Abstract
Nucleolar localization of box C/D small nucleolar (sno) RNAs requires the box C/D motif and, in vertebrates, involves transit through Cajal bodies (CB). We report that in yeast, overexpression of a box C/D reporter leads to a block in the localization pathway with snoRNA accumulation in a specific sub-nucleolar structure, the nucleolar body (NB). The human survival of motor neuron protein (SMN), a marker of gems/CB, specifically localizes to the NB when expressed in yeast, supporting similarities between these structures. Box C/D snoRNA accumulation in the NB was decreased by mutation of Srp40 and increased by mutation of Nsr1p, two related nucleolar proteins that are homologous to human Nopp140 and nucleolin, respectively. Box C/D snoRNAs also failed to accumulate in the NB, and became delocalized to the nucleoplasm, upon depletion of any of the core snoRNP proteins, Nop1p/fibrillarin, Snu13p, Nop56p and Nop5p/Nop58p. We conclude that snoRNP assembly occurs either in the nucleoplasm, or during transit of snoRNAs through the NB, followed by routing of the complete snoRNP to functional sites of ribosome synthesis.
Keywords: Cajal body/nucleolus/snoRNA/trafficking
Introduction
The nucleus is a highly organized cellular compartment and >10 structures have been defined by electron or light microscopy in higher eukaryotes (reviewed in Matera, 1999). The nucleus of lower eukaryotes like yeasts appears less complex, and it is unclear whether the domains observed in higher eukaryotes are absent, perhaps being functionally replaced by other structures, or whether they have simply not been identified yet. While the role of the nuclear structures is not always known precisely, they are thought to perform specialized functions. The most conspicuous nuclear domain is the nucleolus, which is found in all eukaryotic cells and is the major site of ribosome synthesis. Another organelle that has been the focus of recent attention is the Cajal or coiled body (CB; Gall, 2000). CBs are found in vertebrates, invertebrates (Yannoni and White, 1997) and plants (Beven et al., 1995), but have not previously been identified in yeast. While their exact function is unknown, CBs are frequently located close to the nucleolus (Bohmann, 1995; see Matera, 1999 for review) and are likely to be involved in the biogenesis of small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs; Bohmann, 1995; Samarsky et al., 1998; Matera, 1999; Narayanan et al., 1999b).
A major role of the nucleus is the synthesis of many types of RNA, all of which undergo nuclear processing, followed by transport to either the nuclear pore complex (NPC) for cytoplasmic export, or to specific sub-nuclear locations. Indeed, several post-transcriptional processing reactions require RNA factors, notably snRNAs and snoRNAs, which must be localized correctly. In addition to this initial post-transcriptional targeting event, recent experiments have suggested that the steady-state localization of these ribonucleoprotein particles (RNPs) is highly dynamic, and involves a constant and rapid flux of molecules in and out of the compartment in which they concentrate (Phair and Misteli, 2000). Despite their fundamental importance, the mechanisms responsible for targeting RNA to specific destinations within the nucleus are poorly understood.
Most of our knowledge of the mechanisms in intra-nuclear RNA movement stems from analyses of nuclear mRNAs. Analyses of the movement of either bulk polyadenylated RNAs or of specific mRNAs suggest that they move through the nucleoplasm as single molecules by simple diffusion (Femino et al., 1998; Politz and Pederson, 2000). Moreover, pre-mRNAs are believed to assemble into RNP particles and undergo processing—capping, splicing and polyadenylation—at the transcription sites (Zhang et al., 1994; Custodio et al., 1999). During pre-mRNA splicing, export factors of the REF family bind to the matured molecule (Zhou et al., 2000). These recruit TAP, which allows docking of the complex onto the NPCs and subsequent transport to the cytoplasm (see Cullen, 2000 for a review). These and other results suggest a simple general model, in which RNAs assemble into RNP structures and are processed close to the site of transcription. During assembly and processing, the RNAs associate with localization factors that can anchor the RNP to its target compartment, which is reached by diffusion.
A complication to this simple model is that RNAs do not always move directly from their transcription site to their final destinations. Newly synthesized snRNAs are proposed to traffic through the CBs prior to association with the NPCs and nuclear export (Matera, 1999; Smith and Lawrence, 2000). Following re-import, the snRNAs again co-localize with CBs and with nucleoli before being detected in the speckles, their final destination (Sleeman and Lamond, 1999). Other nuclear and cytoplasmic RNAs, including pre-tRNAs, the RNA component of RNase P, signal recognition particle (SRP) and telomerase, transiently localize to the nucleolus before reaching their final destinations (Bertrand et al., 1998; Pederson, 1998; Narayanan et al., 1999a). The transient association of these RNPs with nuclear substructures most likely reflects the localization of specific steps in their biogenesis (Pederson, 1998).
In order to understand the mechanisms of intra-nuclear RNA transport better, we are studying the nucleolar targeting of snoRNAs. Most snoRNAs select sites of rRNA modification by base pairing to the pre-rRNAs. Based on conserved sequence elements, the snoRNAs are grouped into two families, termed box C/D and box H/ACA, which direct rRNA 2′-O-methylation or pseudouridine formation, respectively (Bachellerie and Cavaillé, 1997; Tollervey and Kiss, 1997). The box C/D motifs associate with four common snoRNP proteins: Nop1p/fibrillarin, Nop56p, Nop5p/Nop58p and Snu13p (Tyc and Steitz, 1989; Wu et al., 1998; Lafontaine and Tollervey, 1999, 2000; Watkins et al., 2000).
Previous studies have shown that the box C/D motif is both necessary and sufficient for snoRNA processing, stability and nucleolar localization in vertebrates and yeast (Lange et al., 1998; Samarsky et al., 1998; Narayanan et al., 1999a and references therein). Additionally, the box C/D motif localizes the snoRNAs to CBs (Samarsky et al., 1998), and this is probably an intermediate step on their way to the nucleolus (Narayanan et al., 1999b). In the present study, we took advantage of the yeast model system to carry out a genetic analysis of the roles of the snoRNP proteins in localizing the box C/D snoRNAs to the nucleolus. We report that correct localization requires all four core box C/D proteins, as well as additional nucleolar factors. We further show that the nucleolar transport pathway involves a novel sub-nuclear structure, the nucleolar body (NB), which shares similarities with CBs.
Results
Overexpression of artificial box C/D snoRNAs induces a specific block in the nucleolar localization pathway
The characterization of the various pathways for nuclear RNA export has greatly benefited from competition experiments, in which large amounts of a particular RNA are introduced into cells in order to specifically saturate and block a given pathway. To determine whether box C/D snoRNA localization is also a saturable process dependent on trans-acting factors, and to analyse at which step localization would be blocked, we overexpressed an artificial snoRNA in yeast. U14/MS2x2 contained two binding sites for the phage MS2 coat protein, flanked by the box C/D motif of yeast U14 snoRNA. Because the box C/D motif is both necessary and sufficient for RNA processing and nucleolar targeting, all other sequences of U14 were removed (see Supplementary data, available at The EMBO Journal Online). To stabilize this snoRNA further, the terminal stem of the box C/D motif was extended from 9 to 15 or 22 bp (Huang et al., 1992). U14/MS2x2 was cloned within the actin intron on a multicopy plasmid, and transcription was driven by a strong constitutive promoter (Figure 1A). As controls, we used the same RNA expressed from a centromeric vector, and a similar intronic RNA lacking boxes C and D. Northern blot analysis revealed that the artificial snoRNA was processed and accumulated as expected (see Figure 3C). A minor band of slower mobility, corresponding to unspliced precursors, was also detected. The RNA 5′ end was determined by primer extension (see Supplementary data). The major 5′ end corresponded to the authentic U14 site, but minor forms extended by few nucleotides were also detected, most likely because the long terminal stem slowed final trimming. We have also tested the functionality of U14/MS2x2 as a methylation guide, and found that this snoRNA was indeed capable of efficiently methylating a target RNA at the expected site (see Supplementary data).
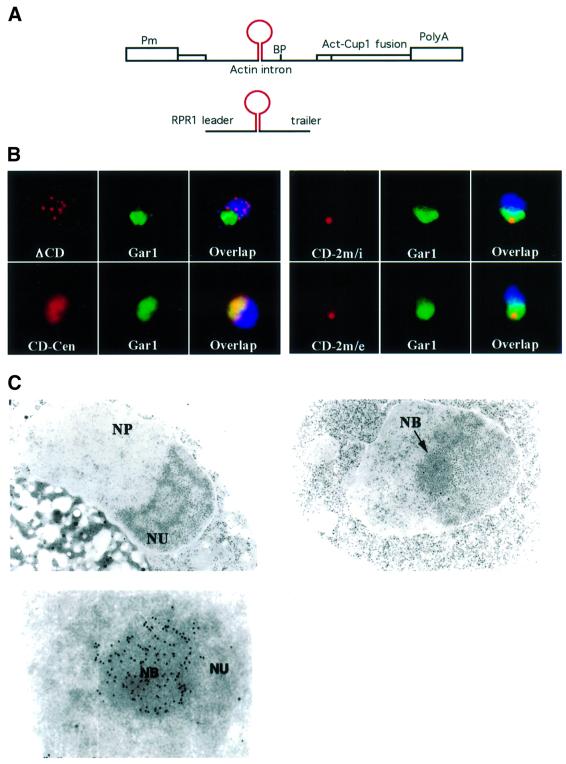
Fig. 1. Overexpressed artificial box C/D snoRNA accumulates in the NB. (A) Schematic of the expression vectors used. The snoRNA is represented by a stem–loop in red. Pm, GPD promoter; BP, branch point of the actin intron; RPR1 leader and trailer, promoter and terminator of the RNase P gene. (B) Localization of the snoRNA reporters by light microscopy. Each field is 5 × 5 µm. The snoRNA (red) was detected by fluorescent in situ hybridization, the nucleolus was labelled with a Gar1–GFP fusion protein (green) and the DNA was stained with DAPI (blue). The artificial snoRNA was expressed from either the RNase P gene (RPR1) on a 2 µ plasmid (CD-2m/e), or the actin intron on a 2 µ plasmid (CD-2m/i), or on a centromeric vector (CD-Cen). A similar RNA lacking boxes C and D was expressed from the actin intron on a 2 µ plasmid, and used as a control (ΔCD). (C) Localization of the snoRNA reporter by electron microscopy. The nucleolus (NU) of different yeast strains (upper left panel: CD-Cen; upper right panel: CD-2m/i) was composed of a fibrillar and a granular component. Upon snoRNA overexpression (CD-2m/i), an additional structure of spherical shape was obvious in the nucleolus (the NB). This fibrillar body exhibited strong immunogold labelling after in situ hybridization using a BrdU-labelled MS2 oligonucleotide probe (lower panel). NP, nucleoplasm.
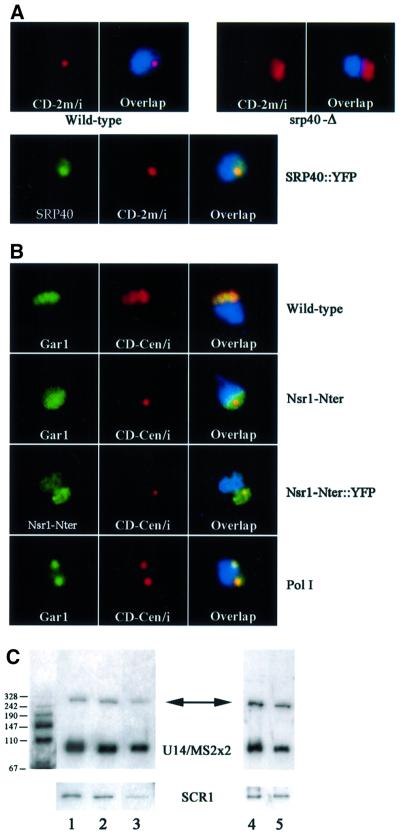
Fig. 3. Antagonistic effects of Srp40p and Nsr1p on snoRNA trafficking. (A) Srp40p is required to accumulate box C/D snoRNA in the NB. The multicopy artificial snoRNA (red) was detected in either wild-type cells (Wild-type), or in isogenic cells deficient in SRP40p (srp40-Δ). SRP40p was fused to YFP (green) and detected in the NB (SRP40::YFP). (B) Nsr1p is required to distribute box C/D snoRNA in the entire nucleolus. The centromeric artificial snoRNA (red) was detected in either wild-type cells (Wild-type), in isogenic cells expressing the N-terminal domain of NSR1p (Nsr1-Nter), or following inactivation of RNA polymerase I (Pol I). The nucleolus was visualized with a Gar1–GFP fusion protein (Gar1, green), and the localization of the N-terminal domain of Nsr1p was visualized through a fusion with YFP (Nsr1-Nter::YFP, green). (C) SnoRNA expression levels in Srp40p and Nsr1p-deficient strains. Northern blots were probed with sequences specific for the artificial snoRNA (upper panels), or for SCR1 RNA for normalization (lower panels). The band corresponding to the mature snoRNA is indicated (91 bases, U14/MS2x2) and an arrow points to the precursor (∼250 bases). The molecular weight markers are shown on the left. Nsr1 (left panel) and Srp40 (right panel) strains were expressing U14/MS2x2 from a centromeric or 2 µ plasmid, respectively. Lane 1, wild-type cells; lane 2, Nsr1-Nter; lane 3, Nsr1-Δ; lane 4, Srp40-Δ; lane 5, wild type.
Localization of the snoRNA was determined by fluorescent in situ hybridization with an oligo probe specific for the MS2 sequences, while the nucleolus was visualized with a functional Gar1p–GFP (green fluorescent protein) fusion (Trumtel et al., 2000; Figure 1B). The artificial snoRNA expressed from the centromeric vector localized correctly to the nucleolus (Figure 1B, CD-Cen). In contrast, the intronic RNA lacking boxes C and D was seen in dispersed foci (3–10) in the nucleoplasm (Figure 1B, ΔCD). These are likely to be sites of transcription and processing, as the use of a chromosomal reporter reduced the number of foci to one (data not shown). The artificial snoRNA expressed from the multicopy vector was also localized to the nucleolus, but surprisingly was strongly concentrated in a single sub-nucleolar region (Figure 1B, CD-2m/i). A similar concentration was seen with an artificial snoRNA expressed from a highly active RNA polymerase III promoter (Figure 1A and B, CD-2m/e; Good and Engelke, 1994). Exonucleases can gain direct access to the termini of this transcript, eliminating the requirement for pre-mRNA splicing to produce mature snoRNA molecules. Thus, sub-nucleolar concentration did not depend on a particular snoRNA maturation pathway, but was dependent on snoRNA expression levels.
The sub-nucleolar localization of the overexpressed box C/D snoRNAs was further analysed by electron microscopy (Figure 1C, upper right panel). The snoRNA-overexpressing cells showed normal overall morphology, with dense fibrillar and granular components, but contained an additional structure, which we called the NB (Figure 1C). This body was spherical, and in most cells was present at a single copy per nucleus. Notably, the NB was contiguous with the dense fibrillar compartment, in which the snoRNAs are normally located. To demonstrate that the NB corresponded to the site of snoRNA concentration observed in fluorescence microscopy, in situ hybridization was performed at the ultrastructural level (Figure 1C, lower panel). Strong enrichment of the immunogold label was seen in the NB, showing it to be the site of snoRNA accumulation.
We conclude that overexpression of the artificial snoRNA blocked a late step in the localization pathway. Rather than becoming dispersed in the dense fibrillar region of the nucleolus, the snoRNAs were restricted to the NB.
The NB is involved in the trafficking of endogenous box C/D snoRNA
To determine whether transport of endogenous snoRNAs also involves transit through the NB, we utilized in situ hybridization to examine the localization of the box C/D snoRNAs U14 and U3 in the presence of the artificial snoRNA (Figure 2A). U14 and U3 localized throughout the nucleolus in wild-type cells, but became concentrated in the NB in cells overexpressing U14/MS2x2 (Figure 2A, U3 and U14). U14 largely co-localized with the artificial snoRNA reporter whereas a substantial pool of the U3 snoRNA was visible outside the NB, reflecting differential sensitivity to retention in the NB. The common C/D snoRNP proteins Nop1p and Nop5/Nop58p also became highly enriched in the NB (shown for Nop1p in Figure 2A), indicating that many or all box C/D snoRNAs are co-localized. Retention in the NB appeared to be specific for box C/D snoRNAs, as localization of a box H/ACA snoRNA, snR10, and an associated protein, Gar1p, was unaffected (data not shown and Figure 1B, right panels).
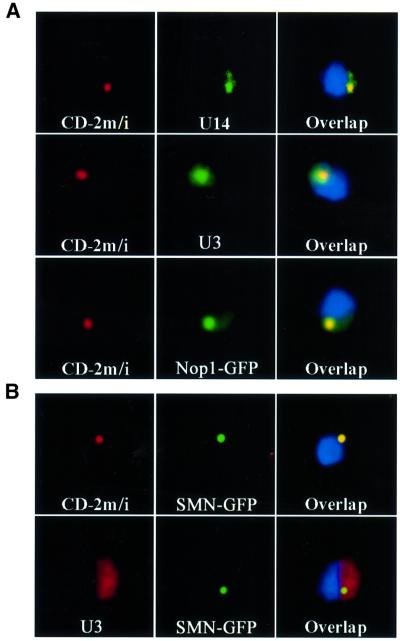
Fig. 2. The NB is a structure hosting several box C/D snoRNAs, and is related to CB. (A) Overexpression of an artificial snoRNA leads to accumulation of endogenous snoRNA in the NB. Yeast cells overexpressing the multicopy, artificial snoRNA were simultaneously labelled for the artificial snoRNA (red), and for endogeneous U14 (green, U14), U3 (green, U3) or Nop1–GFP (green, Nop1). (B) Human SMN localizes in the NB. Human SMN (green) was fused to GFP, and expressed in either wild-type cells (U3), or in cells overexpressing the artificial snoRNA (CD-2m/i). The NB was detected with a probe against the artificial snoRNA (red, CD-2m/i), and the nucleolus with a probe against U3 (red, U3).
We conclude that an excess of the artificial snoRNA saturates the localization machinery, and that the NB represents an intermediate site in the localization of many, and possibly all, box C/D snoRNAs.
The NB shows similarities to the CB
In vertebrates, U3 snoRNA transits through the CB en route to the nucleolus (Narayanan et al., 1999b), suggesting possible similarities to the NB. To test this hypothesis, we expressed a CB marker in yeast. The archetypal human CB marker is p80 coilin, but database searches have failed to identify homologues in yeast, Drosophila or Caenorhabditis elegans. In contrast, another CB marker, the survival of motor neuron protein (SMN), has homologues in many eukaryotes, including the fission yeast Schizosaccharomyces pombe (Hannus et al., 2000), although there is no obvious homologue in Saccharomyces cerevisiae. SMN was originally reported to localize in nucleoplasmic structures called gems (gemini of CB; Liu and Dreyfuss, 1996), but gems co-localize with CB in the vast majority of cells (Carvalho et al., 1999). As the distinction, if any, is unclear, we will use the term ‘CB’ to refer to the gems/CB compartment. Human SMN was fused to GFP and the fusion protein was shown to co-localize with the endogeneous protein in mammalian cells (data not shown). To avoid artifacts due to overexpression in yeast cells, the fusion was expressed at low levels, from a centromeric vector, with transcription driven at the basal level of the inducible MET25 promoter. When co-expressed in yeast with the artificial snoRNA, SMN–GFP was seen as a small dot in the nucleus of living cells. Fixation and hybridization with the MS2 probe showed that SMN–GFP co-localized with the NB (Figure 2B, CD-2m). In cells that did not express the artificial snoRNA, SMN–GFP was still concentrated in a small nucleolar region, as shown by double labelling with a probe specific for U3 (Figure 2B, U3). This suggests that the NB is a CB-like domain, which exists even in the absence of snoRNA overexpression.
Srp40p and Nsr1p have antagonistic activities on the localization of box C/D snoRNAs to the NB
In order to identify the trans-acting factors required for box C/D snoRNA localization, we analysed the localization of U14/MS2x2 in several mutant strains. The mammalian Nopp140 protein associates with both box H/ACA and box C/D snoRNAs, and localizes in CBs and nucleoli (Yang et al., 2000). Strains lacking the yeast homologue of Nopp140, Srp40p, are viable but mildly impaired in growth (Meier, 1996). Deletion of the SRP40 gene did not affect the nucleolar localization of U14/MS2x2 expressed from a centromeric vector (data not shown). However, overexpression of U14/MS2x2 in the -Δ strain did not lead to accumulation in the NB (Figure 3A, srp40-Δ). Northern blot analysis showed similar levels of U14/MS2x2 in the srp40-Δ strain and the isogenic control (Figure 3C). Srp40p was fused to yellow fluorescent protein (YFP), and the fusion protein was shown to be present in the NB in strains overexpressing U14/MS2x2 (Figure 3A, SRP40::YFP). This suggested that Srp40p is required in the NB to retain box C/D snoRNAs.
Nopp140 is a member of a family of nucleolar proteins containing a serine-rich domain interspersed with basic clusters of amino acids, which also includes vertebrate nucleolin (Sicard et al., 1998). Strains lacking the putative yeast homologue of nucleolin, Nsr1p, are viable but impaired in growth and 40S ribosomal subunit synthesis (Lee et al., 1991; Lin et al., 1994). Two Nsr1p-deficient strains were tested for their effects on snoRNA localization. The nsr1-Δ strain lacks the gene entirely (Lin et al., 1994), while the Nsr1-Nter strain lacks the C-terminal region but retains the Srp40p-like serine-rich domain (Lee et al., 1991). In the nrs1-Δ strain expressing U14/MS2x2 snoRNA from a centromeric vector, ∼25% of the cells showed snoRNA accumulation in the NB, as compared with 10% in the control (data not shown). This phenotype was exacerbated by expression of the N-terminal fragment of Nsr1p, with 60% of cells accumulating U14/MS2x2 in the NB (Figure 3B, Nsr1-Nter). As judged by northern blot analysis, this effect was not due to an alteration of snoRNA levels (Figure 3C), and it appeared specific for box C/D snoRNAs as the localization of Gar1p–GFP was unaffected (Figure 3B, Nsr1-Nter).
We conclude that Nsr1p promotes the dispersal of box C/D snoRNAs from the NB to the nucleolus and that its N-terminal domain shows some dominant activity in snoRNA retention. This domain was fused to YFP, and we observed that it was slightly enriched in the NB, although it could also be detected throughout the nucleus (Figure 3B, Nsr1-Nter::YFP).
The core box C/D snoRNP proteins, Snu13p, Nop1p, Nop56p and Nop5/58p, are all required for the nucleolar localization of box C/D snoRNA
Box C/D snoRNAs have been shown to associate tightly with four core snoRNP proteins: Nop1p/fibrillarin, Nop56p, Nop5/58p and Snu13p (Wu et al., 1998; Lafontaine and Tollervey, 1999, 2000; Watkins et al., 2000). Furthermore, the box C/D motif is necessary and sufficient to promote assembly with all four proteins in vitro (Newman et al., 2000; Watkins et al., 2000). It was, therefore, likely that one or more of these proteins would play a role in box C/D snoRNA localization. To investigate this possibility, we constructed Gal-dependent alleles for each of the corresponding genes.
Endogenous U14 was concentrated in the nucleolus of wild-type cells, but was delocalized to the nucleoplasm following depletion of either Nop1p or Nop56p (Figure 4A). Depletion of Nop1p was more effective in delocalizing U14; following Nop1p depletion, U14 was evenly distributed between the nucleolus and the nucleoplasm, whereas it remained largely nucleolar following Nop56p depletion. To rule out the possibility that these effects could be an indirect consequence of a block in ribosome biogenesis, we analysed the localization of U14/MS2x2 in an RNA polymerase I mutant that stopped rRNA synthesis. In such mutants, inactivation of RNA polymerase I affects the nucleolar ultrastructure globally, and this leads to nucleolar segregation (Oakes et al., 1993; Trumtel et al., 2000). In our case, this resulted in the concentration of both Gar1p and the snoRNA reporter in two small areas of the nucleolus, but importantly without mislocalization of snoRNA in the nucleoplasm (Figure 3C).
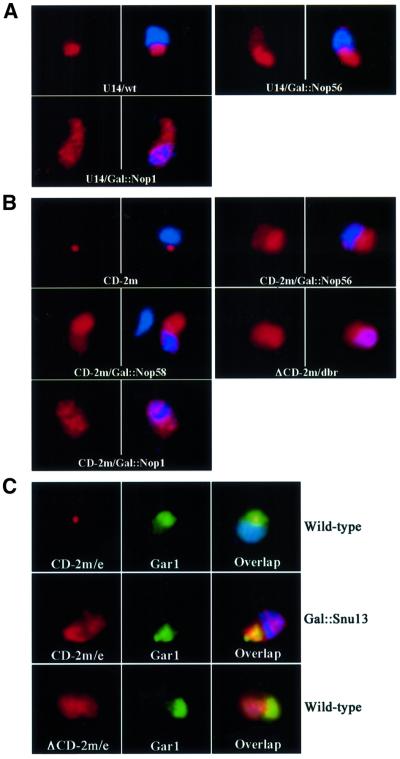
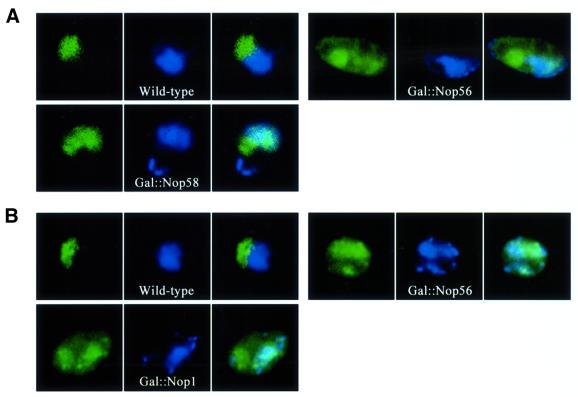
Fig. 4. Nop1p, Nop56p, Nop5/58p and Snu13p are all required for efficient localization of box C/D snoRNAs in the nucleolus. (A) Mislocalization of endogenous U14 in Nop1p- and Nop56p-depleted strains. U14 snoRNA (red) was detected in wild-type cells (U14/wt), or following depletion of Nop56p (U14/Gal::Nop56) or Nop1p (U14/Gal::Nop1). In the right image of each panel, the DNA has been stained with DAPI. The nucleolus is visible as the nuclear region that stains poorly with DAPI. (B) Mislocalization of the artificial snoRNA reporter in strains depleted for Nop1p, Nop56p or Nop58p. The multicopy, intronic, artificial snoRNA (red) was detected in wild-type cells (CD-2m), or following depletion of Nop5p/Nop58p (CD-2m/Gal::Nop58), Nop1p (CD-2m/Gal::Nop1) or Nop56p (CD-2m/Gal::Nop56). For comparison, a similar intronic RNA lacking boxes C and D was stabilized by inactivating the gene for debranching enzyme (ΔCD-2m/dbr). (C) Mislocalization of the artificial snoRNA in strains depleted for Snu13p. The artificial snoRNA (red, CD-2m/e) was expressed from the RPR1 gene on a 2 µ plasmid, and detected in wild-type strains (Wild-type), or following depletion of Snu13p (Gal::Snu13). A similar RNA lacking boxes C and D was used as a control (red, ΔCD-2m/e; Wild-type). The nucleolus was visualized with a Gar1–GFP fusion protein (Gar1, green).
The steady-state levels of endogenous box C/D snoRNAs are strongly affected on depletion of either Nop58p or Snu13p (Lafontaine and Tollervey, 1999; Watkins et al., 2000), precluding similar analysis for these proteins. Long terminal stems were, however, shown to bypass some requirements for snoRNA stability (Huang et al., 1992), and the terminal stem of U14/MS2x2 (15 or 22 bases) stabilized this RNA in the absence of Nop5p/Nop58p (Figure 4B and data not shown). In addition, expression of U14/MS2x2 from a multicopy plasmid allowed us to test whether the core box C/D proteins were required to localize the snoRNA to the NB before its dispersal throughout the nucleolus. Depletion of Nop1p, Nop5/Nop58p or Nop56p delocalized the snoRNA reporter to the nucleoplasm, and the effects were again more pronounced for Nop1p (Figure 4B). The NB was not visible after depletion of any of the proteins, indicating that each is required for snoRNA accumulation in this compartment.
Snu13p binds directly the box C/D motif, but also binds to the first stem–loop of U4 snRNA and is, therefore, essential for pre-mRNA splicing (Watkins et al., 2000). The intronic construct was, therefore, not suited for analysing Snu13p, and the snoRNA reporter was expressed from the RNA polymerase III promoter. The analysis of Snu13p is further complicated by the fact that depletion of the protein affects not only box C/D snoRNA accumulation, but also destabilizes box H/ACA snoRNAs and snRNAs at late time points (Watkins et al., 2000). However, when GAL::snu13 cells are shifted to glucose for only 6–8 h, the effects are specific for box C/D snoRNAs (Watkins et al., 2000). In wild-type cells, the overexpressed artificial snoRNA localized in the NB (Figure 4C, wild type). In contrast, 6 h following the glucose shift, ∼20% of the GAL::snu13 cells showed altered distribution of the snoRNA reporter: accumulation in the NB was lost and a large fraction of the snoRNA became delocalized in the nucleoplasm (Figure 4C, GAL::snu13). In the same cells, the localization of Gar1p–GFP was unaffected, indicating that the mislocalization was specific for the box C/D class of snoRNAs. The resulting snoRNA localization resembled that of the ΔCD RNA expressed from the RNase P promoter (Figure 4C, ΔCD), suggesting that the localization activity of the box C/D motif was inactivated.
A complete set of core snoRNP proteins was thus required for nucleolar accumulation of the box C/D snoRNAs, indicating that full assembly of the snoRNP is necessary for nucleolar targeting or retention. The loss of snoRNA accumulation in the NB following snoRNP protein depletion further suggests that they are required for snoRNA trafficking through this structure, prior to final localization within the nucleolus.
Nop1p and Nop5/Nop58p require other core box C/D snoRNP proteins to localize in the nucleolus
As the box C/D snoRNAs were not accumulated in the nucleolus in the absence of the intact snoRNP, we determined whether this was also the case for the snoRNP proteins. The localization of Nop1p was analysed in strains depleted for Nop56p or Nop5p/Nop58p. Immunofluorescence showed that in these cases, Nop1p became delocalized to the nucleoplasm, and even to the cytoplasm in some cells (Figure 5A). In a similar way, we analysed the localization of Nop5p/Nop58p, and we observed that it became delocalized following depletion of either Nop1p or Nop56p (Figure 5B). These results showed that Nop1p and Nop5p/Nop58p do not accumulate in the nucleolus in the absence of the other components of the snoRNP, suggesting that only the proteins assembled within an intact snoRNP can accumulate into the nucleolus.
Fig. 5. Nucleolar localization of Nop1p and Nop5/Nop58p requires other box C/D core proteins. (A) Nop1p is mislocalized in strains depleted for Nop5p/Nop58p or Nop56p. Nop1p (green) was detected by immunofluorescence in wild-type cells (Wild-type), or following depletion of Nop5p/Nop58p (Gal::Nop58) and Nop56p (Gal::Nop56). (B) Nop5p/Nop58p is mislocalized in strains depleted for Nop1p or Nop56p. Nop5/Nop58p (green) was detected by immunofluorescence in wild-type cells (Wild-type), or following depletion of either Nop1p (Gal::Nop1) or Nop56p (Gal::Nop56).
Discussion
An intact snoRNP is required for nucleolar localization of box C/D snoRNAs
It was shown previously that the box C/D motif is necessary and sufficient to localize an artificial snoRNA to the yeast nucleolus (Samarsky et al., 1998), but the role of snoRNP proteins in this process was not known. The box C/D motif is bound directly by Snu13p (Watkins et al., 2000), and this is proposed to provide a platform for assembling the other core snoRNP proteins: Nop1p/fibrillarin, Nop56p and Nop5/58p. As some of these proteins are required for snoRNA accumulation (Lafontaine and Tollervey, 1999, 2000; Watkins et al., 2000), we analysed the localization of a snoRNA reporter that was stabilized by extending its terminal stem (Huang et al., 1992). By individually depleting each of the proteins, we found that they were all required for normal localization of the box C/D reporter, and that their absence led to mislocalization of the snoRNA in the nucleoplasm. These effects are likely to reflect a direct requirement for snoRNA localization, rather than an indirect consequence of blocking nucleolar function, because we could stop ribosome biogenesis by inactivating RNA polymerase I without releasing snoRNA in the nucleoplasm.
The effects of depleting the proteins were not equivalent. Depletion of Nop1p or Snu13p resulted in an almost even distribution within the nucleus, while depletion of Nop56p and Nop5p/Nop58p had less severe effects: a majority of the snoRNA still localized to the nucleolus in these cases. This is most likely due to the formation of incompletely assembled snoRNP particles with residual affinity for the nucleolus. Indeed, the binding of Nop5/Nop58p to snoRNAs is independent of either Nop1p or Nop56p (Lafontaine and Tollervey, 2000), and we observed that the simultaneous depletion of Nop56p and Nop5/Nop58p resulted in a more severe phenotype (data not shown).
The fact that all snoRNP proteins were required for snoRNA localization, although to different extents, suggests that only completely assembled snoRNP can accumulate efficiently in the nucleolus. This is further supported by the observation that Nop1p and Nop5/Nop58p become delocalized to the nucleoplasm when other snoRNP components are depleted (Wu et al., 1998 and this work). Thus, snoRNPs are unlikely to possess a simple nucleolar localization signal. Their incorporation in the nucleolus probably requires multiple interactions involving several snoRNP components. Complementarity to the pre-rRNA is not required, however, as this is absent from the snoRNA reporter. Studies on the localization of nucleolar proteins have also failed to identify small sequence motifs that confer nucleolar targeting. Instead, one or several domains of the proteins were found to be required and it has generally proved difficult to resolve nucleolar localization from nucleolar function (Snaar et al., 2000 and references therein).
A current model for nucleolar formation proposes that the components self-associate by multiple weak, transient interactions, around sites of recruitment to nascent pre-rRNAs (Lewis and Tollervey, 2000). Photobleaching experiments in human cells indicate that nucleolar fibrillarin exchanges rapidly with a nucleoplasmic pool (Phair and Misteli, 2000). Although not specifically tested, it is likely that the snoRNPs with which fibrillarin is associated are similarly exchanged. In this model, depletion of individual snoRNP components will weaken association with the nucleolar components, altering the equilibrium between the nucleolar-associated and nucleoplasmic pools of snoRNP.
The box C/D snoRNA localization pathway involves a novel sub-nuclear structure and the activities of Srp40p and Nsr1p
Overexpression of an artificial snoRNA induced a block in the localization pathway. The snoRNAs became concentrated in a sub-nucleolar domain, which we termed the NB. Double localization experiments demonstrated that endogenous box C/D snoRNAs are also retained in the NB, although to a lesser extent than the artificial snoRNA. The endogenous snoRNAs contain functional sequences in addition to the box C/D motif, which may partially rescue the block in snoRNA localization (Lange et al., 1998). The block appears to be specific for box C/D snoRNAs as a box H/ACA snoRNP protein Gar1p localized normally when the artificial snoRNA was overexpressed.
Electron microscopy showed that the NB is contiguous with the dense fibrillar region of the nucleolus. This is believed to be the region in which the box C/D snoRNP function in pre-rRNA processing and modification. While we cannot formally exclude the possibility that the NB normally acts as a storage site for snoRNA produced in excess, the data are most consistent with the NB acting as a structure through which snoRNPs transit, either prior to recruitment to the dense fibrillar compartment, or during a cyclic process within the nucleolus. Overexpression of the snoRNA reporter may overload the capacity of the snoRNA trafficking machinery, leading to accumulation in the NB.
The accumulation of box C/D snoRNAs in the NB was influenced by the activities of two nucleolar proteins, Nsr1p and Srp40p. Accumulation of the snoRNAs in the NB was lost in the absence of Srp40p, but was stimulated by the absence of Nsr1p. These effects appear specific for the box C/D class of snoRNAs, as the localization of Gar1p was normal in these strains. Both Srp40p and Nsr1p were detected in the NB, suggesting a direct role in snoRNA trafficking. Nopp140, the mammalian homologue of Srp40p, has been shown to immunoprecipitate box C/D snoRNPs, and this interaction likely involves its central domain, a highly charged serine-rich region interspersed with clusters of basic amino acids (Isaac et al., 1998; Yang et al., 2000). Both Nsr1p and Srp40p share a similar domain, and it appears to be instrumental in the routing process as its expression in a truncated Nsr1p construct increases accumulation of snoRNA in the NB.
The C-terminal RNA binding domain of Nsr1p and Gar2p is highly homologous to that of nucleolin, which is reported to mediate its binding at multiple locations on the pre-rRNA (Ginisty et al., 2000). It was therefore proposed that one function of these proteins is to bring processing factors to rRNA precursors (Sicard et al., 1998). If this model is correct, absence of Nsr1p might delay association of the snoRNPs with the pre-rRNA and lead to accumulation in the NBs. Mammalian Nopp140 has been shown to nucleate assembly of large structures containing many components of the CB (Isaac et al., 1998), and the depletion of Srp40p might therefore inhibit NB formation and/or snoRNP association with the structure.
A relationship between the NB and the CB
In Xenopus oocytes, it has been reported that newly synthesized box C/D snoRNAs transiently localize to the CBs prior to distribution within the nucleolus (Narayanan et al., 1999b). This suggested that the yeast NB could be the functional counterpart of the vertebrate CB. In support of this hypothesis, we show that human SMN, a well-characterized CB marker (Carvalho et al., 1999), localized to the NB when expressed in yeast. Moreover, SMN localized to, and therefore labelled, the NB in the absence of snoRNA overexpression, suggesting that a NB also exists in the wild-type yeast nucleolus. Our observation of nucleolar localization for the yeast NB is not inconsistent with the localization of vertebrate CBs, which are often located close to the nucleolus, and even in some cell lines within nucleoli (Matera, 1999).
Electron microscopy showed that the NB enlarges when the artificial snoRNA is overexpressed. In vertebrates, CBs are dynamic structures that can vary greatly in size (Platani et al., 2000) and are not even detectable in all cell lines, or in all cells in a given cell line. Similar to the situation in yeast, we have observed that in mammalian cells in which CB are small or absent, their formation can be induced by overexpressing snRNAs (Tuma and Roth, 1999; C.Verheggen and E.Bertrand, unpublished results). Thus, the NB and CB can be envisioned as kinetic intermediates along biogenesis pathways of small RNPs, which accumulate when downstream steps become rate limiting. Possibly, the formation of a body facilitates further processing by co-localizing the RNA with processing enzymes and by acting as a store to block diffusion of snoRNP in excess. In vertebrates, snRNAs also localize to CBs, probably during their maturation pathway (Sleeman and Lamond, 1999). Owing to their low abundance, we have not yet been able to visualize endogenous snRNAs in S.cerevisiae, but snRNA-associated proteins have been reported to be present within the nucleolus of the fission yeast S.pombe (Potashkin et al., 1990).
We propose that the nucleolar localization pathway of box C/D snoRNAs is at least partially conserved from yeast to humans and involves a related intermediate, the CB in vertebrates, and the NB in yeast.
A model for box C/D snoRNA localization to the nucleolus
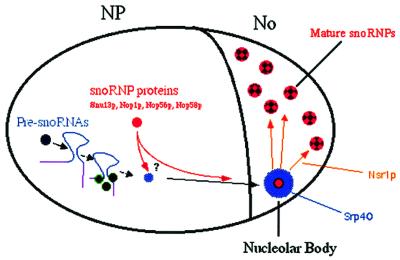
Depletion of the core snoRNP proteins leads to the loss of accumulation of box C/D snoRNA into the NB. This suggests that snoRNPs are already assembled at this point (see Figure 6). However, it is unclear whether snoRNAs are bound by the core proteins at earlier steps. Thus, it could be that snoRNP assembly takes place in the vicinity of the snoRNA transcription site, during its transport toward the nucleolus, or in the NB itself. In this last hypothesis, unassembled snoRNA would need to be stabilized by non-snoRNP proteins on their way toward the nucleolus. Interestingly, it has recently been shown that precursors of U3 snoRNA are not bound by the core snoRNP proteins, but are stabilized by Lhp1p, the yeast homologue of La (Kufel et al., 2000). Localization of these precursors within the yeast nucleus should help to resolve this question. Within the NB, the snoRNAs may interact with Srp40p, and the fully assembled, functional snoRNP would finally be handed over to Nsr1p for delivery to the rRNA precursors.
Fig. 6. A model for the snoRNA trafficking pathway towards the nucleolus (see text).
Finally, the importance of understanding the mechanisms of nucleolar RNA routing is emphasized by the recent discovery that many classes of cellular RNA are found in transient association with this compartment (reviewed in Pederson, 1998). Whether some or all of these transit through the NBs or related structures remains to be determined.
Materials and methods
Strains
The S.cerevisiae strains used in this study are described in Table I. Novel strains were generated by homologous recombination of PCR products as described previously (Lafontaine and Tollervey, 1996). When needed, plasmids were introduced into these strains by the PEG–lithium procedure, and transformants were selected on appropriate drop-out media. For glucose depletion, yeast cells were grown exponentially in selection media containing 2% galactose and 2% raffinose, washed in yeast nitrogen base (YNB), and further grown in selection media containing 4% glucose. For each strain, growth time following the glucose shift was similar to what was described previously. Gal::Snu13 was shifted for 6–9 h (Watkins et al., 2000), Gal::Nop1 was shifted for 18–24 h (Schimmang et al., 1989), and Gal::Nop56 and Gal::Nop58 were shifted for 13–16 h (Lafontaine and Tollervey, 1999, 2000). Northern and/or western blotting confirmed that depletion occurred following transfer to glucose (data not shown). In each case, the phenotypes were similar during this time frame. The polymerase I thermosensitive strain NOY504 was grown at 25°C, and shifted to 37°C for 2 h before fixation.
Table I.
| Strain | Genotype | Reference |
|---|---|---|
| YDL401 | MATa, his3Δ200, leu2Δ1, trp1, gal2, galΔ108, ura3-52 | Lafontaine and Tollervey (1996) |
| YDL527-1 | as YDL401 but HIS3::GAL10::NOP56 | Lafontaine and Tollervey (2000) |
| YDL522-17 | as YDL401 but HIS3::GAL10::NOP58 | Lafontaine and Tollervey (1999) |
| YDL539 | as YDL 401, but dbr1Δ::TRP1 | this study |
| D255 | BWG1-7A (MATa, ura3-52, leu2-3,112, ade1-100, his4-519), but URA3pGAL10::NOP1Gal::Nop1 | Schimmang et al. (1989) |
| YPH99 | MATa, trp1Δ63, his3Δ200, ura3-52, lys2-801, ade2-101, leu2Δ1 | Watkins et al. (2000) |
| YSNU13GAL | as YPH499, but HIS3::GAL1::SNU13 | Watkins et al. (2000) |
| ΔNSR1 | as YPH499, but MATα, nsr1Δ::HIS3 | Lin et al. (1994) |
| W303-1a | MATa, ade2-1, ura3-1,his3-11,15, trp1-1, leu2-3,112, can1-100 | Meier (1996) |
| TMY20 | as W303-1A but srp40Δ::URA3 | Meier (1996) |
| WLY353 | as W303-1A, but NSR1(amino acid 1-186)::HIS3 | Lee et al. (1991) |
| NOY504 | rpa12::LEU2, ade2-101, trp1-1, leu2-3, 112, ura3-1, his3-11, CAN1-100 |
Plasmid construction
The MS2-tagged, artificial snoRNA was generated by PCR, and cloned into pIIIA-MS2x2 at the EcoRI sites, and into pG14AC at the XhoI site of the actin intron. Its sequence is shown as Supplementary data. The mutant snoRNA was identical, but lacked boxes C and D.
Gar1–GFP expression plasmids (pZut3, pZut4) and Nop1–GFP expression plasmids were described previously (Trumtel et al., 2000). YFP-tagged versions of SRP40 and NSR1-Nter were generated as follows. pZut4 was modified to replace GFP by YFP, and SRP40 and NSR1-Nter sequences were then PCR amplified and cloned in-frame in place of Gar1, to generate Y4pZut-SRP40 and Y4pZut-NSR1-Nter, respectively. SMN–GFP was expressed from the centromeric vector pUG35, under the control of the inducible MET25 promoter.
Electron microscopy
For morphological analysis, strains were fixed for 3 h at room temperature in 1.6% glutaraldehyde in 0.1 M Sörensen’s buffer pH 7.4, acetylated and embedded in Epon.
For in situ hybridization, strains were fixed for 17.5 h at room temperature in 8% formaldehyde in 0.1 M Sörensen’s buffer, dehydrated through graded acetone solutions and embedded in Lowicryl K4M. Hybridization was performed by floating the ultrathin sections for 3.5 h at 37°C on 50 µl of the hybridization buffer (10% dextran sulfate and 50% deionized formamide in 2× sodium salt citrate: 0.3 M NaCl, 0.03 M sodium citrate pH 7) containing 1 pmol of BrdU-labelled MS2 oligonucleotide probe and 5 µg of salmon sperm DNA. After washing in phosphate-buffered saline (PBS) buffer (0.14 M NaCl, 6 mM Na2HPO4, 1.5 mM KH2PO4 pH 7.4), hybrids were detected by means of an indirect immunogold labelling procedure using a monoclonal anti-BrdU antibody as previously published (Thiry, 1999). Finally, the ultrathin sections were mounted on nickel grids and stained with uranyl acetate and lead citrate before examination in a Jeol CX 100 II transmission EM operated at 60 kV.
The specificity of the reaction was tested in several ways. First, when the probe was omitted from the hybridization medium, no labelling occurred. In a second control, no label was seen when the primary antibody was omitted. Finally, when a yeast strain devoid of sites for the phage MS2 was used, no labelling was present.
Fluorescence in situ hybridization and probes
Yeast cells were processed for in situ hybridization as described previously (Samarsky et al., 1998), except that oxalyticase was replaced by zymolase (0.2 mg/ml).
The probes were synthesized with modified nucleotides to incorporate primary amines, and then conjugated with Alexa-488 (Molecular Probes), Cy3 or Cy5 (Amersham). The MS2 and U14 probes were described previously (Samarsky et al., 1998), and the U3 probe was an amino-modified DNA oligonucleotide of the following sequence: TXCTATAGAAATGATCCXATGAAGTACGTCGACTXA. Amino-allyl Ts are represented by an X.
Immunolocalization and antibodies
Yeast cells were prepared as for in situ hybridization studies, and then rehydrated in PBS. Anti-Nop1p (Aris and Blobel, 1988) and anti-mouse–fluorescein isothiocyanate (FITC) antibodies were diluted 1/200 in PBS containing 1% bovine serum albumin (BSA). Anti-Nop5 antibody (Wu et al., 1998) was used diluted 1/80 in PBS, 1% BSA. Binding of primary and secondary antibody was allowed for 1 h at 37°C, and cells were washed twice for 30 min at room temperature.
Image acquisition and processing
Images were acquired on a DMRA microscope equipped for epifluorescence, and with a 100× PlanApo objective and a 1.6× eyepiece. Digital images were recorded with a 12-bit C4795-NR CCD camera (Hamamatsu). Both the camera and the microscope were controlled by the software Metamorph (Universal Imaging). When necessary, Maximal Likelihood Estimation deconvolution was performed with the software Huygens (Bitplane, Zurich, Switzerland), on stacks of 20–40 images taken with a Z-step of 0.1 µm. Maximal image projections of the resulting stacks were then converted to 8-bit images and colourized with Photoshop.
RNA extraction and northern blot analysis
Total RNA from yeast cells was isolated using an acidic phenol/glass bead procedure. Northern analysis was performed as described previously (Samarsky et al., 1998). Radiolabelled hybridization probes were generated by random priming on DNA fragments containing either repeated MS2 sites, or the SCR1 sequence.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are very grateful to Tom Meier for the gift of the srp40 strains and related plasmids, to Michèle Caizergues-Ferrer for the idea of nsr1, and for the gift of these strains, to Nick Watkins, Christiane Branlant and Reinhart Lurhrmann for the gift of the Gal::Snu13 strain. Nop1 and Nop5 antibodies were a gift of J.Aris. We also thank Pierre-Emmanuel Gleizes for the gift of pZut3 and pZut4 plasmids, and H.Grosshans and Ed Hurt for the Nop1–GFP plasmid. C.V. was supported by a fellowship from ARC, J.M. by a fellowship from MNRT, and D.L.J.L. is a Chercheur qualifié du Fonds National de la Recherche Scientifique Belge (FNRS). This work was supported by the ARC (grant 9043 to E.B.), the CNRS (grant ACI to E.B.) and AFM (grant to R.B.).
References
- Aris J. and Blobel,G. (1988) Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J. Cell Biol., 107, 17–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachellerie J. and Cavaillé,J. (1997) Guiding ribose methylation of rRNA. Trends Biochem. Sci., 22, 257–261. [DOI] [PubMed] [Google Scholar]
- Bertrand E., Houser-Scott,F., Kendall,A., Singer,R. and Engelke,D. (1998) Nucleolar localization of early tRNA processing. Genes Dev., 12, 2463–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beven A., Simpson,G., Brown,J. and Shaw,P. (1995) The organization of spliceosomal components in the nuclei of higher plants. J. Cell Sci., 108, 509–518. [DOI] [PubMed] [Google Scholar]
- Bohmann K., Ferreira,J.A. and Lamond,A.I. (1995) Mutational analysis of p80 coilin indicates a functional interaction between coiled bodies and the nucleolus. J. Cell Biol., 131, 817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho T., Almeida,F., Calapez,A., Lafarga,M., Berciano,M. and Carmo-Fonseca,M. (1999) The spinal muscular atrophy disease gene product, SMN: a link between snRNP biogenesis and the Cajal (coiled) body. J. Cell Biol., 147, 715–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B. (2000) Nuclear RNA export pathways. Mol. Cell. Biol., 20, 4181–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custodio N., Carmo-Fonseca,M., Geraghty,F., Pereira,H., Grosveld,F. and Antoniou,M. (1999) Inefficient processing impairs release of RNA from the site of transcription. EMBO J., 18, 2855–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femino A., Fay,F., Fogarty,K. and Singer,R. (1998) Visualization of single RNA transcripts in situ. Science, 280, 585–590. [DOI] [PubMed] [Google Scholar]
- Gall J.G. (2000) Cajal bodies: the first 100 years. Annu. Rev. Cell Dev. Biol., 16, 273–300. [DOI] [PubMed] [Google Scholar]
- Ginisty H., Serin,G., Ghisolfi-Nieto,L., Roger,B., Libante,V., Amalric,F. and Bouvet,P. (2000) Interaction of nucleolin with an evolutionarily conserved pre-ribosomal RNA sequence is required for the assembly of the primary processing complex. J. Biol. Chem., 275, 18845–18850. [DOI] [PubMed] [Google Scholar]
- Good P. and Engelke,D. (1994) Yeast expression vectors using RNA polymerase III promoters. Gene, 151, 209–214. [DOI] [PubMed] [Google Scholar]
- Hannus S., Buhler,D., Romano,M., Seraphin,B. and Fischer,U. (2000) The Schizosaccharomyces pombe protein Yab8p and a novel factor, Yip1p, share structural and functional similarity with the spinal muscular atrophy-associated proteins SMN and SIP1. Hum. Mol. Genet., 9, 663–674. [DOI] [PubMed] [Google Scholar]
- Huang G., Jarmolowski,A., Struck,J. and Fournier,M. (1992) Accumulation of U14 small nuclear RNA in Saccharomyces cerevisiae requires box C, box D and a 5′, 3′ terminal stem. Mol. Cell. Biol., 12, 4456–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac C., Yang,Y. and Meier,T. (1998) Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J. Cell Biol., 142, 319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel J., Allmang,C., Chanfreau,G., Petfalski,E., Lafontaine,D. and Tollervey,D. (2000) Precursors to the U3 small nucleolar RNA lack small nucleolar RNP proteins but are stabilized by La binding. Mol. Cell. Biol., 20, 5415–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D. and Tollervey,D. (1996) One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res., 24, 3469–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D. and Tollervey,D. (1999) Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA, 5, 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D. and Tollervey,D. (2000) Synthesis and assembly of the box C + D small nucleolar RNPs. Mol. Cell. Biol., 20, 2650–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T., Borovjagin,A., Maxwell,E. and Gerbi,S. (1998) Conserved boxes C and D are essential nucleolar localization elements of U14 and U8 snoRNAs. EMBO J., 17, 3176–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W., Xue,Z. and Melese,T. (1991) The NSR1 gene encodes a protein that specifically binds nuclear localization sequences and has two RNA recognition motifs. J. Cell Biol., 113, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. and Tollervey,D. (2000) Like attracts like: getting RNA processing together in the nucleus. Science, 288, 1385–1389. [DOI] [PubMed] [Google Scholar]
- Lin J. and Zakian,V. (1994) Isolation and characterization of two Saccharomyces cerevisiae genes that encode proteins that bind to (TG1-3)n single strand telomeric DNA in vitro. Nucleic Acids Res., 22, 4906–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q. and Dreyfuss,G. (1996) A novel nuclear structure containing the survival of motor neurons protein. EMBO J., 15, 3555–3565. [PMC free article] [PubMed] [Google Scholar]
- Matera A. (1999) Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol., 9, 302–309. [DOI] [PubMed] [Google Scholar]
- Meier T. (1996) Comparison of the rat nucleolar protein Nopp140 with its yeast homolog SRP40. J. Biol. Chem., 271, 19376–19384. [PubMed] [Google Scholar]
- Narayanan A., Lukowiak,A., Jady,B., Dragon,F., Kiss,T., Terns,R. and Terns,M. (1999a) Nucleolar localization signals of box H/ACA small nucleolar RNAs. EMBO J., 18, 5120–5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A., Speckmann,W., Terns,R. and Terns,M. (1999b) Role of the Box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol. Biol. Cell, 10, 2131–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman D., Kuhn,J., Shanab,G. and Maxwell,S. (2000) Box C/D snoRNA-associated proteins: two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA, 6, 861–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes M., Nogi,Y., Clarcj,M. and Nomura,M. (1993) Structural alterations of the nucleolus in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Mol. Cell. Biol., 13, 2441–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. (1998) The plurifunctional nucleolus. Nucleic Acids Res., 26, 3871–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair R.D. and Misteli,T. (2000) High mobility of proteins in the mammalian cell nucleus. Nature, 404, 604–609. [DOI] [PubMed] [Google Scholar]
- Platani M., Goldberg,I., Swedlow,J. and Lamond,A. (2000) In vivo analysis of Cajal body movement, separation and joining in live human cells. J. Cell Biol., 151, 1561–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz J. and Pederson,T. (2000) Movement of mRNA from transcription site to nuclear pores. J. Struct. Biol., 129, 252–257. [DOI] [PubMed] [Google Scholar]
- Potashkin J.A., Derby,R.J. and Spector,D.L. (1990) Differential distribution of factors involved in pre-mRNA processing in the yeast cell nucleus. Mol. Cell. Biol., 10, 3524–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarsky D., Fournier,M., Singer,R. and Bertrand,E. (1998) The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J., 17, 3747–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmang T., Tollervey,D., Kern,H., Frank,R. and Hurt,E. (1989) A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J., 8, 4015–4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard H., Faubladier,M., Noaillac-Depeyre,J., Leger-Sylvestre,I., Gas,N. and Caizergues-Ferrer,M. (1998) The role of the Schizosaccharomyces pombe gar2 protein in nucleolar structure and function depends on the concerted action of its highly charged N-terminus and its RNA-binding domains. Mol. Biol. Cell, 9, 2011–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman J. and Lamond,A. (1999) Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr. Biol., 9, 1065–1074. [DOI] [PubMed] [Google Scholar]
- Smith K. and Lawrence,J. (2000) Interactions of U2 gene loci and their nuclear transcripts with Cajal (coiled) bodies: evidence for PreU2 within Cajal bodies. Mol. Biol. Cell, 11, 2987–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaar S., Wiesmeijer,K., Jochemsen,A., Tanke,H. and Dirks,R. (2000) Mutational analysis of fibrillarin and its mobility in living human cells. J. Cell Biol., 151, 653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiry M. (1999) Ultra structural methods for nucleic acid detection by immunocytology. Prog. Histochem. Cytochem., 34, 87–160. [DOI] [PubMed] [Google Scholar]
- Tollervey D. and Kiss,T. (1997) Function and synthesis of small nucleolar RNA. Curr. Opin. Cell Biol., 9, 337–342. [DOI] [PubMed] [Google Scholar]
- Trumtel S., Leger-Sylvestre,I., Gleizes,P.-E., Teulieres,F. and Gas,N. (2000) Assembly and functional organization of the nucleolus: ultra structural analysis of Saccharomyces cerevisiae mutants. Mol. Biol. Cell, 11, 2175–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuma R.S. and Roth,M.B. (1999) Induction of coiled body-like structures in Xenopus oocytes by U7 snRNA. Chromosoma, 108, 337–344. [DOI] [PubMed] [Google Scholar]
- Tyc K. and Steitz,J.A. (1989) U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J., 8, 3113–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins N. et al. (2000) A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell, 103, 457–466. [DOI] [PubMed] [Google Scholar]
- Wu P., Brockenbrough,J., Metcalfe,A., Chen,S. and Aris,J. (1998) Nop5 is a small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. J. Biol. Chem., 273, 16453–16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Isaac,C., Wang,C., Dragon,F., Pogacic,V. and Meier,T. (2000) Conserved composition of mammalian Box H/ACA and Box C/D small nucleolar/RNPs and their interaction with the common factor Nopp140. Mol. Biol. Cell, 11, 567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannoni Y. and White,K. (1997) Association of the neuron-specific RNA binding domain-containing protein ELAV with the coiled body in Drosophila neurons. Chromosoma., 105, 332–341. [DOI] [PubMed] [Google Scholar]
- Zhang G., Taneja,K., Singer,R. and Green,M. (1994) Localization of pre-mRNA splicing in mammalian nuclei. Nature, 372, 809–812. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Luo,M., Straesser,K., Katahira,J.E.H. and Reed,R. (2000) The protein Aly links pre-messenger RNA splicing to nuclear export in metazoans. Nature, 407, 401–405. [DOI] [PubMed] [Google Scholar]