacts with the histone fold domain of H3, suggesting that there may be additional sites of interaction between the chromo domain and H3 (Nielsen et al., 2001). HP1 interaction with methyl-K9 H3 is essential for epigenetic control of heterochromatin assembly in vivo (Bannister et al., 2001; Nakayama et al., 2001).
In our study, we have focused on elucidating the specificity of methyl-K9 recognition using a series of H3 tail peptides described in Materials and methods. Here, we use Drosophila HP1 as a model to characterize HP1 specificity for the H3 tail. Using an antibody specific for methyl-K9 H3, we show that this methylation ‘histone code’ and HP1 protein co-localize in situ on Drosophila polytene chromosome squashes. Through in vitro studies by NMR spectroscopy, fluorescence anisotropy and isothermal titration calorimetry, we characterize the physical determinants of this interaction and map the binding interface. In addition, we use similar in vitro studies to address the significance of point mutations that abolish (V26M) and alter (Y24F) gene silencing function of HP1 in vivo. The physical interactions described here for Drosophila HP1 may serve as general determinants of the specificity of HP1 chromo domains for the methylated H3 tail. However, sequence diversity observed in non-HP1 chromo domains may enable them to achieve different types of chromatin docking interactions. In support, we show that the chromo domain of Esa1 histone acetyltransferase does not interact with the methyl-K9 H3 tail, but it has avidity for the unmodified H3 tail.
Results
In vivo binding studies
To detect patterns of H3 K9 methylation in vivo, we generated an antibody that specifically recognizes methylated K9 of histone H3. Microsequencing and mass spectrometry data indicate that the dimethylated form of a single lysine predominates in histone H3 in vivo (Borun et al., 1972; Duerre and Chakrabarty, 1975; Strahl et al., 1999). Therefore, a synthetic peptide containing this form of lysine modification was used to generate a polyclonal antibody in rabbits. The specificity of the resulting antiserum (hereafter referred to as α-methyl-K9 H3) was assayed by enzyme-linked immunosorbent assay (ELISA). As shown in Figure 1A, α-methyl- K9 H3 recognized methyl-K9 H3 peptide with little cross-reactivity to control peptides, methyl-K4 H3 or unmodified H3. Addition of methyl-K9 H3 peptide to α-methyl-K9 H3 prior to the ELISA abolished recognition of methyl-K9 H3. By comparison, in a parallel control, the same concentration of methyl-K4 H3 peptide did not block recognition of the methyl-K9 H3 peptide. Together, these data demonstrate that α-methyl-K9 H3 specifically recognizes methylated K9 in the context of the H3 tail.
To investigate the in vivo presence of methyl-K9 H3 in Drosophila and human cells, whole-cell extracts were examined by western blot analysis using α-methyl-K9 H3. Histone-enriched extracts were made from MCF7 (human) and S2 (Drosophila) cell lines, and equal amounts of protein were immunoblotted with α-methyl-K9 H3, which showed that methyl-K9 H3 was present in both extracts (Figure 1B). As a negative control, recombinant histone H3 was not detected by α-methyl-K9 H3. Pre-absorption with methyl-K9 H3 peptide abolished α-methyl-K9 H3 recognition of H3, whereas reactivity was not lost with pre-absorption with methyl-K4 H3 peptide (data not shown). These results indicate that K9 methylation occurs in both human and Drosophila H3, and can be detected successfully by our antibody.
To investigate the in vivo significance of HP1 recognition of methyl-K9 H3 in Drosophila, we probed polytene chromosome squashes with α-methyl-K9 H3 antibodies, and detected a chromocentric staining pattern (Figure 1D). This pattern of staining co-localized with α-HP1 staining, suggesting that methyl-K9 H3 and HP1 both reside at the chromocenters (Figure 1D and E). As with ELISA and western analysis, pre-absorption with methyl-K9 H3 peptide was able to ablate α-methyl-K9 H3 specificity, while antisera pre-absorbed with methyl-K4 H3 peptide still exhibited chromocentric staining.
In vitro binding studies
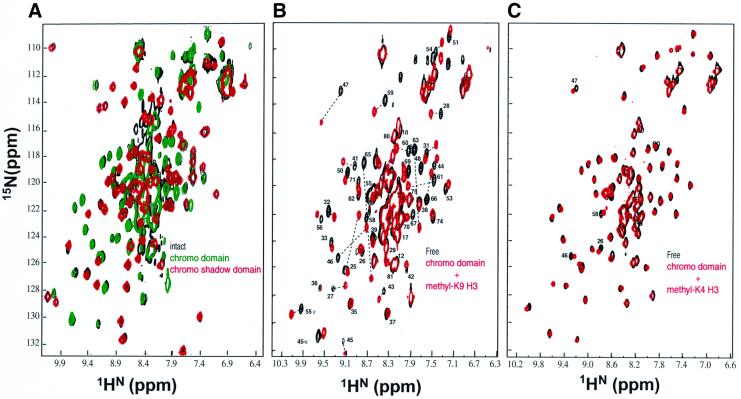
The HP1 family of proteins (Eissenberg and Elgin, 2000) have an N-terminal chromo domain and a related C-terminal chromo shadow domain. The chromo domain is also present in the Drosophila Polycomb protein, which represses expression of homeotic genes. The chromo domain consists of a monomeric three-stranded antiparallel β-sheet that is flanked by a C-terminal α-helix (Ball et al., 1997; Horita et al., 2001). The chromo shadow domain is dimeric, and consists of a three-stranded antiparallel β-sheet that is flanked by two C-terminal α-helices, which form the symmetric dimer interface (Brasher et al., 2000). We prepared recombinant constructs of intact HP1, chromo and chromo shadow domains, and collected the corresponding fingerprint [1H–15N]-HSQC NMR spectra. In agreement with studies on mammalian HP1β (Brasher et al., 2000), we find, using gel filtration studies, that the intact protein and the chromo shadow domain are dimeric, whereas the chromo domain is monomeric. As shown in Figure 2A, each set of the chromo and chromo shadow domain resonances superimposes remarkably well on the spectrum of the intact protein. These data demonstrate that the chromo and chromo shadow domains are independent of each other within the intact protein while tethered via their 50 residue flexible linker, and that these two domains do not interact significantly within the context of the intact protein.
Fig. 2. Two-dimensional [15N–1H]-HSQC NMR spectra demonstrating the specific interaction of the HP1 chromo domain with methyl-K9 H3 peptide. (A) Superposition of the chromo (amino acids 1–84 in green cross-peaks) and chromo shadow (amino acids 132–206 in red cross-peaks) domains spectra on the intact HP1 (amino acids 1–206 in black cross-peaks). In order to improve the NMR spectra of the chromo shadow domain (20 kDa dimer) and intact (50 kDa dimer) proteins, we incorporated 50% perdeuteration of non-labile hydrogens in the samples. This was achieved by growing E.coli in the media containing 50% D2O–50% H2O. (B) Superposition of the chromo domain free (black cross-peaks) and in complex with methyl-K9 H3 peptide (red cross-peaks); stoichiometric binding was achieved at the protein to peptide concentration ratio of 1:4. (C) Superposition of the chromo domain free (black cross-peaks) and in the presence of saturating amount of methyl-K4 H3 peptide (red cross-peaks).
We then collected NMR spectra of HP1 in the presence of H3 tail peptides. No chemical shift changes in HP1 were detected when the unmodified H3 tail was used. In contrast, chemical shift perturbations were detected only for the chromo domain upon addition of methyl-K9 (very large shifts) and methyl-K4 (insignificant shifts) H3 peptides. Figure 2B and C shows the spectra for the chromo domain in the presence of saturating amounts of each modified peptide. Incidentally, titration of methyl-K9 H3 peptides corresponding to residues 1–15 and 1–20 of histone H3 showed exactly the same pattern of chemical shift perturbation, suggesting that H3 residues 15–20 do not contribute to binding. When we titrated methylated lysine amino acids alone (monomethyl, dimethyl and trimethyl forms of H-Lys-OH) to final protein:amino acid ratios of 1:10, no changes in the HSQC spectrum of the chromo domain were detected. This implies that methyl-lysine is only recognized by HP1 when it is presented in the context of the H3 tail sequence.
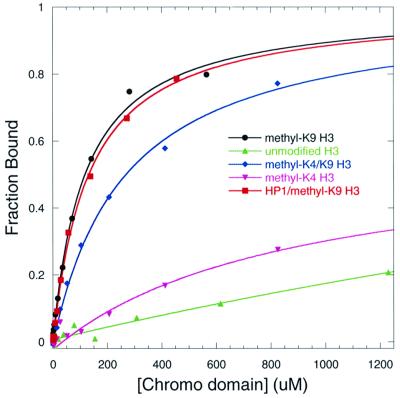
We carried out solution binding studies, and found that a weak binding interaction occurs between the HP1 chromo domain and the methyl-K9 H3 tail. Fluorescence anisotropy studies indicate that the intact protein and the chromo domain have similar dissociation constants (KD of ∼100 µM, Figure 3; Table I). Therefore, the chromo domains of the dimeric intact protein are independent of each other, and are each able to bind the peptide in a 1:1 complex, a finding in close agreement with the NMR data shown in Figure 2A. Moreover, using this assay, we show that methyl-K4 H3 tail interacts very weakly (KD = 1.9 ± 0.5 mM) while the unmodified H3 tail does not interact with the chromo domain (Figure 3). In addition, an H3 tail containing both methyl-K4 and methyl-K9 modifications can still bind with a 2.5-fold weaker affinity (KD = 268 ± 25 µM) than that containing only methyl-K9 modification.
Fig. 3. Binding of the HP1 chromo domain to fluorescein-labeled H3 tail peptides as measured by fluorescence polarization. Curves represent the best fit to the data as described in Materials and methods. The fraction bound corresponds to the normalized anisotropy. Binding assays were performed for the HP1 chromo domain with unmodified H3 (triangles), methyl-K9 H3 (circles), methyl-K4 H3 (crosses) and methyl-K4/methyl-K9 H3 (diamonds). The binding assay performed for intact HP1 with methyl-K9 H3 is shown with squares.
Table I. Thermodynamic parameters involved in interaction of Drosophila HP1 with methyl-K9 H3 peptide.
| Temperature (°C) | KD (fluorescence µM) | KD (ITC µM) | ΔH (ITC kcal/mol) | N (ITC) | ΔG (kcal/mol) | TΔS (kcal/mol) |
|---|---|---|---|---|---|---|
| 25 | 120 ± 12 | 105 ± 24 | –11.7 ± 2.4 | 0.90 ± 0.11 | –5.4 | –6.3 |
| (133 ± 11) | ||||||
| 15 | 80 ± 8 | 59 ± 8 | –10.6 ± 0.5 | 0.96 ± 0.02 | –5.6 | –5.0 |
| (91 ± 5) |
Binding results were obtained from fluorescence anisotropy (fluorescence) and isothermal titration calorimetry (ITC). Fluorescence anisotropy of the fluorescein-labeled peptide was used to determine the apparent dissociation constant KD (when a 1:1 binding stoichiometry is assumed) for the chromo domain and intact HP1 (values in parentheses) at 15 and 25°C. Using ITC, the single site binding constant KD, the heat of binding ΔH and the number of sites N were independently measured variables at 15 and 25°C. The free energy and entropy changes for binding were then calculated using the following relationships: ΔG = –RT ln KD and ΔG = ΔH – TΔS, respectively. In each case, parameters are reported as the mean (± average deviation from the mean) obtained from two independent titration experiments.
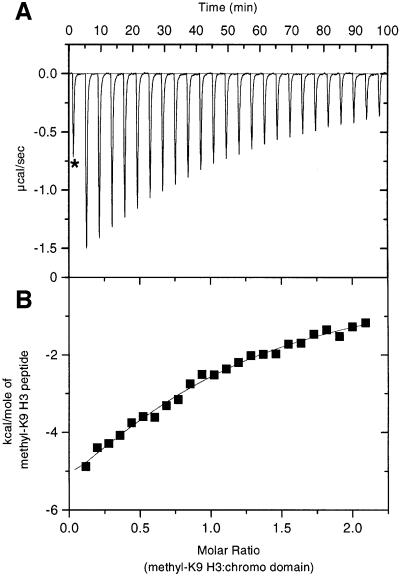
Using isothermal titration calorimetry (ITC), we have obtained additional thermodynamic parameters that characterize the interaction of the chromo domain with methyl-K9 H3 tail. By titrating the peptide into the chromo domain sample, we find that the binding is exothermic and has a large enthalpic contribution (ΔH = –11.6 kcal/mol at 25°C; Figure 4). Binding also has a modest Gibbs free energy (ΔG = –5.4 kcal/mol) that is consistent with the small cost in entropy (TΔS = –6.3 kcal/mol). Therefore, burial of the polar surface is probably important for binding. In support, when fluorescence binding assays were performed under destabilizing conditions for polar interactions (0.5 M NaCl was added to the buffer), the affinity dropped 4-fold (KD = 396 ± 60 µM). In contrast, when binding assays were performed under stabilizing conditions for basic interactions (the pH was raised from 6 to 8), a 2-fold enhancement was detected (KD = 55 ± 7 µM) (data not shown).
Fig. 4. Binding of the HP1 chromo domain to methyl-K9 H3 peptide as measured by isothermal titration calorimetry at 25°C. (A) Raw data for injections of the peptide into the chromo domain as described in Materials and methods. The asterisk corresponds to the first injection, which was excluded from the analysis. (B) Integrated heats of injections, with the solid line corresponding to the best fit of the data using the MicroCal software.
ITC studies show that binding is more exothermic at higher temperature, and comparison of average ΔHs at 25 and 15°C suggests a small negative ΔCp of association (–0.12 kcal/mol; although within the experimental error of ΔHs). This result is qualitatively in agreement with the idea that binding probably occurs in the absence of significant conformational changes in the protein, and that there is a decrease in exposure of hydrophobic groups to water when the complex forms. By comparison, interaction of the SH2 module with phosphotyrosyl peptide has a ΔCp of –0.1 kcal/mol, and peptide binding occurs with minimal conformational rearrangements (for a review see Kuriyan and Cowburn, 1997).
Nature of the binding interface
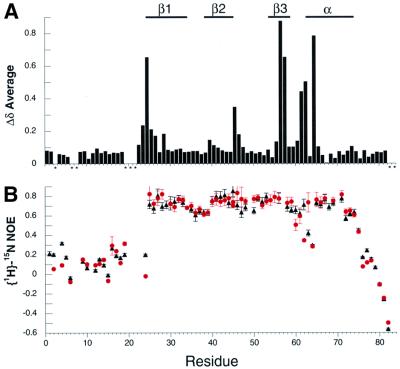
In order to identify the chromo domain surface of interaction with methyl-K9 H3 tail, we collected a series of three-dimensional NMR spectra (see Materials and methods) and determined the sequential assignment for the free and complex forms. Upon completing the assignment, we noted that a number of backbone nuclei show unusually large chemical shift perturbations upon methyl-K9 H3 binding (Figure 2B). Figure 5A illustrates the weighted average chemical shift difference, Δδave, for each amino acid upon methyl-K9 H3 peptide binding calculated by using the relationship 0.25[(ΔHN)2 + (ΔN/5)2 + (ΔCα/2)2 + (ΔC′/2)2]1/2, where Δi is the chemical shift difference for resonance i in the free and complexed states (Grzesiek et al., 1996). The resulting chemical shift perturbations are consistent with the formation of a specific complex in which only selected residues participate in peptide binding. The chemical shift changes in the 13C′ and 13Cα resonances do not result in changes in the secondary structure elements, as judged by the chemical shift index analysis and the analysis of the nuclear Overhauser effect (NOE; data not shown).
Fig. 5. NMR-detected differences between the complex (with methyl-K9 H3 peptide) and free chromo domain. (A) Backbone chemical shift differences between the chromo domain bound to methyl-K9 H3 peptide and free chromo domain are tabulated in the form of weighted average chemical shift difference, Δδave, as described in the text; the asterisks correspond to the sites with no available data. Secondary structure elements are shown at the top. (B) 15N relaxation data measured at 60.8 MHz for 1 mM free (triangles) and complex (circles) chromo domain.
The {1H}–15N NOE values for the backbone 15N nuclei in the free and complex forms of the chromo domain are shown in Figure 5B. The {1H}–15N NOE values are influenced by the internal dynamics of individual amides as well as the overall rotational diffusion of the protein (Kay et al., 1989). In the free protein, the central portion of the polypeptide (region of {1H}–15N NOE ∼0.8) has restricted internal motion indicative of a highly defined structure, with the exception of the mobile turn region that connects β3 to the α-helix. In contrast, residues 1–24 and 76–82 at the N- and C-termini ({1H}–15N NOE ≤0.5) are under substantial motion lacking a well-defined structure. Upon complex formation with the peptide, the protein shows essentially no change in the pattern and quantity of {1H}–15N NOE values. However, the turn connecting β3 to the α-helix adopts a new pattern of mobility at residues 62 and 63, consistent with implicating this region in H3 tail recognition. Overall, these results imply that structural perturbations caused by peptide binding in the core region of the chromo domain are minimal, and in close agreement with the small ΔCp of binding measured by calorimetry.
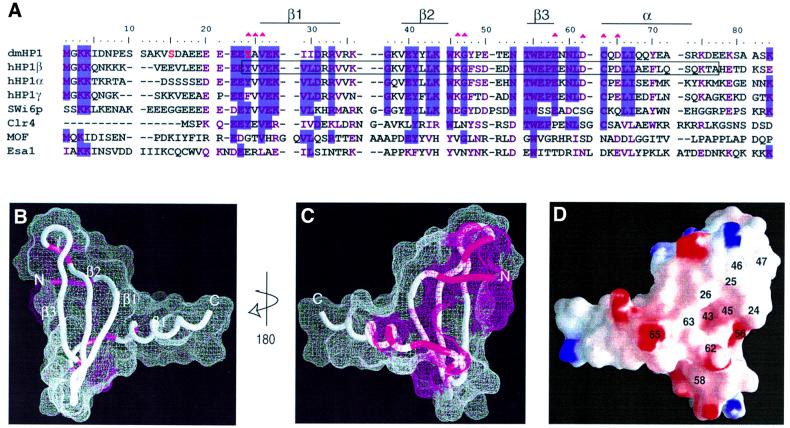
To map the peptide-binding surface, we relied on the existing three-dimensional structure of the 70% identical chromo domain from mammalian HP1β (Ball et al., 1997). Figure 6B and C illustrates the sites of chemical shift perturbations with Δδave ≥0.2 caused by methyl-K9 H3 peptide binding. This analysis suggests that a concave binding interaction surface exists only on one face of the β-sheet that is not comprised of residues in contiguous primary sequence (Figure 6A). The floor of this concave surface is formed by conserved hydrophobic residues L43 and W45. In addition, the electrostatic charge distribution at the methyl-K9-binding surface of HP1 is shown in Figure 6D. A region of negatively charged surface, generated by conserved residues E56 and D62, surrounded by highly conserved residues Y24, V26, L43, W45 and C63, coincides with the NMR-detected site of interaction shown in Figure 6C. The side chain of the methylated lysine residue in the H3 tail contains a positive charge with the two methyl groups. These may form favorable interactions near the L43/W45 dip of the chromo domain. Methyl-K9 and methyl-K4 modifications occur in the context of different primary sequences (RTKQT for K4 versus ARKST for K9), which are probably very important for their recognition by different chromatin-regulating factors.
Fig. 6. Map of the binding surface of the chromo domain for methyl-K9 H3 peptide. (A) The amino acid sequence of the Drosophila HP1 (dmHP1) chromo domain (as prepared for this study) is shown in an alignment (using CLUSTALW) with other chromo domains: human HP1 types α, β and γ, S.pombe Swi6p (amino acids 59–145), Clr4 (amino acids 1–72), human MOF (amino acids 370–456) and S.cerevesiae Esa1 (amino acids 11–97). Secondary structure elements present in dmHP1 are marked. Residues that participate in peptide binding are marked by magenta triangles. Residues in red correspond to the sites of phosphorylation in dmHP1. The boxed sequence in HP1β corresponds to the range of the three-dimensional structure of the core of this chromo domain (Ball et al., 1997) that is used to prepare (B)–(D). (B and C) Two surface views of the HP1 chromo domain related by 180° rotation. The magenta surface corresponds to residues demonstrating large perturbations, Δδave ≥0.2 (Figure 5A). (D) The surface electrostatic potential for the methyl-lysine-binding face. The residue numbers corresponding to magenta regions in (C) are labeled. In addition, residues L43, W45 and E56 were labeled in the putative binding pocket. (B)–(D) were prepared using GRASP (Nicholls, 1993).
Characterization of mutants that affect the gene silencing function of HP1
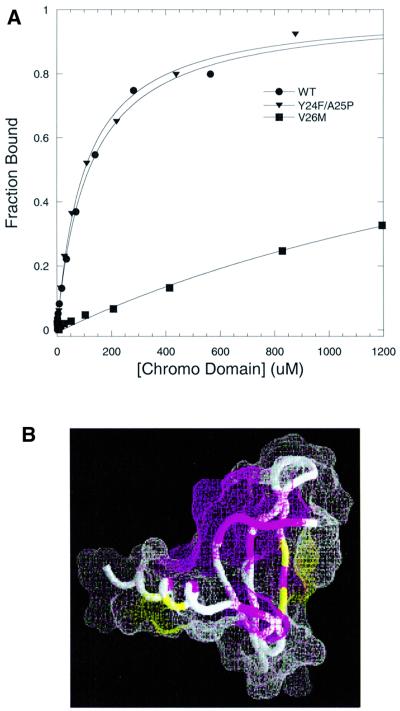
A natural missense variant of Drosophila HP1, V26M, was shown to disrupt its gene silencing function (Platero et al., 1995). As shown in Figure 6, V26 is an extremely conserved residue that is implicated at the surface of the chromo domain for binding methyl-K9 H3 tail. We carried out fluorescence binding assays for the V26M chromo domain, and found that methyl-K9 H3 binding is completely ablated (Figure 7A). If we assume that the overall structure of the V26M variant is similar to that of the wild-type (the elution profile from the size-exclusion Superdex 75 column is identical to that of the wild-type), then an explanation for the abolished binding may be that a bulkier methionine at the interface of H3 tail binding (Figure 6D) precludes the packing of methyl-K9 on the chromo domain. However, analysis of the [1H–15N]-HSQC spectrum of the mutant led to the conclusion that the structure of the V26M chromo domain is severely destabilized (Figure 7B). This is indicated by the heavy amount of chemical exchange detected for the backbone amides of a set of non-contiguous residues surrounding the site of the mutation. The backbone amide signals disappear on the HSQC spectrum, indicative of structural heterogeneity at the putative surface of interaction for methyl-K9 H3 (Figure 7B). Moreover, significant chemical shift perturbations occur in regions distant from the site of mutation. These findings emphasize that a well-defined surface of the HP1 chromo domain is necessary for the recognition of the H3 tail.
Fig. 7. Effect of point mutations in the specificity of the HP1 chromo domain for methyl-K9 H3 peptide. (A) Fluorescence polarization assays are illustrated for wild-type (circles), V26M (squares) and Y24F/A25P (triangles); measurements were performed at 25°C in 50 mM sodium phosphate pH 6 and 25 mM NaCl buffer. (B) The NMR chemical shift perturbations in backbone 1HN and 15N nuclei of the V26M mutant are marked on the structure (Ball et al., 1997) using the same view as in Figure 6C. Regions in magenta correspond to the sites where NMR signals disappear (residues 25–28, 43, 55, 62, 66 and 70) and regions in yellow (residues 54, 56 and 68) correspond to the sites with weighted chemical shift perturbations in the range 0.1 < Δδ < 0.2. Regions shown in white do not show any significant perturbation upon mutation.
The Y24F/A25P mutation also antagonizes the silencing activity of HP1 in vivo (Platero et al., 1995). Using specific antibodies for each of the human HP1 isoforms, it has been shown that HP1 proteins segregate in distinct regions of the human nucleus (Minc et al., 1999). Their isoform-specific HP1 localization is phosphorylation dependent and dynamic during the cell cycle. In Drosophila, HP1 is also multiply phosphorylated, and hyper-phosphorylation is correlated with heterochromatin assembly during development (Zhao et al., 2001). Several in vivo sites of phosphorylation have been characterized and mapped to S15 and Y24 in the chromo domain of Drosophila HP1. Residue Y24 is especially conserved and noteworthy. While mammalian HP1α and β isoforms conserve this putative tyrosine phosphorylation site, HP1γ does not, and at this position contains phenylalanine. Due to the involvement of Y24 in methyl-K9 H3 binding, we performed a fluorescence binding assay for the Y24F/A25P variant, and found that the binding affinity (KD = 102 ± 9) agrees closely with that of the wild-type (Table I). Moreover, the structure assessment of this mutant by NMR analysis of the backbone amides showed that this mutation causes small chemical shift perturbations (Δδ <0.1), which are limited to the site of mutation, thus maintaining essentially wild-type structure. Therefore, recombinant chromo domains containing either F24 or unphosphorylated Y24 behave similarly. Due to the significance of polar interactions for binding to methyl-K9 H3, it is plausible that in vivo phosphorylation of HP1 isoforms that contain Y24 may serve as an enhancer of the interaction with the positively charged methyl-K9 in histone H3.
Not all chromo domains bind methyl-K9 H3 tail
A recent study on the histone acetyltransferase MOF, which contains a chromo domain, reported that chromo domains might also be protein–RNA binding modules (Akhtar et al., 2000). In vivo association of MOF with the male X chromosome was suggested to depend on interaction of MOF with roX2 RNA. Removal of the chromo domain or point mutations in the chromo domain of MOF abolished in vitro RNA binding. Figure 6A shows that a poor sequence homology exists between the chromo domains of MOF and HP1 in the region implicated in methyl-K9 H3 binding, suggesting that chromo domains may well have diverse functions in eukaryotes.
Esa1 and MOF contain closely related chromo domains (Figure 6A) and belong to the MYST family of histone acetyltransferases (Mizzen et al., 1998; Sterner and Berger, 2000). To test if the chromo domain of Esa1 behaves similarly to that of HP1, we prepared a recombinant construct of the Esa1 chromo domain, and carried out binding studies to assess its affinity for histone H3 tail. Fluorescence binding assays were performed under conditions similar to those described for HP1 at 25°C and pH 8, and are shown in Figure 8; whereas the HP1 chromo domain is stable at pH 6–8, the Esa1 chromo domain is more stable at pH 8.
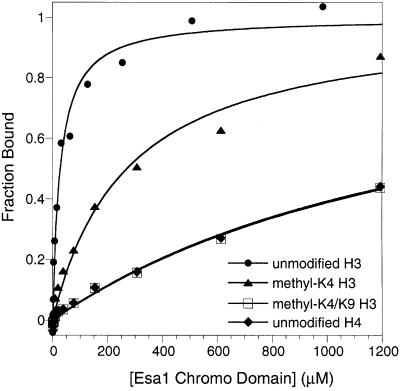
Fig. 8. Fluorescence polarization assays for interaction of the Esa1 chromo domain with fluorescein-labeled unmodified H3 (circles), methyl-K4 H3 (triangles), methyl-K4/methyl-K9 H3 (open squares) and unmodified H4 (diamonds). Measurements were performed at 25°C in 50 mM sodium phosphate pH 8 and 25 mM NaCl buffer. Curves represent the best fit to the data as described in Materials and methods. Fraction bound corresponds to the normalized anisotropy.
The Esa1 chromo domain shows avidity for H3 tails in a manner distinct from that of the HP1 chromo domain. The Esa1 chromo domain has a strong avidity for unmodified H3 tail (KD = 28 ± 6 µM). The presence of methyl-K4 modification reduces this interaction 10-fold (KD = 270 ± 68 µM) and, unlike HP1, the Esa1 chromo domain does not prefer the H3 tail with methyl-K9 or both methyl-K4 and methyl-K9 modifications (KD = 1.5 ± 0.4 mM for binding to methyl-K4/methyl-K9; see Figure 8). Because in vitro acetyltransferase activity of Esa1 modifies specific lysines in both histone H3 and H4 tails, we also performed binding assays with H4 tail peptides, but detected no significant avidity for unmodified H4 (KD = 1.5 ± 0.4 mM; see Figure 8) or acetyl-K12 H4 tails. Thus, one plausible role for the chromo domain of Esa1 may be to associate with the nucleosomes using the unmethylated H3 as a molecular handle, and this may be important for Esa1 to perform its acetylation function. Alternatively, the chromo domain of Esa1, like that of MOF, may bind preferentially to RNA (or methylated RNA) in ways that remain to be defined (Akhtar et al., 2000).
Discussion
We demonstrate, using a methyl-K9 H3-specific antibody, that this methylation mark and HP1 protein co-localize in situ on Drosophila polytene chromosome squashes. Using NMR spectroscopy, large chemical shift perturbations are detected on the spectrum of HP1 subsequent to methyl-K9 H3 peptide binding that are not detected with either unmodified or methyl-K4 H3 peptides. These perturbations occur in the chromo domain of HP1 and suggest a high degree of selectivity in binding of methylated peptide. This property of HP1 may play an important role in recognizing methyl-K9 for the ‘off’ state of transcription in heterochromatin, distinguishing it from methyl-K4, which has been associated with the ‘on’ state of transcription (e.g. in active macronuclei in Tetrahymena; Strahl et al., 1999).
Using two independent binding assays (ITC and fluorescence anisotropy), we extend the in vitro pull-down assays that were published recently (Bannister et al., 2001; Lachner et al., 2001). Our in-solution binding data reach remarkably different conclusions regarding the affinity constant for the interaction of the chromo domain with the H3 tail, and provide insights into the selectivity of methyl-K9 binding (Figure 3). It has been shown that in vivo the dimethylated form of a single lysine predominates in histone H3 (Borun et al., 1972; Duerre and Chakrabarty, 1975; Strahl et al., 1999). Indeed, our antibody shows that dimethyl lysine is present in vivo (Figure 1). Therefore, we used H3 peptides (residues 1–15) containing dimethyl-K9 or both dimethyl-K4 and dimethyl-K9 modifications in our binding assays; KD = 100 µM or KD = 268 µM, respectively. However, Bannister et al. (2001) used H3 peptide (residues 1–16) containing both trimethyl-K4 and trimethyl-K9 modifications for their binding assay with the chromo domain (HP1β chromo domain residues 10–80, see Figure 6A). We report a KD value 1000-fold weaker than the 70 nM reported by Bannister et al., which was obtained through solid-phase binding studies using the technique of surface plasmon resonance. Our findings are consistent with the dissociation constants reported for the bromodomain; a single bromodomain for a single acetyl-lysine-containing H4 tail peptide of >300 µM (Dhalluin et al., 1999; Hudson et al., 2000), and a double bromodomain for a multiple acetyl-lysine-containing H4 tail in the range 1–5 µM (Jacobson et al., 2000).
The key residues that participate in defining the putative methyl-K9-binding pocket are mapped on the three-dimensional structure and are in agreement with mutations that abolish silencing in vivo. The known K9 H3-specific methyltransferases from yeast to human all contain chromo domains that are closely related to the HP1 chromo domain. The structure of the chromo domain of S.pombe methyltransferase Clr4 has been reported recently (Horita et al., 2001). The putative methyl-K9 H3-binding surface is conserved in sequence (Figure 6A) as well as the surface profile, suggesting that methyl-K9 H3 could also serve as a molecular handle for Clr4.
Finally, we show that a distantly related chromo domain from yeast Esa1 does not have an affinity for methyl-K9, while it can associate with the unmodified histone H3 tail. This suggests that chromo domains may have diverse functions, with those of the HP1 family targeting the methyl-K9-containing histone H3. Accordingly, and in keeping with emerging findings that the bromodomain serves as a chromatin-targeting module (Winston and Allis, 1999), we favor the view that appropriate methylation marks dictate the recruitment of distinct chromo domain-containing factors and, in turn, contribute to gene activation or silencing. In sum, our structural and binding studies provide considerable insight into the nature of histone H3 recognition by the chromo domain.
Materials and methods
Synthetic peptides
Peptides were synthesized at the Baylor College of Medicine Protein Core Facility (Houston, TX). The primary sequence of each peptide is given below. In each case, an asterisk corresponds to the site of post-translational modification (dimethylated lysine in H3 peptides and acetylated lysine in H4 peptide), and underlining corresponds to a non-native residue: unmodified H3, NH2-ARTKQTARKSTGGKAPRKQLC-COOH; methyl-K9 H3, NH2-ARTKQTARK*STGGKAY-COOH; methyl-K4 H3, NH2-ARTK*QTARKSTGGKAPRKQLC-COOH; methyl-K4/methyl-K9 H3, NH2-ARTK*QTARK*STGGKAY-COOH; unmodified H4, NH2-SGRGKGGKGLGKGGAKRHRY-COOH; and acetyl-K14 H4, NH2-SGRGKGGKGLGK*GGAKRHC–COOH.
Immunoassays
Polytene chromosome preparation was as described previously (Bone et al., 1994). Salivary glands from third-instar larvae were dissected in 0.7% NaCl and fixed for 5 min in phosphate-buffered saline (PBS) at pH 7.2, containing 3.7% formaldehyde, 0.1% Triton X-100 and 0.2% NP-40 according to standard procedures. The slides were then washed in PBS for 30 min, in PBST (PBS plus 0.2% Triton X-100) for 30 min, and blocked in PBST with 2% bovine serum albumin (BSA) for 30 min. Primary and secondary antibody incubation was for 1 h at 30°C in a humidified chamber. Both primary antibodies were applied simultaneously. Rabbit α-methyl-K9 H3 polyclonal antibodies were applied at a concentration of 1:500 and mouse C1A9 anti-HP1 monoclonal antibodies at a concentration of 1:400. Both secondary antibodies were applied simultaneously. Both Cy3-conjugated donkey anti-mouse and fluorescein isothiocyanate (FITC)-conjugated anti-rabbit antibodies (Jackson Labs) were applied at a concentration of 1:100. Counterstaining of the chromosomes was for 5 min in 1 µg/ml 4′,6-diamidino-2-phenylindole (DAPI) followed by a quick wash in water. ELISA and preparation of histone-enriched acid-soluble extracts were as described previously (Cheung et al., 2000).
Cloning, expression and protein purification
The coding regions of intact (amino acids 1–206), chromo (amino acids 1–84) and chromo shadow (amino acids 132–206) of Drosophila HP1 (SWISS-PROT accession code: P05205) were subcloned into NdeI– BamHI sites of pET16b vector (Novagen) and expressed in BL21(DE3) Escherichia coli with an N-terminal His tag. Variants of the chromo domain were prepared by subcloning the DNA of Drosophila HP1 corresponding to point mutations V26M and Y24F/A25P (Platero et al., 1995). A similar strategy was used to subclone the coding region of the chromo domain (amino acids 1–111) of Saccharomyces cerevisiae Esa1 (SWISS-PROT accession code: Q08649).
The proteins were purified by Ni-affinity chromatography (Qiagen) followed by gel filtration with Superdex 75 (Pharmacia) under native conditions. To avoid loss in yield, the N-terminal His tag was not removed. Uniformly 15N- and 15N,13C-labeled proteins were prepared by growth in minimal media containing 15NH4Cl and/or [13C]glucose as the sole sources of nitrogen and carbon, respectively; yield ∼10 mg of pure protein per liter of minimal medium. Sample purity was confirmed by SDS–PAGE and mass spectrometry (Mr of intact HP1, observed = 25 707, calculated = 25 706; Mr of HP1 chromo domain, observed = 12 396, calculated = 12 395.4; Mr of HP1 chromo shadow domain, observed = 10 943, calculated = 10 941.9; Mr of Esa1 chromo domain, observed = 15 437.1, calculated = 15 569.4).
NMR spectroscopy
NMR data were collected with a Bruker AVANCE (600 MHz) spectrometer for samples containing 0.5–1 mM protein in 50 mM sodium phosphate pH 6, 25 mM NaCl, 1 mM sodium azide and 0.1 mM dithiothreitol (DTT) in 95% H2O–5% D2O at 25°C. Stoichiometric binding of proteins to methyl-K9 H3 peptide was established by two-dimensional heteronuclear [1H–15N] data. Spectra were processed using NMRPipe (Delaglio et al., 1995) and analyzed with NMRView (Johnson and Blevins, 1994). Water signal suppression was achieved with flip-back pulses (Grzesiek and Bax, 1993). Sequential assignments of the backbone were made with data derived from three-dimensional HNCA, HN(CO)CA and HNCO (Grzesiek and Bax, 1992). Backbone assignments and the secondary structure analysis were completed with data derived from three-dimensional 15N-edited NOESY (mixing time = 100 ms) and TOCSY (Marion et al., 1989).
The {1H}–15N steady-state heteronuclear NOEs for the backbone 15N nuclei of the free and complex chromo domains were measured by collecting two-dimensional [15N–1H]-HSQC NMR spectra with the water flip-back method (Grzesiek and Bax, 1993). These spectra were collected in an interleaved manner, one with a recovery delay of 6 s (reference spectrum) and the other with a recovery delay of 3 s followed by 3 s of proton saturation (NOE spectrum). The {1H}–15N NOE value for each residue was calculated as the ratio of the cross-peak intensity NOE/reference, and error was estimated from the baseline noise in the two spectra.
Binding assays
All protein concentrations were determined by absorbance spectroscopy using predicted extinction coefficients (for intact HP1, ε280 = 26 030 M–1 cm–1; for HP1 chromo domain, ε280 = 17 780 M–1 cm–1; for Esa1 chromo domain, ε280 = 16 620 M–1 cm–1). Peptide concentrations were estimated from the mass, except in tyrosine-containing peptides, where absorbance spectroscopy using the extinction coefficient, ε280 = 1280 M–1 cm–1, was used. In preparations of cysteine-containing peptides, an ∼10-fold excess amount of TCEP was used to prevent covalent oligomerization.
Fluorescence anisotropy binding assays (Klotz, 1997) were performed on a Beacon 2000 fluorescence polarization system (PanVera) as follows. Peptides were N-terminally labeled with fluorescein succinimidyl ester. A 100 µg aliquot of peptide was added to a reaction solution containing 1.2 mM fluorescein compound and 100 mM potassium phosphate pH 7 to obtain a final reaction volume of 50 µl at 37°C. After 1 h, 5 µl of 1 M Tris–HCl pH 8 was added to quench the reaction. After quenching for 30 min at room temperature, the reaction mixture was passed over a 1 ml G15 Sephadex (Pharmacia) spin column (pre-equilibrated with 50 mM sodium phosphate pH 6, 25 mM NaCl buffer) to remove free fluorescein and hydroxy succinimide. Fractions containing the peptide were determined by measuring polarization. Binding assays were performed in 50 mM sodium phosphate buffer, and in the presence of ∼100 nM fluorescein-labeled peptide. Protein was titrated in the nanomolar– millimolar concentration range. Fluorescence polarization (P) values were converted to anisotropy (A) values by the equation: A = 2P/(3 – P); anisotropies of the labeled free peptides were typically ∼0.008. Binding curves were analyzed by non-linear least-squares fitting of the data using KaleidaGraph (Synergy Software). Data were fitted using the equation A = [Af – (Ab – Af)][protein]/(KD + [protein]), where Af and Ab represent the anisotropy of the free and bound peptides, respectively.
Thermodynamic parameters for methyl-K9 H3 peptide binding to the chromo domain were determined by standard ITC methods conducted at 25 ± 0.1 and 15 ± 0.1°C using a VP-ITC instrument from MicroCal (Northampton, MA) (Wiseman et al., 1989). The HP1 chromo domain was dialyzed against 50 mM sodium phosphate pH 6, 25 mM NaCl buffer. Lyophilized peptides were dissolved in the same exchange buffer. Exothermic heats of reaction (µcal/s) were measured by automated sequencing of 25 injections of H3 peptide (1 mM), each 10 µl, spaced at 2 min intervals, into 1.41 ml of chromo domain (90 µM). The heats of dilution were obtained by titrating the identical peptide sample into a cell containing sample buffer at each temperature, and subtracted from the raw data prior to analysis. Binding curves were analyzed by non-linear least-squares fitting of the data using the MicroCal software.
Acknowledgments
Acknowledgements
We are grateful to Drs M.F.Summers and J.H.Bushweller for access to the ITC instrument. We thank Drs R.L.Biltonen and F.Rastinejad for critical reading of the manuscript. This work was supported by startup funds from the University of Virginia School of Medicine (to S.K.) and an NIH grant GM53512 (to C.D.A.). S.K. was a special fellow of the Leukemia and Lymphoma Society.
References
- Akhtar A., Zink,D. and Becker,P.B. (2000) Chromodomains are protein–RNA interaction modules. Nature, 407, 405–409. [DOI] [PubMed] [Google Scholar]
- Ball L.J. et al. (1997) Structure of the chromatin binding (chromo) domain from mouse modifier protein 1. EMBO J., 16, 2473–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A.J., Zegerman,P., Partridge,J.F., Miska,E.A., Thomas,J.O., Allshire,R.C. and Kouzarides,T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–124. [DOI] [PubMed] [Google Scholar]
- Bone J.R., Lavender,J., Richman,R., Palmer,M.J., Turner,B.M. and Kuroda,M.I. (1994) Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes Dev., 8, 96–104. [DOI] [PubMed] [Google Scholar]
- Borun T.W., Pearson,D.W. and Paik,W.K. (1972) Studies of histone methylation during the HeLa S-3 cell cycle. J. Biol. Chem., 247, 4288–4298. [PubMed] [Google Scholar]
- Brasher S.V. et al. (2000) The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J., 19, 1584–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P., Tanner,K.G., Cheung,W.L., Sassone-Corsi,P., Denu,J.M. and Allis,C.D. (2000) Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell, 5, 905–915. [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek,S., Vuister,G.W., Zhu,G., Pfeifer,J. and Bax,A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX PIPES. J. Biomol. NMR, 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Dhalluin C., Carlson,J.E., Zeng,L., He,C., Aggarwal,A.K. and Zhou,M.-M. (1999) Structure and ligand of a histone acetyltransferase bromodomain. Nature, 399, 491–496. [DOI] [PubMed] [Google Scholar]
- Duerre J.A. and Chakrabarty,S. (1975) Methylated basic amino acid composition of histones from various organs from the rat. J. Biol. Chem., 250, 8457–8461. [PubMed] [Google Scholar]
- Eisseberg J.C. (1989) Position effect variegation in Drosophila: towards genetics of chromatin assembly. BioEssays, 11, 14–17. [DOI] [PubMed] [Google Scholar]
- Eissenberg J.C. and Elgin,S.C.R. (2000) The HP1 protein family: getting a grip on chromatin. Curr. Opin. Genet. Dev., 10, 204–210. [DOI] [PubMed] [Google Scholar]
- Eissenberg J.C., James,T.C., Foster-Hartnett,D.M., Hartnett,T., Ngan,V. and Elgin,S.C. (1990) Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc. Natl Acad. Sci. USA, 87, 9923–9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanti L., Giovinazzo,G., Berloco,M. and Pimpinelli,S. (1998) The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol. Cell, 2, 527–538. [DOI] [PubMed] [Google Scholar]
- Grzesiek S. and Bax,A. (1992) Improved 3D triple-resonance NMR techniques applied to a 31 kDa protein. J. Magn. Reson., 96, 432–440. [Google Scholar]
- Grzesiek S. and Bax,A. (1993) The importance of not saturating H2O in protein NMR. Application to sensitivity enhancement and NOE measurement. J. Am. Chem. Soc., 115, 12593–12593. [Google Scholar]
- Grzesiek S., Bax,A., Clore,G.M., Gronenborn,A.M., Hu,J.-S., Kaufman,J., Palmer,I., Stahl,S.J. and Wingfield,P.T. (1996) The solution structure of HIV-1 Nef reveals an unexpected fold and permits delineation of the binding surface for the SH3 domain of Hck tyrosine protein kinase. Nature Struct. Biol., 3, 340–345. [DOI] [PubMed] [Google Scholar]
- Horita D.A., Ivanova,A.V., Altieri,A.S., Klar,A.J.S. and Byrd,R.A. (2001) Solution structure, domain features and structural implications of mutants of the chromo domain from the fission yeast histone methyltransferase Clr4. J. Mol. Biol., 307, 861–870. [DOI] [PubMed] [Google Scholar]
- Hudson B.P., Martinez-Yamout,M.A., Dyson,H.J. and Wright,P.E. (2000) Solution structure and acetyl-lysine binding acitivity of the GCN5 bromodomain. J. Mol. Biol., 304, 355–370. [DOI] [PubMed] [Google Scholar]
- Jacobson R.H., Ladurner,A.G., King,D.S. and Tjian,R. (2000) Structure and function of a human TAFII250 double bromodomain module. Science, 288, 1422–1425. [DOI] [PubMed] [Google Scholar]
- James T.C. and Elgin,S.C. (1986) Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell. Biol., 6, 3862–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T. (2001) Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol., 11, 266–273. [DOI] [PubMed] [Google Scholar]
- Johnson B.A. and Blevins,R.A. (1994) NMRview: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR, 4, 603–614. [DOI] [PubMed] [Google Scholar]
- Kay L.E., Torchia,D.A. and Bax,A. (1989) Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry, 28, 8972–8979. [DOI] [PubMed] [Google Scholar]
- Klotz I.M. (1997) Ligand–Receptor Energetics. John Wiley & Sons, New York, NY.
- Kuriyan J. and Cowburn,D. (1997) Modular peptide recognition domains in eukaryotic signaling. Annu. Rev. Biophys. Biomol. Struct., 26, 259–288. [DOI] [PubMed] [Google Scholar]
- Lachner M., O’Carroll,D., Rea,S., Mechtler,K. and Jenuwein,T. (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature, 410, 116–120. [DOI] [PubMed] [Google Scholar]
- Marion D., Driscoll,P.C., Kay,L.E., Wingfield,P.T., Bax,A., Gronenborn,A.M. and Clore,G.M. (1989) Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heteronuclear 1H–15N Hartmann–Hahn-multiple quantum coherence and nuclear Overhauser–multiple quantum coherence spectroscopy: application to interleukin 1β. Biochemistry, 28, 6150–6156. [DOI] [PubMed] [Google Scholar]
- Minc E., Allory,V., Worman,H.J., Courvalin,J.C. and Buendia,B. (1999) Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma, 108, 220–234. [DOI] [PubMed] [Google Scholar]
- Mizzen C. et al. (1998) Signaling to chromatin through histone modifications: how clear is the signal? Cold Spring Harb. Symp. Quant. Biol., 68, 469–481. [DOI] [PubMed] [Google Scholar]
- Nakayama J.-I., Rice,J.C., Strahl,B.D., Allis,C.D. and Grewal,S.I.S. (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science, 292, 110–113. [DOI] [PubMed] [Google Scholar]
- Nicholls A. (1993) GRASP: Graphical Representation and Analysis of Surface Properties. Columbia University, New York, NY.
- Nielsen A.L., Oulad-Abdelghani,M., Ortiz,J.A., Remboutsika,E., Chambon,P. and Losson,R. (2001) Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell, 7, 729–739. [DOI] [PubMed] [Google Scholar]
- Ohe Y. and Iwai,K. (1981) Human spleen histone H3. Isolation and amino acid sequence. J. Biochem., 90, 1205–1211. [DOI] [PubMed] [Google Scholar]
- Owen D.J., Ornaghi,P., Yang,J., Lowe,N., Evans,P.R., Ballario,P., Neuhaus,D., Filetici,P. and Travers,A.A. (2000) The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase Gcn5p. EMBO J., 19, 6141–6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platero J.S., Hartnett,T. and Eissenberg,J.C. (1995) Functional analysis of the chromo domain of HP1. EMBO J., 14, 3977–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S. et al. (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature, 406, 593–599. [DOI] [PubMed] [Google Scholar]
- Rice J.C. and Allis,C.D. (2001) Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell Biol., 13, 263–273. [DOI] [PubMed] [Google Scholar]
- Spofford J.B. (1976) Position effect variegation in Drosophila. In Ashburner,M. and Novitski,E. (eds), Genetics and Biology of Drosophila. Vol. 1c. Academic Press, London, pp. 955–1019.
- Sterner D.E. and Berger,S.L. (2000) Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev., 64, 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Strahl B.D., Ohba,R., Cook,R.G. and Allis,C.D. (1999) Methylation of histone H3 at lysine 4 is highly conserved and correlated with transcriptionally active nuclei in Tetrahymena. Proc. Natl Acad. Sci. USA, 96, 14967–14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B.M. (2000) Histone acetylation and an epigenetic code. BioEssays, 22, 836–845. [DOI] [PubMed] [Google Scholar]
- Weiler K.S. and Wakimoto,B.T. (1995) Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet., 29, 557–605. [DOI] [PubMed] [Google Scholar]
- Winston F. and Allis,C.D. (1999) The bromodomain: a chromatin-targeting module? Nature Struct. Biol., 6, 601–604. [DOI] [PubMed] [Google Scholar]
- Wiseman T., Williston,S., Brandts,J.F. and Lin,N.L. (1989) Rapid measurement of binding constant and heats of binding using a new titration calorimeter. Anal. Biochem., 179, 131–137. [DOI] [PubMed] [Google Scholar]
- Wolffe A.P. and Guschin,D. (2000) Review: chromatin structural features and targets that regulate transcription. J. Struct. Biol., 129, 102–22. [DOI] [PubMed] [Google Scholar]
- Zhao T., Heyduk,T. and Eissenberg,J.C. (2001) Phosphorylation site mutations in heterochromatin protein 1 (HP1) reduce or eliminate silencing activity. J. Biol. Chem., 276, 9512–9518. [DOI] [PubMed] [Google Scholar]