Abstract
The requirement for small molecule transport systems across the peroxisomal membrane has previously been postulated, but not directly proven. Here we report the identification and functional reconstitution of Ant1p (Ypr128cp), a peroxisomal transporter in the yeast Saccharomyces cerevisiae, which has the characteristic sequence features of the mitochondrial carrier family. Ant1p was found to be an integral protein of the peroxisomal membrane and expression of ANT1 was oleic acid inducible. Targeting of Ant1p to peroxisomes was dependent on Pex3p and Pex19p, two peroxins specifically required for peroxisomal membrane protein insertion. Ant1p was essential for growth on medium-chain fatty acids as the sole carbon source. Upon reconstitution of the overexpressed and purified protein into liposomes, specific transport of adenine nucleotides could be demonstrated. Remarkably, both the substrate and inhibitor specificity differed from those of the mitochondrial ADP/ATP transporter. The physiological role of Ant1p in S.cerevisiae is probably to transport cytoplasmic ATP into the peroxisomal lumen in exchange for AMP generated in the activation of fatty acids.
Keywords: adenine nucleotide transporter/mitochondrial carrier family/peroxisomes/Saccharomyces cerevisiae/YPR128c
Introduction
Peroxisomes are ubiquitous organelles of eukaryotic cells, which harbor a variable number of oxidative enzymatic reactions, such as the α- and β-oxidation of fatty acids (for review see van den Bosch et al., 1992). The importance of peroxisomes for cellular function is emphasized by a number of severe inherited diseases in man that are caused by peroxisomal dysfunction (Lazarow and Moser, 1995; Gould and Valle, 2000).
Peroxisomal metabolism requires a continuous flux of metabolites and cofactors across the peroxisomal membrane. The peroxisomal membrane has long been thought to be freely permeable to small solutes (Van Veldhoven et al., 1987). This view was supported by direct permeability measurements of plant and mammalian peroxisomes using patch–clamp techniques and the identification of porin-like channels that mediate diffusion of metabolites across the membrane of plant peroxisomes (Lemmens et al., 1989; Reumann, 2000). Recent data, however, indicate that yeast peroxisomes are impermeable to NAD(H), NADP(H) and acetyl-CoA in vivo (van Roermund et al., 1995; Henke et al., 1998). Moreover, it was shown that the peroxisome membrane of mammals is impermeable even to protons (Dansen et al., 2000). The recognition of the peroxisomal membrane as a barrier for hydrophilic solutes strongly argues in favor of the existence of specific shuttle systems that catalyze the transport of substrates across the peroxisomal membrane.
To date, only a few peroxisomal membrane proteins with similarities to known small-molecule transport systems have been identified. Foremost of these are four different ATP-binding cassette (ABC) half transporters in mammals (Shani et al., 1997) and their yeast counterparts Pat1p (Pxa2p)/Pat2p (Pxa1p), which are probably involved in acyl-CoA transport (Hettema et al., 1996; Shani and Valle, 1996). Other potential peroxisomal transporters fall into the class of the mitochondrial carrier family (MCF) (Walker, 1992; Palmieri, 1994). This group of transporters is composed of three tandem-repeated modules of ∼100 amino acids with conserved sequence features. Each module is made of two hydrophobic transmembrane α-helices joined by a large hydrophilic loop. Peroxisomal members of this family include human Pmp34p (Wylin et al., 1998), the related Pmp47p of Candida boidinii (McCammon et al., 1990; Jank et al., 1993) and a human Ca2+-dependent solute carrier (Weber et al., 1997). Recently, genetic evidence for a function of Pmp47p in the utilization of medium-chain fatty acids has been provided (Nakagawa et al., 2000). However, transport activity and substrate specifity has not been proven experimentally for any of the putative peroxisomal carriers.
Here we report the identification and functional characterization of Ant1p, a peroxisomal member of the family of mitochondrial carrier proteins in Saccharomyces cerevisiae. Ant1p was overexpressed, purified, reconstituted into phospholipid vesicles and shown to represent a novel type of adenine nucleotide transporter. Consistent with this function, deficiency of Ant1p did result in an inability to utilize medium-chain fatty acids, a process that is dependent on intra-peroxisomal ATP. We also show that C.boidinii Pmp47p is a true ortholog of Ant1p and discuss our findings in terms of transport processes across the peroxisomal membrane.
Results
Identification of Ant1p in a reverse genetic screen for peroxisomal membrane proteins
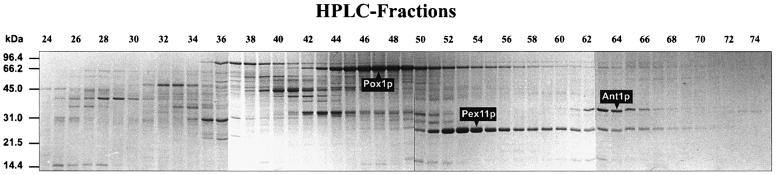
In an attempt to identify peroxisomal membrane proteins, peroxisomes were purified from oleic acid-induced S.cerevisiae cells and subjected to a set of consecutive extraction steps. Proteins remaining in the membranous fraction after high salt treatment were solubilized by SDS and separated by reversed-phase high-peformance liquid chromatography (HPLC) (Erdmann and Blobel, 1995). Proteins present in individual HPLC fractions were further separated by SDS–PAGE. This strategy yielded ∼30 discernible protein bands of distinct size and intensity (Figure 1). The two most prominent bands represent Pox1p and Pex11p. The identity of another abundant protein with an apparent molecular weight of 35 kDa (indicated by an arrow in Figure 1) was determined by partial protein sequencing. The sequences obtained from three internal peptides generated by digestion of the purified protein with Lys-C protease match to a single open reading frame in the yeast genome database, YPR128c, and correspond to predicted amino acids 115–141, 169–181 and 182–202. The deduced amino acid sequence of YPR128c, designated here as Ant1p, gives rise to a molecular weight of 35 kDa, which fits well with the size of the isolated protein. Database searches revealed that the protein belongs to the family of structurally related MCF proteins, ranging from yeast to man (Palmieri, 1994).
Fig. 1. Purification and identification of Ant1p. High-salt extracted peroxisomal membranes were solubilized in SDS and proteins contained therein were separated by preparative reversed-phase HPLC. The protein identified by peptide sequencing as Ant1p (Ypr128cp) is indicated. Molecular weight standards are indicated on the left.
Ant1p is a peroxisomal membrane protein
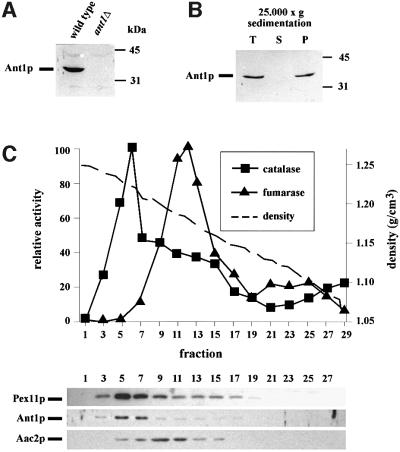
To underscore the finding of Ant1p being present in a peroxisomal membrane preparation, the location of native Ant1p was examined by subcellular fractionation studies. For that, an antibody directed against a bacterially expressed fragment of Ant1p was generated, which detected a band of 35 kDa in a protein extract of an oleic acid-induced wild-type strain, but not in that of an ant1Δ deletion strain (Figure 2A), demonstrating that the antibody is mono-specific for Ant1p. Organelles were isolated from whole-cell lysates of oleic acid-induced wild-type cells by centrifugation at 25 000 g. The specific antibody detected Ant1p exclusively in the organellar pellet fraction, which contains mainly mitochondria and peroxisomes (Figure 2B). To determine the distribution of Ant1p between mitochondria and peroxisomes, whole-cell lysates were fractionated by sucrose density gradient centrifugation. The gradient fractions were analyzed for enzymatic activities of the mitochondrial and peroxisomal marker enzymes fumarase and catalase, respectively. Immunoblot analysis of the fractions demonstrated that Ant1p comigrated with both the peroxisomal membrane protein Pex11p as well as with catalase at a density of 1.22 g/cm3, whereas the mitochondrial carrier Aac2p was found to peak at a density of 1.18 g/cm3 (Figure 2C). This result indicated that Ant1p is a peroxisomal protein, which was in agreement with a previous report by Geraghty et al. (1999).
Fig. 2. Ant1p is localized to peroxisomes. (A) Immunological detection of Ant1p. Equal amounts of whole-cell lysates of oleic acid-induced wild-type (UTL-7A) as well as ant1Δ cells were separated by SDS–PAGE and blotted onto a nitrocellulose filter. Antibodies directed against Ant1p were applied and immunoreactive complexes were visualized with the ECL system. (B) Subcellular fractionation analysis. Cell-free extracts of oleate-induced wild-type cells (T) were separated by differential centrifugation into a 25 000 g pellet containing mainly peroxisomes and mitochondria (P) and a supernatant fraction (S). Equal portions of each fraction were analyzed by immunoblotting using anti-Ant1p antibodies. (C) Immunological detection of Ant1p in a sucrose density gradient. Cell-free extracts of oleate-induced wild-type cells (UTL-7A) were separated on a continuous sucrose density gradient (20–53%). The resulting fractions were immunologically analyzed for the distribution of Ant1p, the peroxisomal membrane protein Pex11p and mitochondrial Aac2p. The same fractions were also analyzed for the enzymatic activities of peroxisomal catalase (squares) and mitochondrial fumarase (triangles), presented in each case as the percentage of the peak fraction. The fractions’ densities are illustrated as a hatched line in the same diagram.
The sub-peroxisomal location of Ant1p was analyzed by extracting a 25 000 g organellar pellet with low salt, high salt or with 100 mM Na2CO3 pH 11. The peroxisomal matrix enzyme Fox3p was already extractable by low salt, the membrane-associated Fat2p (Pcs60p) was susceptible to high-salt extraction, whereas the integral peroxisomal membrane protein Pex3p was resistant to all treatments. Ant1p was completely resistant to both low- and high-salt extraction, yet could be partially released from membranes by alkaline treatment (Figure 3A). This latter observation did not hold true for MCF proteins in general, as Aac2p remained exclusively in the membranous fraction upon carbonate extraction. Treatment of such a 25 000 g organellar pellet with proteinase K did result in the degradation of Ant1p in the absence of Triton X-100, whereas the matrix enzyme Fox3p was protected under these conditions (Figure 3B). This property of Ant1p is in line with the topology of a protein harboring multiple transmembrane spans.
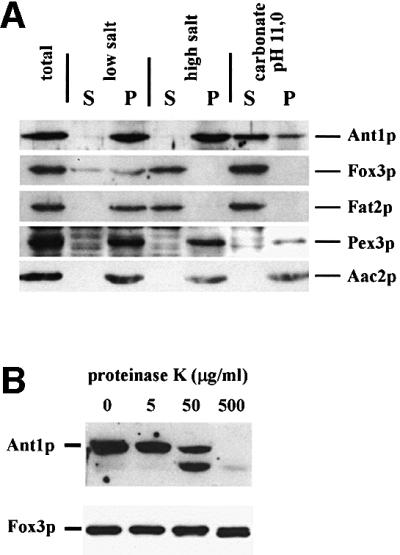
Fig. 3. Ant1p is a peroxisomal membrane protein. (A) Sub-peroxisomal fractionation analysis. A 25 000 g pellet of an oleic acid-induced wild-type strain (UTL-7A) was divided into three parts and treated with either 10 mM Tris–HCl pH 8 (low salt), 10 mM Tris–HCl pH 8/500 mM KCl (high salt) or 100 mM Na2CO3 pH 11 (carbonate). After 30 min incubation, each sample was separated into a pellet (P) and a supernatant (S) fraction by a 200 000 g centrifugation step. Proportionate volumes of the resulting fractions were subjected to immunoblotting using antibodies directed against Ant1p, Pex3p, Fat2p/Pcs60p, Fox3p and Aac2p. (B) Protease protection assay. Organelles isolated from a wild-type strain were split into four parts and incubated for 30 min with increasing concentrations of proteinase K. Reactions were stopped by the addition of 4 mM PMSF and trichloroacetic acid, separated by SDS–PAGE and analyzed by immunoblotting.
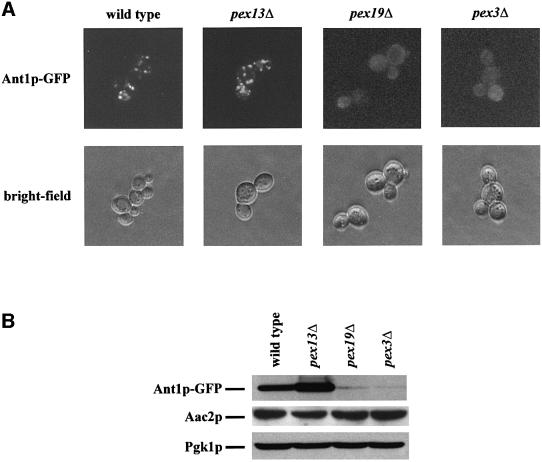
Targeting of Ant1p was further investigated by fluorescence microscopy using a green fluorescent protein (GFP) fusion protein. Since the tagged protein was able to restore growth of an ant1Δ mutant on lauric acid (see below), the fusion protein is expected to maintain the same subcellular localization as the native protein. To exclude the possibility of overexpression artefacts, the fusion protein was expressed from a single genomic copy under the control of its native promoter. In an oleic acid-induced wild-type strain a punctate fluorescence pattern was observed. However, in pex19Δ and pex3Δ mutants, which entirely lack peroxisomes but contain mitochondria, fluorescence was largely diffuse (Figure 4A). According to subcellular fractionation studies using flotation gradients, the very weak additional punctate staining pattern seen in pex19Δ cells is likely to be due to a mistargeting of Ant1p to mitochondria (data not shown) as observed for other peroxisomal membrane proteins (Hettema et al., 2000). This observation indicated that the punctate fluorescence seen in the wild-type strain indeed represented the peroxisomal compartment. As expected for a peroxisomal membrane protein, a punctate fluorescence pattern was observed in the pex13Δ mutant, which is known to be defective for peroxisomal matrix protein import, but still contains peroxisomal membrane ghosts (Erdmann and Blobel, 1996). Since a number of peroxisomal membrane proteins are degraded in the absence of peroxisomes (Sacksteder et al., 2000), the amount of the Ant1p fusion protein in these pex mutant strains was determined by immunoblot analysis. Protein abundance was drastically lowered in the peroxisome-deficient pex19Δ and pex3Δ strains, whereas cytosolic phosphoglycerate kinase (Pgk1p) and mitochondrial Aac2p were present in comparable amounts (Figure 4B). Thus it may well be that the diffuse fluorescence pattern observed in the pex19Δ and pex3Δ mutants originated in part from GFP-containing degradation products of the fusion protein. These data clearly establish that Ant1p targeting to peroxisomes depends on the route for peroxisomal membrane proteins.
Fig. 4. Peroxisomal location of Ant1p depends on the membrane protein targeting route. (A) Localization of an Ant1p–GFP fusion protein. The wild-type UTL-7A and the otherwise isogenic pex13Δ, pex19Δ and pex3Δ strains expressing Ant1p–GFP under oleic acid-induction conditions were examined for GFP fluorescence. Structural integrity of the cells is documented by bright-field microscopy. (B) Stability of the Ant1p–GFP fusion protein in pex mutants. The same strains as in (A) were induced for 14 h in rich oleate-containing medium. Whole-cell extracts of these samples were analyzed for the amount of Ant1p–GFP, mitochondrial Aac2p and cytosolic Pgk1p by immunological detection.
Ant1p is required for growth on medium-chain fatty acids
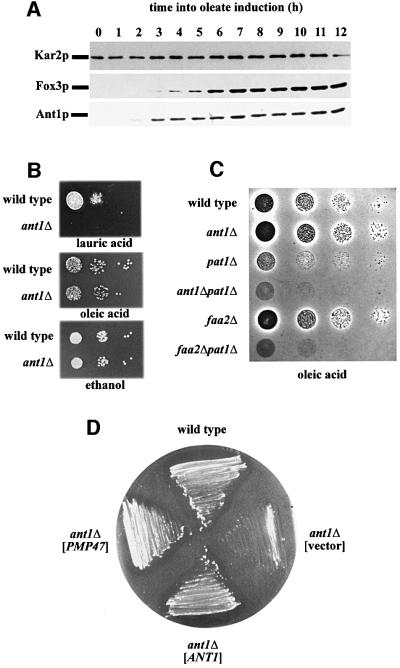
The amount of Ant1p was analyzed at various time points upon shifting cells from low glucose to oleic acid-containing medium. Ant1p expression increased similarly to the inducible Fox3p control (Figure 5A), indicative of Ant1p being involved in fatty acid β-oxidation. To elucidate the potential role of Ant1p in the breakdown of fatty acids, wild-type and ant1Δ strains were grown on plates containing ethanol, the long-chain fatty acid oleic acid (C18:1) and the medium-chain fatty acid lauric acid (C12) as sole carbon source. Many laboratory yeast strains do not utilize medium-chain fatty acids efficiently. Therefore, growth assays were carried out in the genetic background of a segregant of the diploid strain FY1679 (Winston et al., 1995), which proved best suited for growth on lauric acid. The strain deleted in ANT1 was not affected for growth on ethanol or oleic acid, but failed to form colonies on lauric acid (Figure 5B). The involvement of Ant1p in the utilization of fatty acids was further investigated by analyzing a pat1Δ ant1Δ strain for growth on oleic acid. Pat1p is required for the transport of activated long-chain fatty acids across the peroxisomal membrane. However, residual growth on oleate can be observed in a pat1Δ strain due to an alternative, albeit inefficient, transport of oleic acid as free fatty acid (Hettema et al., 1996). The latter route is predominant for medium-chain fatty acids and depends on an intra-peroxisomal ATP-consuming activation of imported free fatty acids via the acyl-CoA synthetase Faa2p. Blocking both routes as in a pat1Δ faa2Δ strain will result in a complete loss of fatty acid utilization and in an inability to use fatty acids as an energy source (Hettema et al., 1996). Most interestingly, on oleic acid plates, a pat1Δ ant1Δ double deletion was detrimental to the cells and no zones of clearing appeared, thus resembling the pat1Δ faa2Δ strain (Figure 5C). Faa2p and Ant1p are therefore likely to be involved in the same pathway of fatty acid metabolism.
Fig. 5. Ant1p is involved in medium-chain fatty acid utilization. (A) Kinetics of Ant1p expression under oleic acid-induction conditions. Wild-type strain UTL-7A grown in minimal 0.3% glucose-containing medium was transferred to oleate-containing medium and aliquots were removed at the time points indicated. Whole-cell extracts of these samples were analyzed for the amount of Ant1p, the oleic acid-inducible Fox3p and the constitutively expressed Kar2p by immunological detection. (B) Growth behavior of an ant1Δ mutant on various carbon sources. Serial dilutions of wild-type strain FY1679α and the otherwise isogenic ant1Δ mutant were spotted on plates containing ethanol, oleic acid or lauric acid, and incubated for 2–7 days at 30°C. (C) Analysis of an ant1Δ pat1Δ double mutant. The gene deletion strains indicated (in the genetic background of FY1679α) were similarly tested for growth on oleic acid. (D) Complementation test with C.boidinii Pmp47p. Transformants of the ant1Δ mutant, expressing either Pmp47p from C.boidinii [ant1Δ (PMP47)] from plasmid pRS-315-24-47, Ant1p [ant1Δ (ANT1)] or the empty vector [ant1Δ (vector)] were streaked on lauric acid plates and incubated for 7 days at 30°C.
MCF proteins have been identified as peroxisomal proteins in mammals (Pmp34p) (Wylin et al., 1998) and C.boidinii (Pmp47p) (Jank et al., 1993). The latter protein has previously been implicated in medium-chain fatty acid metabolism (Nakagawa et al., 2000) and shown to be targeted to peroxisomes when expressed in S.cerevisiae (McCammon et al., 1990). In addition, a phylogenetic analysis of Pmp47p and all MCF members of S.cerevisiae linked Pmp47p most closely to Ant1p (not shown). We therefore tested whether heterologous expression of Pmp47p could restore growth of an ANT1 deletion strain on lauric acid plates. Plasmid-borne copies of either ANT1 or PMP47 caused the deletion strain to grow and zones of clearing appeared (Figure 5D). This result indicated that the two peroxisomal MCF members, Ant1p and Pmp47p, are true orthologs, required to transport a compound that is mandatory for the peroxisomal degradation of medium-chain fatty acids.
Overexpression and purification of Ant1p
To test directly the transport properties of Ant1p, a reconstitution assay was required. All attempts to express the protein heterologously in Escherichia coli failed, probably due to a disparate codon usage. A His6-epitope-tagged version of the protein was therefore ectopically expressed from the GAL1 promoter in S.cerevisiae (Figure 6A, lane 2). The protein was purified by affinity chromatography from a solubilized organellar preparation. As shown in Figure 6A (lane 4), no protein bands were detected in addition to tagged Ant1p in a gel stained with silver nitrate, indicating that purification occurred to apparent homogeneity. Approximately 25 µg of purified Ant1p were obtained per liter of culture. For comparison, a 25-fold protein excess of the flow-through of the affinity column is shown in lane 3. The flow-through consisted of the solubilized organellar proteins that did not bind to the Ni-NTA–agarose column. Anti-Ant1p antibodies, but not anti-Aac2p antibodies, were able to detect the purified protein, whereas the opposite was true for Aac2p present in the flow-through (Figure 6B). The identity of the isolated Ant1p was confirmed by mass spectrometric analysis. By the same technique, we could not detect any contamination with the mitochondrial ADP/ATP carrier in the purified sample, in accordance with the western blot analysis (Figure 6B).
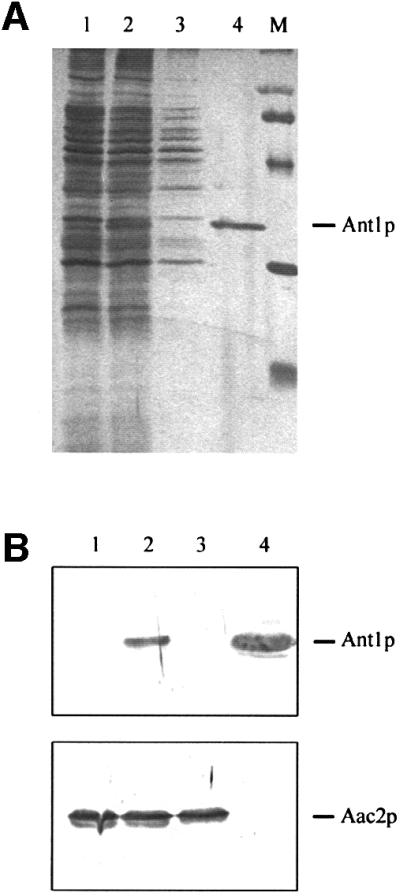
Fig. 6. Purification of the His6-tagged Ant1p. (A) Proteins were separated by SDS–PAGE and stained with silver nitrate. Lanes 1 and 2, organellar pellet protein (8 µg) from wild-type (lane 1) and YPH499-pHPR178 cells (lane 2). Lane 3, Triton-solubilized extract (3.5 µg protein) of the organellar pellet in lane 2 after incubation with Ni-NTA agarose (flow through). Lane 4, His6-tagged Ant1p (0.14 µg) purified from the organellar pellet in lane 2. Lane M, molecular weight markers (97.4, 66.2, 45, 31 and 21.5 kDa). The protein identified by MALDI-TOF mass spectrometry as Ant1p (Ypr128cp) is indicated. (B) 30 µg of organellar pellet protein from wild-type (lane 1) and YPH499-pHPR178 cells (lane 2) were separated by SDS–PAGE, transferred to nitrocellulose, and blotted with antibodies directed against Ant1p and Aac2p. Lane 3, 30 µg of Triton-solubilized extract of the organellar pellet in lane 2 after incubation with Ni-NTA–agarose (flow through). Lane 4, 2 µg of tagged Ant1p purified from the organellar pellet in lane 2.
Ant1p is a novel adenine nucleotide transporter
Upon reconstitution into liposomes, recombinant Ant1p catalyzed an active [14C]ATP/ATP exchange that was completely inhibited by a mixture of pyridoxal 5′-phosphate and bathophenanthroline. No such activity was detected with recombinant Ant1p that had been boiled before incorporation into liposomes nor by reconstitution of two unrelated MCF carriers, Crc1p and Odc1p (Palmieri et al., 1999, 2001), which had been purified from yeast using the same expression vector, nor of a mock control. Furthermore, reconstituted Ant1p did not catalyze the homo-exchanges of phosphate, pyruvate, malonate, succinate, malate, oxoglutarate, ketoisocaproate, citrate, carnitine, ornithine, lysine, arginine, histidine, glutathione, choline, spermine, proline and threonine (external concentration 1 mM, internal concentration 10 mM; data not shown), all being substrates for various known mitochondrial transporters.
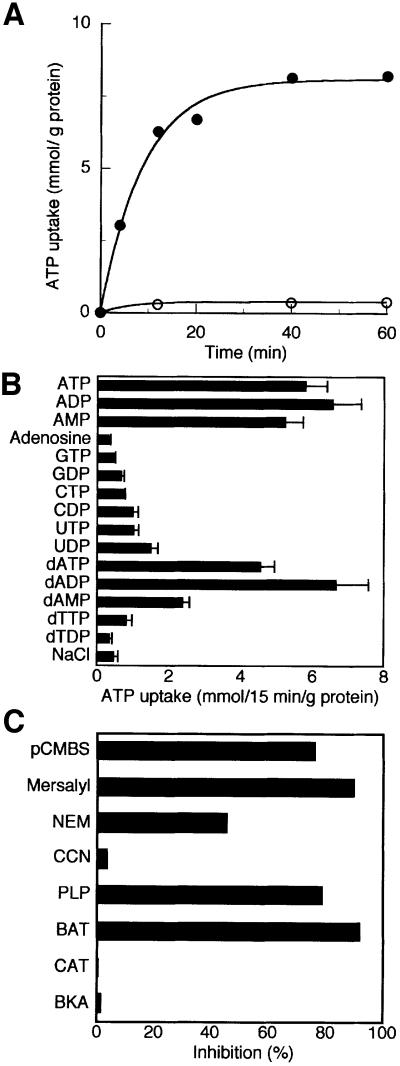
The uptake of 50 µM [14C]ATP into liposomes reconstituted with purified Ant1p and containing 20 mM ATP followed first-order kinetics (rate constant 0.11 min–1; initial rate 0.91 mmol/min/g protein), isotopic equilibrium being approached exponentially (Figure 7A). In contrast, when the proteoliposomes were pre-loaded with GTP instead of ATP, the uptake of ATP was very low.
Fig. 7. Ant1p catalyzes the transport of adenine nucleotides. (A) Time-course of [14C]ATP/ATP exchange in proteoliposomes reconstituted with the recombinant Ant1p. [14C]ATP (50 µM) was added to proteoliposomes containing 20 mM ATP (filled circles) or 20 mM GTP (open circles). (B) Dependence of Ant1p activity on internal substrate. Proteoliposomes were preloaded internally with various substrates (concentration 20 mM). Transport was started by adding 50 µM [14C]ATP and stopped after 15 min. The values are means of at least three experiments. (C) Inhibition of [14C]ATP/ATP exchange by various reagents. Proteoliposomes were preloaded internally with 20 mM ATP. Transport was started by adding 50 µM [14C]ATP and stopped after 15 min. Inhibitors were added 5 min before the labeled substrate. The final concentration of the inhibitors was 2 mM except for mercurials (0.1 mM), N-ethylmaleimide (1 mM) and carboxyatractyloside and bongkrekate (0.02 mM). The extents of inhibition (%) from a representative experiment are reported. Similar results were obtained in at least three independent experiments. pCMBS, p-chloromercuriphenylsulfonate; NEM, N-ethylmaleimide; CCN, α-cyano-4-hydroxycinnamate; PLP, pyridoxal 5′-phosphate; BAT, bathophenanthroline; CAT, carboxyatractylate, BKA, bongkrekic acid.
The substrate specificity of reconstituted Ant1p was examined in detail by measuring the uptake of [14C]ATP into proteoliposomes that had been pre-loaded with various potential substrates (Figure 7B). The highest activities were observed in the presence of internal ATP, ADP and AMP. [14C]ATP was also taken up efficiently by proteoliposomes containing the corresponding deoxy-nucleotides. Much lower activity was found with internal UDP, UTP and CDP. Very low activities were observed with any of the other nucleotides tested as well as with adenosine and (not shown) with GMP, CMP, UMP, dGDP, dCDP, dUDP, with fumarate, succinate, l-malate, malonate, phosphate, oxoglutarate, citrate, sulfate, oxaloacetate, pyruvate, phosphoenolpyruvate, l-carnitine, l-ornithine, l-citrulline, glutamate, aspartate or glutamine. The residual activity in the presence of these substrates was virtually the same as the activity observed in the presence of NaCl. Therefore the substrate specificity of Ant1p is confined to adenine nucleotides.
The [14C]ATP/ATP exchange in proteoliposomes reconstituted with purified Ant1p was inhibited strongly by organic mercurials like mersalyl and p-chloromercuriphenylsulfonate and partly by N-ethylmaleimide (Figure 7C). Bathophenanthroline and pyridoxal 5′-phosphate (inhibitors of many mitochondrial carriers) also had a considerable inhibitory effect. Strikingly, carboxyatractyloside and bongkrekic acid, the specific and powerful inhibitors of the mitochondrial ADP/ATP carrier (Klingenberg, 1979), had no effect on the activity of reconstituted Ant1p (Figure 7C). Also, these inhibitors caused virtually no inhibition when added together at either side of the liposomal membrane (not shown). In addition, no significant inhibition was observed with α-cyano-4-hydroxycinnamate, a powerful inhibitor of the pyruvate carrier (Halestrap, 1975) (Figure 7C).
Discussion
A clear picture of solute transport across the peroxisomal membrane did not evolve earlier, partly because of the lack of reliable in vitro transport studies with isolated peroxisomes, as these become extremely fragile upon isolation. The results reported here demonstrate for the first time the transport properties of a reconstituted peroxisomal carrier protein, thereby circumventing the need for intact peroxisomal preparations.
Ant1p represents an abundant protein of the peroxisomal membrane (see Figure 1), estimated densitometrically to account for ∼5% of total peroxisomal membrane protein of oleic acid-induced cells (data not shown). ANT1 upregulation was observed in the presence of oleate (Figure 5A) or laurate (unpublished data), probably due to a region in the ANT1 promoter that resembles the consensus sequence of an oleate response element (Rottensteiner et al., 1996), albeit with an unusual 14-nucleotide spacing of the inverted CGG triplets. To date, Ant1p is the only peroxisomal member of the mitochondrial carrier family in S.cerevisiae. Peroxisomal targeting and stability of Ant1p were dependent on Pex3p and Pex19p, the two peroxins specifically required for peroxisomal membrane protein insertion. It is therefore likely that Ant1p is specifically recruited by the Pex3p/Pex19p-dependent pathway, which would destine Ant1p to peroxisomes and not to mitochondria.
The transport properties of Ant1p show that it is a novel adenine nucleotide transporter. Besides transporting ATP, ADP and AMP with high efficiency, reconstituted Ant1p also accepts the corresponding deoxynucleotides as substrates. The substrate specificity of Ant1p is distinct from that of any other previously characterized member of the MCF (see Dolce et al., 2001). In particular, Ant1p differs markedly from the mitochondrial ADP/ATP carrier. First, in contrast to Ant1p, the mitochondrial ADP/ATP carrier does not transport AMP (Klingenberg, 1979). Secondly, Ant1p efficiently transports the adenine nucleotide deoxy-nucleotides, which are very poor substrates for the ADP/ATP carrier (Klingenberg, 1979). Thirdly, Ant1p is not inhibited by carboxyatractyloside and bongkrekic acid, the powerful and specific inhibitors of the ADP/ATP carrier (Klingenberg, 1979). Consistent with their different transport properties, Ant1p shares only 13–16% identical amino acids with the yeast ADP/ATP carrier isoforms (for references see Nelson et al., 1998) and they appear to be phylogenetically distant (Nelson et al., 1998).
Ant1p is also quite different from the mammalian deoxynucleotide carrier (Dolce et al., 2001) and its ortholog in yeast (L.Palmieri, unpublished data), as the latter proteins transport all deoxynucleotides whereas Ant1p specificity is confined to the adenine nucleotides.
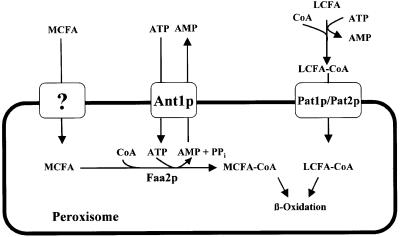
In S.cerevisiae, β-oxidation of fatty acids is confined to peroxisomes. Import of the fatty acid substrates has been postulated to occur via two independent transport systems with partially overlapping substrate specificity (Figure 8) (Hettema and Tabak, 2000). Long-chain fatty acids (>C16) are prefentially activated in the cytosol and subsequently imported into peroxisomes as CoA-derivatives in a process that involves the ABC transporters Pat1p/Pat2p (Verleur et al., 1997). Medium-chain fatty acids are imported as free fatty acid via an as yet uncharacterized transport system that seems to involve Pex11p (van Roermund et al., 2000). The activation of medium-chain fatty acids, catalyzed by the acyl-CoA synthetase Faa2p, occurs inside peroxisomes and consequently depends on the availability of intraperoxisomal ATP. Consistent with this model, the growth defect of cells lacking Ant1p on the medium-chain fatty acid lauric acid (C12) can be explained by Ant1p functioning as a peroxisomal adenine nucleotide transporter, which supplies intraperoxisomal Faa2p with the ATP needed for the activation of medium-chain fatty acids. In the absence of pat1Δ, activation of long-chain fatty acids also depends on Faa2p and Ant1p. This assumption is supported by genetic data showing synthetic lethality of a pat1Δ ant1Δ double knock out on oleic acid medium, which was also observed with the corresponding pat1Δ faa2Δ knock out strain.
Fig. 8. Model for the function of the adenine nucleotide carrier Ant1p in yeast peroxisomal fatty acid metabolism. Long-chain fatty acids (LCFA) are activated in the cytosol and enter the peroxisome as acyl-CoA esters via the heterodimeric ABC transporter Pat1p/Pat2p. Medium-chain fatty acids (MCFA) enter peroxisomes as free fatty acids, which are activated in the peroxisomal lumen via the acyl-CoA synthetase Faa2p. Ant1p catalyzes the exchange of ATP against AMP across the peroxisomal membrane, which is required for the intraperoxisomal activation of medium-chain fatty acids.
It is interesting to note that the hydrolysis of ATP catalyzed by Faa2p yields AMP and pyrophosphate. A peroxisomal adenylate kinase, which could generate ADP from AMP and ATP, has not yet been identified. Consequently, it is likely that a major function of Ant1p is to exchange AMP for ATP across the peroxisomal membrane. This gives a physiological explanation for the diversity in substrate specifity of the peroxisomal and mitochondrial ATP transporters.
Another peroxisomal enzyme likely to depend on the supply of ATP is Pcs60p (Fat2p), an abundant peroxisomal protein of the family of ATP-dependent CoA-ligases that function via a covalent AMP intermediate (Blobel and Erdmann, 1996). The role of Pcs60p in peroxisome metabolism has not yet been specified, but a member of this family has been shown to contain very-long-chain acyl-CoA synthetase activity (Watkins et al., 1998).
Expression of C.boidinii Pmp47p in an ant1Δ mutant restored growth on a medium-chain fatty acid. It is therefore straightforward to postulate that Pmp47p represents the peroxisomal adenine nucleotide transporter of C.boidinii. This assumption is in line with a similar phenotype of a pmp47 mutant, which is also defective for medium-chain fatty acid utilization (Nakagawa et al., 2000). Interestingly, this mutant also fails to grow on methanol because of a misfolding of one of the key enzymes in methanol utilization, dihydroxyacetone synthase (Sakai et al., 1996). In the light of our results, the existence of an ATP-dependent chaperone required for folding a subset of matrix proteins within peroxisomes can now be envisaged.
In humans, peroxisomal medium-chain fatty acyl-CoA synthetase is lacking because these fatty acids are almost exclusively degraded by the mitochondrial β-oxidation system. On the other hand, human peroxisomes contain a very-long-chain fatty acyl-CoA synthetase within peroxisomes (Steinberg et al., 1999) that accepts both very-long-chain fatty acids and the branched-chain fatty acid pristanic acid as a substrate (Steinberg et al., 1999). Cholesterol biosynthesis is another example of an ATP-consuming pathway occurring in peroxisomes. Three consecutive steps of this pathway, catalyzed by mevalonate kinase, phosphomevalonate kinase and mevalonate diphosphate decarboxylase require ATP. These enzymes hydrolyze ATP to ADP and Pi (Olivier and Krisans, 2000), while the peroxisomal acyl-CoA synthetases generate AMP and PPi. Thus, it is tempting to speculate that the transport properties of Ant1p are resembled by those of hPmp34p, the putative ortholog in human peroxisomes where AMP and ADP have to be recycled in the cytosol, and ATP to be imported into peroxisomes.
Materials and methods
Strains, media and growth conditions
Escherichia coli strain DH5α was used for all plasmid amplifications and isolations according to standard protocols (Sambrook et al., 1989). The S.cerevisiae strains used are listed in Table I. ANT1 was deleted in FY1679α by the ‘short flanking homology’ method using the removable loxP-kanMX4-loxP marker as described previously (Güldener et al., 1996). A LEU2-based disruption cassette was used to delete ANT1 in strain UTL-7A. The PAT1 (PXA2) locus in FY1679α was deleted with the help of a kanMX4-based disruption cassette that had been amplified from genomic DNA of strain BY4741ykl188cΔ (EUROSCARF, Frankfurt, Germany) using primer pair RE181/190. Deletion of FAA2 was similarly accomplished using primer pair RE224/225 and genomic DNA of strain BJ1991faa2Δ (Hettema et al., 1996). All gene deletion strains were verified by PCR. Yeast transformation was carried out as described previously (Schiestl and Gietz, 1989).
Table I. Saccharomyces cerevisiae strains used.
| Strain | Description | Source or reference |
|---|---|---|
| (1) UTL-7Aa | MATα leu2-3, 112 ura3-52 trp1 | Erdmann et al. (1989) |
| (2) FY1679 (S288C) | MATa/MATα ura3-52/ura3-52 trp1Δ63/TRP1 | Winston et al. (1995) |
| leu2Δ–1/LEU2 his3Δ200/HIS3 | ||
| (3) FY1679α2 | MATα ura3-52 trp1-Δ63 leu2-Δ1 | Winston et al. (1995) |
| YPH499 | MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 | Sikorski and Hieter (1989) |
| trp1-Δ63 lys2-801 | ||
| (4) UTL-7Apex13Δ1 | pex13Δ::URA3 | Erdmann and Blobel (1996) |
| (5) UTL-7Apex3Δ1 | pex3Δ::LEU2 | Höhfeld et al. (1991) |
| (6) UTL-7Apex19Δ1 | pex19Δ::LEU2 | Götte et al. (1998) |
| UTL-7Aant1Δ1 | ant1Δ::LEU2 | this study |
| yHPR2213 | ant1Δ::Loxp | this study |
| yHPR2223 | pat1Δ::kanMX4 | this study |
| yHPR2233 | pat1Δ::kanMX4 ant1Δ::Loxp | this study |
| yHPR2531 | ANT1–GFP-prA::TRP1 (pHPR129) | this study |
| yHPR2545 | pex3Δ::LEU2 ANT1–GFP-prA::TRP1 (pHPR129) | this study |
| yHPR2563 | faa2Δ::LEU2 | this study |
| yHPR2573 | pat1Δ::kanMX4 faa2Δ::LEU2 | this study |
| yHPR2584 | pex13Δ::URA3 ANT1–GFP-prA::TRP1 (pHPR129) | this study |
| yHPR2596 | pex19Δ::LEU2 ANT1–GFP-prA::TRP1 (pHPR129) | this study |
aParental genotypes, e.g. UTL-7Apex13Δ1 was derived from (1) UTL-7A.
Standard media for cultivating bacteria and yeast strains were prepared according to Sambrook et al. (1989). YNO medium was composed of 0.67% yeast nitrogen base without amino acids, 0.1% yeast extract, amino acids as required, 0.1% oleic acid and 0.05% Tween-40. Rich YPO medium contained 0.1% yeast extract, 2% peptone, 0.2% oleic acid plus 0.02% Tween-80, adjusted to pH 7 with NaOH. Lauric acid plates were made of 0.17% yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate, amino acids as required, 0.1% yeast extract, 0.5% K-phosphate buffer pH 6, 0.07% lauric acid, 0.28% Tween-20 and 2% agar. Oleic acid- and ethanol-containing plates were similarly prepared, but contained 0.1% oleic acid/0.5% Tween-80 or 2% ethanol, respectively. For the analysis of expression levels of Ant1p-GFP-prA, and monitoring its fluorescence, cells were induced in YPO medium as described previously (Rottensteiner et al., 1996). For the isolation of organelles and the kinetics of oleate induction, cells were induced in YNO medium according to Erdmann et al. (1989). For the high-yield expression and purification of recombinant Ant1p, S.cerevisiae YPH499 cells (Sikorski and Hieter, 1989) were transformed with plasmid pHPR178 (see below) and transformants (YPH499-pHPR178 cells) were grown as described previously (Palmieri et al., 2001).
Plasmid constructions
Plasmids and oligonucleotides used are listed in Tables II and III, respectively. Unless otherwise stated, genomic DNA of a S.cerevisiae wild-type strain was used as a template for PCRs. Cloning of PCR-derived fragments into the target vectors was preceded by their subcloning into EcoRV-digested pBluescript SK+. The ANT1 complementing plasmid pWG35/3 was generated by cloning the PCR product of primer pair KU167/178 (containing the ANT1 ORF including its promoter and terminator region) into pRS416 via SacI and HindIII. For the bacterial expression of amino acids 90–210 of Ant1p, pWG35/7 was constructed by inserting a BamHI–SacI-digested ANT1 fragment (primer pair KU206/207) into pET21b (Novagen, Madison, WI). The ANT1 disruption cassette was generated by assembling the ANT1 promoter region (a SacI–XbaI fragment of pWG35/3), the LEU2 marker [an XbaI–BamHI fragment of pJJ283 (Jones and Prakash, 1990)] and the ANT1 terminator region (a BamHI–HindIII fragment of a PCR with primer pair KU168/178) in pBluescript SK+.
Table II. Plasmids used.
| Plasmid | Description | Source or reference |
|---|---|---|
| pHPR104 | PrA in pBluescript SK+ (Stratagene) | this study |
| pHPR129 | ANT1–GFP-prA in YIplac204 | this study |
| pHPR137 | ANT1-prA in YEplac195 | this study |
| pHPR178 | ANT1-His6 in pYES2 (Invitrogen) | this study |
| pWG35/3 | ANT1 in pRS416 | this study |
| pWG35/7 | ANT1 in pET21b (Novagen) | this study |
| pRS-315-24-47 | PMP47 in pRS315 | McCammon et al. (1990) |
| pRS416 | URA3-marked centromeric plasmid | Sikorski and Hieter (1989) |
| YIplac204 | TRP1-marked integrative plasmid | Gietz and Sugino (1988) |
| YEplac195 | URA3-marked episomal plasmid | Gietz and Sugino (1988) |
Table III. Oligonucleotides used.
| Oligonucleotide | Sequence (5′→3′) |
|---|---|
| RE149 | GCGGCCGCATGCTTGCGCAACACGATGAAG |
| RE150 | GCGGCCGCTACCCTGAAAATAAAGATTTTCATTCGCGTCTACTTTCGG |
| RE181 | CCACACCATCATCCTTAT |
| RE190 | AGGCAGTGGGTCGTGACATG |
| RE195 | GAATTCGCGGGTACAGTTTAATT |
| RE196 | GCGGCCGCTAGTGGAAGCCAGCTTGCG |
| RE224 | AGAACATCCTCCGGACCGTC |
| RE225 | TTAATGGTATAAATAGTTGG |
| RE389 | AATTAAGCTTATGTTAACTCTAGAGTCTGC |
| RE390 | AATTCTCGAGTCAGTGATGGTGATGGTGATGAGTGGAAGCCAGCTTGCG |
| KU167 | GCAGGCCTGAGCTCCGCGGGTACAGTTTAATT |
| KU168 | CCCTCGAGGGATCCGAATCCTTACTATTACTG |
| KU178 | ATCCCGGGAAGCTTAATAGTCGACAAGCGCCC |
| KU206 | GCGGATCCGGAATTCGTTTATTTCTTTTGGTAC |
| KU207 | GCGGATCCCCAGGAGCTCGAGTTGGAATGGTCATGGAA |
The Ant1p–GFP-prA fusion protein was constructed as follows: the protein A tag was amplified from plasmid pBS-NOP1-prA (E.Hurt, Heidelberg, Germany) using primer pair RE149/150 and cloned into pBluescript SK+ (pHPR104). The ANT1 open reading frame including its promoter region was generated by a PCR with primer pair RE195/196. Plasmid pHPR137 was cloned by a triple ligation of ANT1pr–ANT1 as an EcoRI–NotI fragment, the prA tag isolated from pHPR104 as a NotI–PstI fragment and EcoRI–PstI-digested YEplac195. The ANT1–GFP-prA fusion (pHPR129) was obtained by a ligation of the EcoRI–HindIII ANT1-prA insert from pHPR137 into YIplac204 and subsequent insertion of a GFP-encoding NotI fragment (Clontech, Palo Alto, CA, USA). The His6-tagged Ant1p was expressed from plasmid pHPR178, which was obtained by cloning the HindIII–XhoI-digested PCR fragment ANT1-His6 (primer pair RE389/390) into pYES2 (Invitrogen, Groningen, The Netherlands).
Biochemical analysis of peroxisomes
The preparation and high-salt extraction of peroxisomal membranes, reversed-phase HPLC separation of peroxisomal membrane proteins and their subsequent sequence analysis has been described previously (Erdmann and Blobel, 1995). Fractionation of yeast lysates and differential centrifugation at 25 000 g was performed according to Erdmann et al. (1989). Peroxisomes were separated from mitochondria in a vertical rotor (SV-288, Sorvall RCB5; DuPont, BadNauheim, Germany) on continuous 20–53% (w/w) sucrose gradients (run for 1.5 h at 48 000 g) or on discontinuous 50–25% (w/w) sucrose flotation gradients (run for 4 h at 48 000 g) as described previously (Hettema et al., 2000).
Antibodies and immunoblot analysis
A fragment of Ant1p comprising amino acids 90–215 was expressed as a His6-tagged fusion protein in E.coli BL21(DE3) (Novagen) and purified under denaturing conditions from inclusion bodies. For the generation of polyclonal antibodies, the isolated protein was used to immunize rabbits (Eurogentech, Serain, Belgium). Monoclonal anti-yeast 3-phosphoglycerate kinase (Pgk1p) antibodies were purchased from Molecular Probes (Eugene, OR, USA). Antibodies against Aac2p, Fox3p, Fat2p/Pcs60p, Kar2p, Pex3p and Pex11p have been described previously (Rose et al., 1989; Erdmann and Blobel, 1995; Blobel and Erdmann, 1996). Immunoreactive complexes were visualized using anti-rabbit or anti-mouse IgG-coupled horseradish peroxidase in combination with the ECL system from Amersham Pharmacia Biotech (Uppsala, Sweden).
Purification and mass spectrometric analysis of Ant1p-His6
Organelles from YPH499-pHPR178 lysates were harvested by centrifugation according to standard procedures (Daum et al., 1982) and solubilized in buffer A (500 mM NaCl, 10 mM PIPES pH 7) containing 1% Triton X-100 (w/v) and 0.1 mM phenylmethylsulfonyl fluoride (PMSF), at a final concentration of 0.8 mg protein/ml. After incubation for 20 min at 4°C, the mixture was spun at 138 000 g for 20 min. The supernatant (0.6 ml) was incubated batchwise for 30 min at 4°C with 0.2 ml Ni-NTA–agarose (Qiagen, Hilden, Germany) that had previously been equilibrated with buffer A. The resin was packed into a column (0.5 cm internal diameter) and washed with 2 ml of the following buffers: B, 500 mM NaCl, 10 mM imidazole, 1% Triton X-100, 5% glycerol, cardiolipin (0.25 mg/ml), 10 mM PIPES pH 7; C, 300 mM NaCl, 10 mM imidazole, 0.8% Triton X-100, 0.1% Triton X-114, 3% glycerol, cardiolipin (0.25 mg/ml), 10 mM PIPES pH 7; D, 100 mM NaCl, 10 mM imidazole, 0.4% Triton X-100, 0.2% Triton X-114, 1.5% glycerol, cardiolipin (0.25 mg/ml), 10 mM PIPES pH 7.5; E, 50 mM NaCl, 10 mM imidazole, 0.1% Triton X-100, 0.4% Triton X-114, glycerol 0.1%, cardiolipin (0.25 mg/ml), 10 mM PIPES pH 7; F, 25 mM NaCl, 20 mM imidazole, 0.4% Triton X-114, cardiolipin (0.25 mg/ml), 10 mM PIPES pH 7. Finally, pure Ant1p was eluted with a buffer containing 25 mM NaCl, 100 mM imidazole, 0.4% Triton X-114, cardiolipin (4 mg/ml) and 10 mM PIPES pH 7. The yield of purified protein was determined by laser densitometry as described previously (Palmieri et al., 1996). Proteins were analyzed by SDS–PAGE in 17.5% gels and either stained or transferred to nitrocellulose membranes for western blotting. The band containing purified Ant1p was excised from a gel stained with Coomassie Blue dye and subsequently digested in-gel with trypsin as described previously (Shevchenko et al., 1996). Digest supernatant (1 µl) was used to obtain a peptide mass fingerprint by matrix-assisted laser desorption– ionization mass spectrometry (MALDI-MS) using a fast evaporation/nitrocellulose matrix.
Reconstitution of Ant1p into liposomes
Purified Ant1p was reconstituted by cyclic removal of the detergent with a hydrophobic column (Palmieri et al., 1995). Three hundred microliters of purified protein (about 1 µg of protein) were mixed with 100 µl of 10% egg-yolk phospholipids (Fluka) in the form of sonicated liposomes, 0.8 mg of cardiolipin, 90 µl of 10% Triton X-114, 20 mM ATP (except where otherwise indicated), 10 mM PIPES pH 7 and water to a final volume of 700 µl. This mixture was recycled 13 times through an Amberlite column (Superchrom, Milan, Italy) (3.4 × 0.5 cm) pre-equilibrated with a buffer containing 10 mM PIPES pH 7 and the substrate at the same concentration as in the starting mixture. All operations were performed at 4°C, except the passages through Amberlite, which were carried out at room temperature.
Transport measurements
External substrate was removed from proteoliposomes on a Sephadex G-75 column pre-equilibrated with buffer G (50 mM NaCl and 10 mM PIPES pH 7). Transport at 25°C was started by adding 50 µM [14C]ATP to the proteoliposomes, and terminated by addition of 30 mM pyridoxal 5′-phosphate and 10 mM bathophenanthroline [the ‘inhibitor-stop’ method (Palmieri et al., 1995)]. In controls, the inhibitors were added with the labeled substrate. The external radioactivity was removed on Sephadex G-75 and the internal radioactivity was measured. The transport activity was the difference between experimental and control values. The initial rate of transport was calculated in mmol/min/g protein from the time course of isotope equilibration (Palmieri et al., 1995). Various other transport activities were also assayed by the inhibitor-stop method.
Miscellaneous
Catalase (EC 1.11.1.6) and fumarate hydratase (fumarase, EC 4.2.1.2) activity measurements and GFP fluorescence microscopy of live cells were performed according to published protocols (Veenhuis et al., 1987; Westermann and Neupert, 2000).
Acknowledgments
Acknowledgements
We thank Joel Goodman, Ed Hurt and Wolf-Hubert Kunau for plasmids and antibodies. Our gratitude goes to Mathias Dreger for the MALDI-MS analysis, and to Xinji Hong for assistance in the lauric acid growth assays. This work was supported by the Deutsche Forschungsgemeinschaft, grant ER178/2-3, and by the Fonds der Deutschen Chem. Industrie (to R.E.) and by MURST-PRIN, MURST L.488/92 CO3 and CO4, MURST-CNR L.95/95, CEGBA, CNR target project on Biotechnology and the European Social Fund (to F.P.). H.R. was supported by a long-term EMBO fellowship (ALTF-255-2000).
References
- Blobel F. and Erdmann,R. (1996) Identification of a yeast peroxisomal member of the family of AMP-binding proteins. Eur. J. Biochem., 240, 468–476. [DOI] [PubMed] [Google Scholar]
- Dansen T.B., Wirtz,K.W., Wanders,R.J. and Pap,E.H. (2000) Peroxi somes in human fibroblasts have a basic pH. Nature Cell Biol., 2, 51–53. [DOI] [PubMed] [Google Scholar]
- Daum G., Gasser,S.M. and Schatz,G. (1982) Import of proteins into mitochondria. Energy-dependent, two-step processing of the intermembrane space enzyme cytochrome b2 by isolated yeast mitochondria. J. Biol. Chem., 257, 13075–13080. [PubMed] [Google Scholar]
- Dolce V., Fiermonte,G., Runswick,M.J., Palmieri,F. and Walker,J.E. (2001) The human mitochondrial deoxynucleotide carrier and its role in the toxicity of nucleoside antivirals. Proc. Natl Acad. Sci. USA, 98, 2284–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann R. and Blobel,G. (1995) Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J. Cell Biol., 128, 509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann R. and Blobel,G. (1996) Identification of Pex13p a peroxisomal membrane receptor for the PTS1 recognition factor. J. Cell Biol., 135, 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann R., Veenhuis,M., Mertens,D. and Kunau,W.H. (1989) Isolation of peroxisome-deficient mutants of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 86, 5419–5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraghty M.T., Bassett,D., Morrell,J.C., Gatto,G.J.,Jr, Bai,J., Geisbrecht,B.V., Hieter,P. and Gould,S.J. (1999) Detecting patterns of protein distribution and gene expression in silico. Proc. Natl Acad. Sci. USA, 96, 2937–2942. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gietz R.D. and Sugino,A. (1988) New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene, 74, 527–534. [DOI] [PubMed] [Google Scholar]
- Götte K., Girzalsky,W., Linkert,M., Baumgart,E., Kammerer,S., Kunau,W.H. and Erdmann,R. (1998) Pex19p, a farnesylated protein essential for peroxisome biogenesis. Mol. Cell. Biol., 18, 616–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S.J. and Valle,D. (2000) Peroxisome biogenesis disorders: genetics and cell biology. Trends Genet., 16, 340–345. [DOI] [PubMed] [Google Scholar]
- Güldener U., Heck,S., Fielder,T., Beinhauer,J. and Hegemann,J.H. (1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res., 24, 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halestrap A.P. (1975) The mitochondrial pyruvate carrier. Kinetics and specificity for substrates and inhibitors. Biochem. J., 148, 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke B., Girzalsky,W., Berteaux-Lecellier,V. and Erdmann,R. (1998) IDP3 encodes a peroxisomal NADP-dependent isocitrate dehydro genase required for the β-oxidation of unsaturated fatty acids. J. Biol. Chem., 273, 3702–3711. [DOI] [PubMed] [Google Scholar]
- Hettema E.H. and Tabak,H.F. (2000) Transport of fatty acids and metabolites across the peroxisomal membrane. Biochim. Biophys. Acta, 1486, 18–27. [DOI] [PubMed] [Google Scholar]
- Hettema E.H., van Roermund,C.W., Distel,B., van den Berg,M., Vilela,C., Rodrigues-Pousada,C., Wanders,R.J. and Tabak,H.F. (1996) The ABC transporter proteins Pat1 and Pat2 are required for import of long-chain fatty acids into peroxisomes of Saccharomyces cerevisiae. EMBO J., 15, 3813–3822. [PMC free article] [PubMed] [Google Scholar]
- Hettema E.H., Girzalsky,W., van Den Berg,M., Erdmann,R. and Distel,B. (2000) Saccharomyces cerevisiae Pex3p and Pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J., 19, 223–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhfeld J., Veenhuis,M. and Kunau,W.H. (1991) PAS3, a Saccharomyces cerevisiae gene encoding a peroxisomal integral membrane protein essential for peroxisome biogenesis. J. Cell Biol., 114, 1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jank B., Habermann,B., Schweyen,R.J. and Link,T.A. (1993) PMP47, a peroxisomal homologue of mitochondrial solute carrier proteins. Trends Biochem. Sci., 18, 427–428. [PubMed] [Google Scholar]
- Jones J.S. and Prakash,L. (1990) Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast, 6, 363–366. [DOI] [PubMed] [Google Scholar]
- Klingenberg M. (1979) The ADP, ATP shuttle of the mitochondrion. Trends Biochem. Sci., 4, 249–252. [Google Scholar]
- Lazarow P.B. and Moser,H.W. (1995) Disorders in peroxisome biogenesis. In Scriver,C.R., Beaudet,C.R., Sly,W.S. and Valle,D. (eds), The Metabolic and Molecular Bases of Inherited Disease, Vol. 2. McGraw-Hill Book Co., New York, NY, pp. 2287–2324.
- Lemmens M., Verheyden,K., Van Veldhoven,P., Vereecke,J., Mannaerts,G.P. and Carmeliet,E. (1989) Single-channel analysis of a large conductance channel in peroxisomes from rat liver. Biochim. Biophys. Acta, 984, 351–359. [DOI] [PubMed] [Google Scholar]
- McCammon M.T., Dowds,C.A., Orth,K., Moomaw,C.R., Slaughter,C.A. and Goodman,J.M. (1990) Sorting of peroxisomal membrane protein PMP47 from Candida boidinii into peroxisomal membranes of Saccharomyces cerevisiae. J. Biol. Chem., 265, 20098–20105. [PubMed] [Google Scholar]
- Nakagawa T., Imanaka,T., Morita,M., Ishiguro,K., Yurimoto,H., Yamashita,A., Kato,N. and Sakai,Y. (2000) Peroxisomal membrane protein Pmp47 is essential in the metabolism of middle-chain fatty acid in yeast peroxisomes and Is associated with peroxisome proliferation. J. Biol. Chem., 275, 3455–3461. [DOI] [PubMed] [Google Scholar]
- Nelson D.R., Felix,C.M. and Swanson,J.M. (1998) Highly conserved charge-pair networks in the mitochondrial carrier family. J. Mol. Biol., 277, 285–308. [DOI] [PubMed] [Google Scholar]
- Olivier L.M. and Krisans,S.K. (2000) Peroxisomal protein targeting and identification of peroxisomal targeting signals in cholesterol biosynthetic enzymes. Biochim. Biophys. Acta, 1529, 89–102. [DOI] [PubMed] [Google Scholar]
- Palmieri F. (1994) Mitochondrial carrier proteins. FEBS Lett., 346, 48–54. [DOI] [PubMed] [Google Scholar]
- Palmieri F., Indiveri,C., Bisaccia,F. and Iacobazzi,V. (1995) Mito chondrial metabolite carrier proteins: purification, reconstitution, and transport studies. Methods Enzymol., 260, 349–369. [DOI] [PubMed] [Google Scholar]
- Palmieri L., Palmieri,F., Runswick,M.J. and Walker,J.E. (1996) Identification by bacterial expression and functional reconstitution of the yeast genomic sequence encoding the mitochondrial dicarboxylate carrier protein. FEBS Lett., 399, 299–302. [DOI] [PubMed] [Google Scholar]
- Palmieri L., Lasorsa,F.M., Iacobazzi,V., Runswick,M.J., Palmieri,F. and Walker,J.E. (1999) Identification of the mitochondrial carnitine carrier in Saccharomyces cerevisiae. FEBS Lett., 462, 472–476. [DOI] [PubMed] [Google Scholar]
- Palmieri L., Agrimi,G., Runswick,M.J., Fearnley,I.M., Palmieri,F. and Walker,J.E. (2001) Identification in Saccharomyces cerevisiae of two isoforms of a novel mitochondrial transporter for 2-oxoadipate and 2-oxoglutarate. J. Biol. Chem., 276, 1916–1922. [DOI] [PubMed] [Google Scholar]
- Reumann S. (2000) The structural properties of plant peroxisomes and their metabolic significance. Biol. Chem., 381, 639–648. [DOI] [PubMed] [Google Scholar]
- Rose M.D., Misra,L.M. and Vogel,J.P. (1989) KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell, 57, 1211–1221. [DOI] [PubMed] [Google Scholar]
- Rottensteiner H., Kal,A.J., Filipits,M., Binder,M., Hamilton,B., Tabak,H.F. and Ruis,H. (1996) Pip2p: a transcriptional regulator of peroxisome proliferation in the yeast Saccharomyces cerevisiae. EMBO J., 15, 2924–2934. [PMC free article] [PubMed] [Google Scholar]
- Sacksteder K.A., Jones,J.M., South,S.T., Li,X., Liu,Y. and Gould,S.J. (2000) PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J. Cell Biol., 148, 931–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y., Saiganji,A., Yurimoto,H., Takabe,K., Saiki,H. and Kato,N. (1996) The absence of Pmp47, a putative yeast peroxisomal transporter, causes a defect in transport and folding of a specific matrix enzyme. J. Cell Biol., 134, 37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schiestl R.H. and Gietz,R.D. (1989) High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet., 16, 339–346. [DOI] [PubMed] [Google Scholar]
- Shani N. and Valle,D. (1996) A Saccharomyces cerevisiae homolog of the human adrenoleukodystrophy transporter is a heterodimer of two half ATP-binding cassette transporters. Proc. Natl Acad. Sci. USA, 93, 11901–11906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani N., Jimenez-Sanchez,G., Steel,G., Dean,M. and Valle,D. (1997) Identification of a fourth half ABC transporter in the human peroxisomal membrane. Hum. Mol. Genet., 6, 1925–1931. [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm,M., Vorm,O. and Mann,M. (1996) Mass spectrometric sequencing of proteins from silver-stained poly acrylamide gels. Anal. Chem., 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg S.J., Wang,S.J., Kim,D.G., Mihalik,S.J. and Watkins,P.A. (1999) Human very-long-chain acyl-CoA synthetase: cloning, topography, and relevance to branched-chain fatty acid metabolism. Biochem. Biophys. Res. Commun., 257, 615–621. [DOI] [PubMed] [Google Scholar]
- van den Bosch H., Schutgens,R.B., Wanders,R.J. and Tager,J.M. (1992) Biochemistry of peroxisomes. Annu. Rev. Biochem., 61, 157–197. [DOI] [PubMed] [Google Scholar]
- van Roermund C.W., Elgersma,Y., Singh,N., Wanders,R.J. and Tabak,H.F. (1995) The membrane of peroxisomes in Saccharomyces cerevisiae is impermeable to NAD(H) and acetyl-CoA under in vivo conditions. EMBO J., 14, 3480–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roermund C.W., Tabak,H.F., van Den Berg,M., Wanders,R.J. and Hettema,E.H. (2000) Pex11p plays a primary role in medium-chain fatty acid oxidation, a process that affects peroxisome number and size in Saccharomyces cerevisiae. J. Cell Biol., 150, 489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Veldhoven P.P., Just,W.W. and Mannaerts,G.P. (1987) Permeability of the peroxisomal membrane to cofactors of beta-oxidation. Evidence for the presence of a pore-forming protein. J. Biol. Chem., 262, 4310–4318. [PubMed] [Google Scholar]
- Veenhuis M., Mateblowski,M., Kunau,W.H. and Harder,W. (1987) Proliferation of microbodies in Saccharomyces cerevisiae. Yeast, 3, 77–84. [DOI] [PubMed] [Google Scholar]
- Verleur N., Hettema,E.H., van Roermund,C.W., Tabak,H.F. and Wanders,R.J. (1997) Transport of activated fatty acids by the peroxisomal ATP-binding-cassette transporter Pxa2 in a semi-intact yeast cell system. Eur. J. Biochem., 249, 657–661. [DOI] [PubMed] [Google Scholar]
- Walker J.E. (1992) The mitochondrial transporter family. Curr. Opin. Struct. Biol., 2, 519–526. [Google Scholar]
- Watkins P.A., Lu,J.F., Steinberg,S.J., Gould,S.J., Smith,K.D. and Braiterman,L.T. (1998) Disruption of the Saccharomyces cerevisiae FAT1 gene decreases very long-chain fatty acyl-CoA synthetase activity and elevates intracellular very long-chain fatty acid concentrations. J. Biol. Chem., 273, 18210–18219. [DOI] [PubMed] [Google Scholar]
- Weber F.E., Minestrini,G., Dyer,J.H., Werder,M., Boffelli,D., Compassi,S., Wehrli,E., Thomas,R.M., Schulthess,G. et al. (1997) Molecular cloning of a peroxisomal Ca2+-dependent member of the mitochondrial carrier superfamily. Proc. Natl Acad. Sci. USA, 94, 8509–8514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B. and Neupert,W. (2000) Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast, 16, 1421–1427. [DOI] [PubMed] [Google Scholar]
- Winston F., Dollard,C. and Ricupero-Hovasse,S.L. (1995) Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast, 11, 53–55. [DOI] [PubMed] [Google Scholar]
- Wylin T., Baes,M., Brees,C., Mannaerts,G.P., Fransen,M. and Van Veldhoven,P.P. (1998) Identification and characterization of human PMP34, a protein closely related to the peroxisomal integral membrane protein PMP47 of Candida boidinii. Eur. J. Biochem., 258, 332–338. [DOI] [PubMed] [Google Scholar]