Abstract
Uroporphyrinogen III synthase, U3S, the fourth enzyme in the porphyrin biosynthetic pathway, catalyzes cyclization of the linear tetrapyrrole, hydroxymethylbilane, to the macrocyclic uroporphyrino gen III, which is used in several different pathways to form heme, siroheme, chlorophyll, F430 and vitamin B12. U3S activity is essential in all organisms, and decreased activity in humans leads to the autosomal recessive disorder congenital erythropoetic porphyria. We have determined the crystal structure of recombinant human U3S at 1.85 Å resolution. The protein folds into two α/β domains connected by a β-ladder. The active site appears to be located between the domains, and variations in relative domain positions observed between crystallographically independent molecules indicates the presence of flexibility that may be important in the catalytic cycle. Possible mechanisms of catalysis were probed by mutating each of the four invariant residues in the protein that have titratable side chains. Additionally, six other highly conserved and titratable side chains were also mutated. In no case, however, did one of these mutations abolish enzyme activity, suggesting that the mechanism does not require acid/base catalysis.
Keywords: cobalamin/congenital erythropoietic porphyria/heme/porphyrin/uroporphyrinogen
Introduction
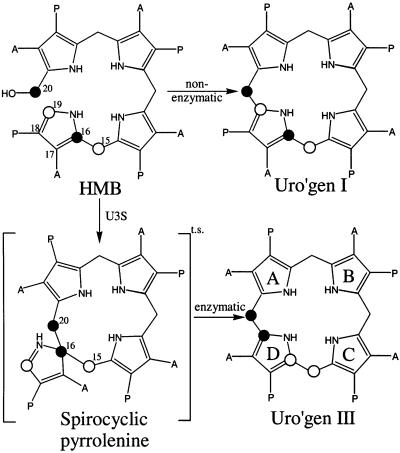
Porphyrins are essential components of a wide range of chemical reactions that are required for life. These macrocyclic tetrapyrroles act as cofactors for a multitude of enzymes that perform a variety of processes within the cell such as methionine synthesis (vitamin B12), oxygen transport (heme), methane synthesis (F430) and photosynthesis (chlorophyll). The biosynthesis of all porphyrins proceeds from the five-carbon precursor δ-aminolevulinic acid, through three enzymatic steps, to the first cyclic precursor uroporphyrinogen III (uro’gen III) (Battersby and Leeper, 1990; Scott, 1993). Uroporphyrinogen III synthase [U3S; EC 4.2.1.75, hydroxymethylbilane hydrolase (cyclizing)] is the enzyme that catalyzes formation of uro’gen III, the branch point for the various sub-pathways leading to the wide diversity of porphyrins. U3S exists as a monomer (Tsai et al., 1987) and catalyzes ring closure of the linear tetrapyrrole hydroxymethylbilane (HMB), with concurrent flipping of the D ring, to form uro’gen III (Battersby, 1978a,b; Pichon et al., 1994b) (see Figure 1). Asymmetry of the D ring is therefore a hallmark of all biologically relevant porphyrins.
Fig. 1. Conversion of HMB to uroporphyinogen I (non-enzymatic) and uro’gen III (enzymatic). A, acetate; P, propionate. The linear tetrapyrrole is cyclized by U3S with inversion of ring D to form the asymmetric product uro’gen III (pyrrole rings labeled A–D). The spirocyclic pyrrolenine has been proposed as a transition state intermediate (Mathewson and Corwin, 1961). The numbering scheme follows that of Pichon et al. (1994b). Although the numbers assigned to the D ring carbon atoms formally change during the reaction, for simplicity we refer to the scheme shown here throughout.
A catalytic mechanism has been proposed for U3S in which rearrangement of the A ring results in the loss of the C20 hydroxyl group to create a carbo-cation at C20 that performs an electrophilic attack on C16 to form a spirocyclic pyrrolenine intermediate (Figure 1). This intermediate resolves in the opposite direction to generate an azafulvene that cyclizes to uro’gen III (Mathewson and Corwin, 1961; Leeper, 1994; Pichon et al., 1994a). This proposed scheme is consistent with carbon radiolabeling experiments that showed that the D ring is flipped (Battersby, 1978b; Battersby et al., 1978), and with the observation that a synthetic spirolactam, differing from the proposed spirocyclic intermediate only by having an amide in place of an imine, is stable and functions as a potent inhibitor of the enzyme with a Ki of 1–2 µM (Stark et al., 1986; Cassidy et al., 1991).
Congenital erythropoetic porphyria (CEP) is transmitted as an autosomal recessive trait in which the activity of U3S is severely reduced. Biochemically the disease is characterized by the accumulation in plasma, tissues and red cells, of excess type I uroporphyrin and coproporphyrin, which are then eliminated by excretion in the urine and feces. Clinically CEP is characterized by a severe photosensitivity with skin fragility, bullous lesions, hypertrichosis and scarring on light-exposed areas. The accumulation of porphyrins in erythrocytes often leads to hemolytic anemia and splenomegaly. Individuals with CEP can be either homozygous for the same mutation or heterozygous for two different mutations and the severity of the disease is inversely related to the remaining enzymatic activity of U3S (Kappas et al., 1995; Desnick et al., 1998).
In an effort to further understand this enzyme, we have determined the crystal structure of recombinant human U3S. The protein folds into a two-domain structure, with each domain consisting of a parallel β-sheet surrounded by α-helices. The two domains are connected by a two-strand anti-parallel β-ladder. We propose that the active site lies between the domains, where many of the surface-exposed conserved residues cluster. An alignment of the two molecules in the asymmetric unit reveals shifts in bridge residues that propagate into larger domain motions. This inherent flexibility may be an essential feature of substrate binding and product release.
Results and discussion
Structure determination
Recombinant human U3S was expressed in Escherichia coli and purified as described in Materials and methods. The structure was determined by multiwavelength anomalous diffraction (MAD) at 2.1 Å resolution (Table I) using a seleniomethionine-substituted U3S mutant (SeU3S) that contained two additional methionine residues that were introduced in place of Leu139 and Leu232 in order to facilitate phase determination. Crystals of U3S belong to space group P1 and contain two molecules in the asymmetric unit. The native structure was refined against 1.85 Å data to an R factor (Rfree) of 20.0% (25.1%) with good geometry (Table II). In general, the Fourier maps are well defined throughout the protein, with the exception of the C-terminal five residues and an unstructured loop (residues 114–118). The final model includes residues 1–113 and 119–260 of each molecule of U3S in the asymmetric unit and 569 water molecules (PDB code 1JR2).
Table I. Data collection statistics.
| SeMet MAD | ||||
|---|---|---|---|---|
| λ1 (peak) | λ2 (edge) | λ3 (remote) | Native | |
| Wavelength (Å) | 0.980271 | 0.98048 | 0.930003 | 0.97 |
| Obs. reflections | 176 828 | 171 818 | 98 977 | 201 469 |
| Unique reflections | 36 129 | 36 164 | 36 085 | 46 295 |
| Highest res. shell (Å) | 2.00–2.03 | 2.00–2.03 | 2.00–2.03 | 1.85–1.88 |
| Completeness (%) | 90.1 (78.0) | 91.4 (79.6) | 89.8 (73.6) | 94.0 (75.8) |
| Rsyma (%) | 7.7 (34.4) | 7.8 (40.9) | 7.7 (34.0) | 4.5 (32.7) |
| Avg. I/σ (I) | 18.4 (3.2) | 17.5 (2.6) | 12.5 (2.0) | 22.5 (2.5) |
| Mosaicity (°) | 0.704 | 0.522 | 0.551 | 0.154 |
Numbers in parentheses are for highest resolution shell.
aRsym = 100 * Σ | I – <I> | / Σ I, where I is the intensity of an individual measurement and <I> is the average intensity from multiple observations.
Table II. Refinement statistics.
| Resolution range (Å) | 58.7–1.85 |
| No. of protein atomsa | 3966 |
| No. of solvent molecules | 569 |
| R factor (%)b | 20.0 |
| Rfree (%)c | 25.1 |
| R.m.s.d. (bond lengths) (Å) | 0.012 |
| R.m.s.d. (bond angles) (°) | 1.639 |
| <B> (Å2) main chain | 30.8 |
| <B> (Å2) side chain | 33.1 |
| <B> (Å2) water molecules | 44.7 |
| No. of φ/ψ angles (%)d | |
| most favored | 94.5 |
| allowed | 5.5 |
aNon-hydrogen atoms only.
bR factor = 100 × Σ (||Fobs| – |Fcalc||) / Σ|Fobs|.
cRfree is the R factor for a selected subset (5%) of the reflections that were not included in previous refinement calculations.
dStereochemistry was assessed with PROCHECK (Laskowski et al., 1993).
Structure of uro’gen III synthase
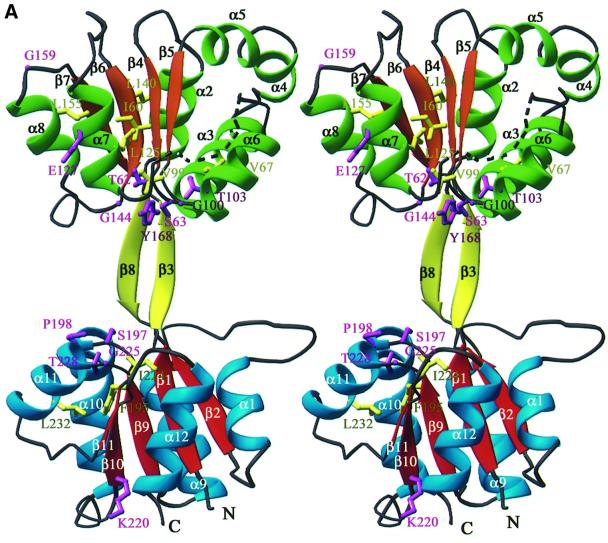
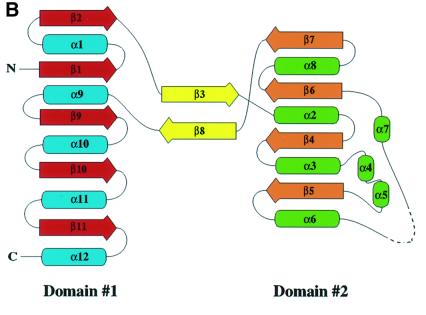
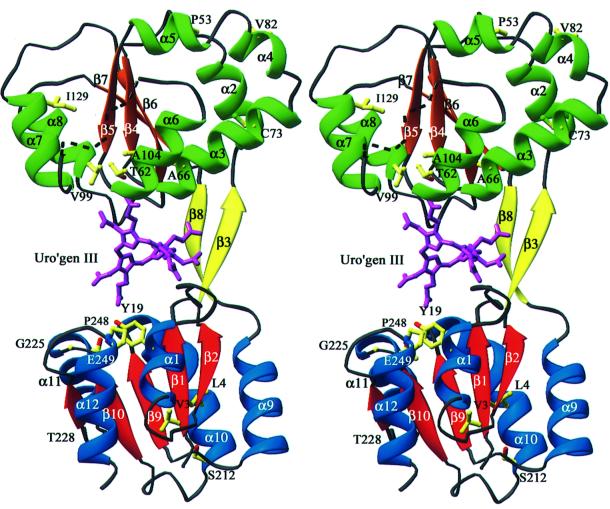
U3S adopts an elongated bi-lobed structure in which the two domains are connected by a two-strand anti-parallel β-sheet (Figure 2). The two domains have similar folds that comprise a parallel β-sheet surrounded by α-helices. Domain 1 (residues 1–35 and 173–260), which belongs to the flavodoxin-like fold family (Murzin et al., 1995), comprises a five-strand parallel β-sheet (strand order 21345) that is surrounded by five α-helices. Domain 2 adopts a DNA glycosylase-like fold, in which a four-strand parallel β-sheet (strand order 2134) is surrounded by seven α-helices. Although the two domains have similar topology, least squares overlap on the β-sheets does not result in close superposition of the α-helices. The C-terminal ends of the β-sheets of each domain face each other and point toward the central intervening opening (Figure 2).
Fig. 2. The U3S structure. Helices and strands of domain 1 are blue and red, helices and strands of domain 2 are green and orange. Strands connecting the two domains are yellow. Secondary structure elements and chain termini are labeled. The dotted line represents the disordered loop (residues 114–118). (A) Ribbon representation of the U3S structure in stereo. The side chains of conserved residues are colored purple if solvent exposed and yellow if buried (see also Figures 3 and 4). (B) Topology diagram. Helices are shown as ellipsoids and strands are shown as arrows. (A) and Figures 4–7 were made using the program Ribbons (Carson, 1991). Secondary structure was defined by DSSP (Kabsch and Sander, 1983).
Comparison with other structures
A structural similarity search (Holm and Sander, 1997) using the isolated domain 1 of U3S identified the vitamin B12 binding domain of methionine synthase (PDB identifier 1BMT) as being one of the most structurally similar proteins. As discussed below under ‘Proposed active site’, the alignment with methionine synthase suggests a location for the tetrapyrrole binding pocket on U3S. The protein most similar to domain 2 is the NAD binding domain of flavohemoglobin (PDB identifier 1CQX), although this structural similarity does not seem to reflect functional similarity, since U3S does not utilize NAD. The entire U3S structure is superficially similar to the l-fucose isomerase, ATC-like and periplasmic binding protein-like fold families.
Evidence for interdomain flexibility
The two independent molecules in the asymmetric unit adopt similar conformations. Least squares overlap on Cα atoms of domain 1, excluding residues 7–19, yielded a root mean square deviation (r.m.s.d.) of 0.44 Å. Residues 7–19 form a loop that adopts different conformations in the two molecules (maximal Cα–Cα displacement of 5.8 Å after superposition) as a result of crystal lattice constraints. Least squares overlap on Cα atoms of domain 2 for the two molecules in the asymmetric unit yields an r.m.s.d. of 0.34 Å.
Although the structure within each domain is highly similar between the two molecules in the asymmetric unit, there is a 13.5° rotational motion of the domains with respect to one another. This corresponds to an ∼2.5 Å increase in the opening of the cleft between the domains. As discussed below, we propose that this cleft contains the enzyme active site. This relative domain motion results from small changes in the φ/ϕ angles of residues 36–41 and 168–172, which comprise the anti-parallel β-sheet that connects the two domains. This distortion does not alter the β-sheet character of these residues or packing interactions between side chains. Opening and closing of the intradomain cleft may be an important feature of catalysis since the more open conformation may facilitate substrate binding and product release, whereas the closed conformation may be required for macrocyclic ring closure.
Proposed active site
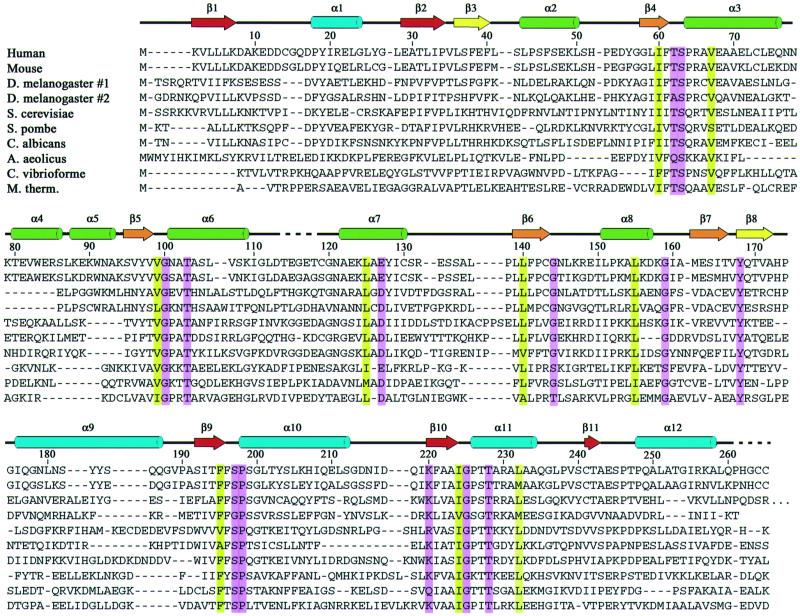
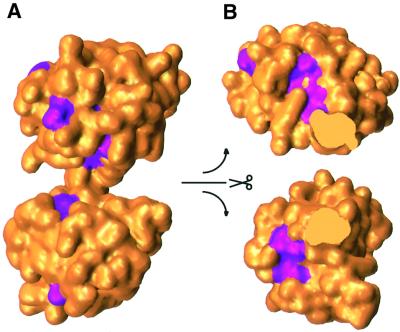
The distribution of conserved residues suggests that the enzyme active site is located in the large open cleft between the two domains. A sequence alignment of U3S from 10 species (Figure 3) revealed that seven residues are invariant and a further 15 residues show conservative substitutions as defined by the Clustal W program (Higgins and Sharp, 1989). Many of these residues are buried within the hydrophobic core of domain 1 or domain 2, where they presumably stabilize the folded conformation. Of 13 surface-exposed residues that are invariant or conserved, 10 line the cleft between the two domains, Thr62, Ser63, Thr103, Gly100, Gly144, Tyr168, Ser197, Pro198, Gly225 and Thr228 (Figure 4). Three other conserved residues, Glu127, Gly159 and Lys220, are distributed on the ‘front’ face of the molecule as viewed in Figure 4.
Fig. 3. Alignment of U3S protein sequences. Alignment was performed using the Clustal method (Higgins and Sharp, 1989). Every 10th residue is indicated using the numbering of the human sequence. Conserved residues are highlighted: surface exposed, magenta; buried, yellow. The color code for secondary structural elements is the same as for Figure 2. Disordered residues that have been omitted from the model are indicated with a dashed line. DDBJ/EMBL/GenBank database entries shown are: human, Homo sapiens (A40483); mouse, Mus musculus (A56838); Drosophila melanogaster (AAF46419 and AAF55222); Saccharomyces cerevisiae (NP_014921); Schizosaccharomyces pombe (P87214); Candida albicans (CAA22001); Aquifex aeolicus (E70452); Chlorobium vibrioforme (Q59335); Methanobacterium thermoautotrophicum (O26268).
Fig. 4. Surface representation of U3S. Conserved residues are colored magenta (see Figure 3). (A) View direction is the same as in Figure 2. No conserved residues are visible when the molecule is viewed from the opposite (back) direction. (B) The two domains of U3S are shown independently, split open at the center to expose the cleft surface.
In an effort to identify groups essential for U3S activity, we mutated the titratable, invariant or conserved residues listed above and determined the activities of each mutant protein (Table III). In addition, Thr227 and Arg65 are defined as non-conserved, but lie in the proposed active site cleft and were therefore also targeted for analysis. None of these residues is absolutely required for activity of the enzyme, and only three diminish the activity, with the least active mutant retaining >30% of wild-type activity. This suggests that the enzymatic mechanism does not include acid/base catalysis.
Table III. Mutant U3S activities.
| Mutation | U3S activitya |
|---|---|
| Wild type | 100 ± 3.2 |
| Thr62→Ala | 97.8 ± 7.0 |
| Ser63→Ala | 97.1 ± 7.2 |
| Arg65→Ala | 74.1 ± 3.4 |
| Thr103→Ala | 63.5 ± 1.1 |
| Glu127→Ala | 100.6 ± 10.6 |
| Tyr168→Phe | 49.1 ± 1.3 |
| Ser197→Ala | 101.1 ± 3.7 |
| Lys220→Ala | 109.3 ± 4.3 |
| Thr227→Ala | 115.2 ± 9.3 |
| Thr228→Ala | 32.2 ± 3.1 |
aExpressed as a percentage of wild-type control.
The three point mutants that exhibit greatest reduction in activity (Thr103Ala, Tyr168Ala and Thr228Ala) all lack a hydroxyl group compared with the wild-type sequence. These residues may function as hydrogen bond donors and acceptors for the variety of carboxylate side chains or pyrrole nitrogens of the substrate. It is interesting to note that only one of the conserved residues is positively charged, suggesting that the enzyme does not make extensive use of charge–charge interactions with the carboxylate side chains. Instead, a potentially larger number of weaker interactions may develop between ligand and polar or hydrophobic side chains. The robustness of enzyme activity to our mutational analysis may indicate that no one mutation will disrupt HMB binding when a large number of other contacts are left intact.
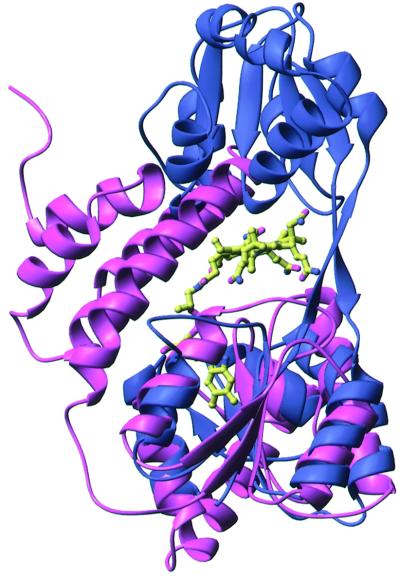
As described above, the vitamin B12 binding domain of methionine synthase (1BMT) aligns well with domain 1 of U3S (Figure 5; r.m.s.d. of 3.2 Å over 105 of 125 Cα atoms). Like U3S, methionine synthase comprises two domains: a C-terminal domain that resembles domain 1 of U3S and an N-terminal domain that is formed by five helices (Drennan et al., 1994). A model of a putative U3S–product complex was generated by aligning U3S domain 1 with the methionine synthase structure and positioning an energy-minimized model (QUANTA/CHARMM; Molecular Simulations Inc.) of the U3S product, uro’gen III, in the space equivalent to that occupied by vitamin B12 in methionine synthase. Slight adjustments were made to the inherently flexible propionate side chain in order to alleviate unreasonable contacts, and the resultant model is seen in Figure 6. Although we do not propose specific residue contacts, this modeling exercise supports the proposal that this cleft could house the U3S active site. Initial attempts to cocrystallize and soak U3S crystals with either a linear bilane inhibitor (Phichon et al., 1994b) or the spirolactam inhibitor (Stark et al., 1986; Cassidy et al., 1991) have failed.
Fig. 5. Superposition of U3S (blue) and methionine synthase (pink). View direction is rotated by 90° about the vertical axis compared with Figure 2(A). Methionine synthase is aligned with domain 1 of U3S. Vitamin B12, bound to methionine synthase, is shown in standard atom colors as a ball-and-stick representation.
Fig. 6. Model of uro’gen III bound to U3S and location of the clinical mutations. View direction of this stereo image is similar to that in Figure 5. The side chains of residues identified as clinical mutations are shown in standard atom colors. In purple, an energy minimized model of the product, uro’gen III, has been aligned with the cobalamin molecule shown in Figure 5 and adjusted to avoid steric clashes. Note that uro’gen III is inherently very flexible and the model here is intended only to illustrate the plausibility of binding in the U3S cleft.
Location of clinical mutations
We examined the location and possible structural ramifications of the reported clinical mutations of U3S (Takamura et al., 1997; Desnick et al., 1998 and references therein; Saval and Tirado, 1999; Rogounovitch et al., 2000); see Table IV and Figure 6. Only two of these residues, Thr62 and Thr228, both of which are conserved, are positioned near the proposed active site cleft. We find that the purified recombinant Thr62Ala mutant retains 100% of wild-type activity (Table III). This is in contrast to earlier studies by Warner et al. (1992), where no activity was reported for the same mutant in a recombinant crude lysate. This apparent discrepancy may result from the failure of Warner et al. (1992) to verify expression of recombinant protein in their experiment. Mutation of Thr228 to a methionine side chain would probably affect local tertiary structure since the larger side chain would clash structurally with nearby residues. In addition, the loss of the Thr228 hydroxyl or Cγ group appears to be important for catalysis because mutation to alanine reduces activity to ∼30% of that of the wild-type protein. The remaining clinical mutations appear likely to disrupt tertiary structure (Table IV). The resultant unstable proteins may be insoluble or have a reduced half-life resulting in a clinical phenotype.
Table IV. Clinical U3S single-residue mutations.
| Mutation | Location | Effect |
|---|---|---|
| Val3→Phe | β1 | Larger residue inserted into hydrophobic core |
| Leu4→Phe | β1 | Larger residue inserted into hydrophobic core |
| Tyr19→Cys | α1 | Loss of buried packing interaction |
| Pro53→Leu | α2 | Expected to disrupt packing |
| Thr62→Ala | β4–α3 | Solvent exposed to active site cleft |
| Ala66→Val | α3 | Larger residue is expected to disrupt packing |
| Cys73→Arg | α3 | Charged residue inserted into hydrophobic core |
| Val82→Phe | α4 | Large hydrophobic exposed to solvent |
| Val99→Ala | β5–α6 | Loss of packing interactions |
| Ala104→Val | α6 | Larger residue is expected to disrupt packing |
| Ile129→Thr | α7 | Hydrophilic residue inserted into hydrophobic core |
| Ser212→Pro | α10 | Disruption of helix, packing constraints |
| Gly225→Ser | β10–α11 | Disruption of tight turn |
| Thr228→Met | α11 | Larger residue could disrupt packing |
| Pro248→Gln | α12 | Tight turn, hydrophilic residue into hydrophobic packing environment |
| Glu249→stop | α12 | Truncated protein, loss of helix 12 |
Materials and methods
All chemicals were purchased from Sigma unless otherwise indicated.
Cloning
The U3S gene was PCR amplified from a human HeLa cDNA library using Taq polymerase (Boehringer Mannheim) and primers derived from the human gene sequence of U3S (Tsai et al., 1988). The expression construct encodes the human U3S sequence with a 21-residue N-terminal polyhistidine tag sequence (MGH10SSGHIEGRH) in the pET16b plasmid (Novagen) designated pHt-U3S. The plasmid pHt-U3S-MII, which codes for the double mutant Leu139Met/Leu232Met, was made from pHt-U3S plasmid using the QuikChange site-directed mutagenesis kit (Stratagene). All plasmid constructs were confirmed by sequencing at the University of Utah core facility.
Protein preparation and crystallization
All U3S constructs were expressed in BL21(DE3)pLysS cells using standard growth and isopropyl-β-d-thiogalactoside induction protocols. The enzyme was affinity purified using a Ni2+-charged chelating Sepharose (Pharmacia) column followed by dialysis into 10 mM Tris pH 7.1, 190 mM KCl, 10 mM NaCl, 1.1 mM MgCl2, 4 mM EDTA. A subsequent round of size exclusion chromatography (Pharmacia Sephadex 75) was followed by dialysis into 10 mM Tris pH 7.5 and 1 mM dithiothreitol. The protein was concentrated to 5 mg/ml for crystallization. The selenomethionine-substituted protein (SeU3S) was prepared using the gal– met– auxotroph B834(DE3) cell line and grown in minimal media supplemented with selenomethionine (Ramakrishnan et al., 1993). The best crystals of both native and SeU3S were obtained by adding 2 µl of 5 mg/ml protein to 2 µl of reservoir solution in sitting drop trays at 4°C. The reservoir solution contained 20% 2-methyl-2,4-pentanediol, 100 mM MES pH 6.0.
X-ray data collection
Crystals were suspended in a rayon loop (Hampton), plunged into liquid nitrogen without the need for additional cryoprotectant, and maintained at 100 K during data collection. Data from a single native protein crystal were collected on a Mar345 image plate area detector at beamline 9-1 of the Stanford Synchrotron Radiation Laboratory (SSRL). A total of 247° of data were collected in 1° oscillations. All data were indexed and scaled with the programs Denzo and Scalepack (Otwinowski, 1993). The crystals belonged to spacegroup P1, had cell dimensions of a = 40.0 Å, b = 59.4 Å, c = 62.2 Å, α = 80.6°, β = 73.5°, γ = 88.5°, and contained two molecules in the asymmetric unit
Data for MAD phase determination were collected from a single SeU3S crystal on a Brandeis B4 CCD detector at beamline X-25 of the National Synchrotron Light Source (NSLS), Brookhaven National Labs.
Structure determination
Crystallographic computing was performed using programs from the CCP4 suite (CCP4, 1994) unless otherwise stated. Eight Se sites were located using the program Solve (Terwilliger and Berendzen, 1999) and refined with the program Sharp (de la Fortelle and Bricogne, 1997). Initial phase estimates were refined by solvent flattening and histogram mapping using the program DM (Cowtan and Zhang, 1999). Further improvement in the map quality was obtained by 2-fold non-crystallographic averaging using symmetry operators derived from the selenium positions.
The model was built with the program O (Jones et al., 1991) and refined with Refmac5 (Murshudov et al., 1999) using the stereochemical target values of Engh and Huber (1991). Initial refinement calculations were performed using the SeU3S data from 58.7 to 2.1 Å and included phase restraints, with Fs and SigFs used as output by Sharp and the Henderson–Lattman coefficients used as output by DM. NCS restrains were not used during refinement. Methionines 139 and 232 were then changed to the native leucine residues in the model, and refinement continued against the native data from 58.7 to 1.85 Å, with no phase information, to a final R value of 20.0% and an Rfree of 25.1%.
Activity assay
The activities of the mutant U3S proteins were measured using the following coupled assay using enzymes purified as described above. A reaction mixture containing 100 µl of 20 mM Tris pH 8.2, 280 µl of 20 mM potassium phosphate pH 8.2 and 20 µl of porphobilinogen deaminase (crudely purified protein that has an activity of ∼20 nmol of product produced per 75 µl of protein per hour) was incubated in the dark at 37°C for 2 min. A 20 µl aliquot of porphobilinogen (0.54 mg/ml) was added and the reaction was incubated for an additional 2.5 min. Then 1 µg of either native or mutant protein was added and allowed to react for an additional 5 min. The reaction was stopped by the addition of 80 µl of 6 M HCl and exposed to UV light at room temperature for 30 min. A 25 µl aliquot of the sample was then injected onto a Waters µBondapak C18 column and eluted isocratically from the HPLC with a mobile phase of 13% acetonitrile/87% 1 M ammonium acetate pH 5.16 (v/v), yielding baseline separation of the peaks arising from uro’gen I and uro’gen III. The peaks were monitored using a fluorescence detector with an excitation wavelength of 404 nm and an emission wavelength of 618 nm. The areas of these peaks were integrated and compared with those arising from reactions using native U3S or a non-enzymic control (using 1 µg of bovine serum albumin instead).
Acknowledgments
Acknowledgements
We thank Jim Metheral for a human HeLa cDNA library. This research was carried out in part at the NSLS, Brookhaven National Laboratory, which is supported by the US Department of Energy, Division of Materials Sciences and Division of Chemical Sciences. Part of this research was conducted at the SSRL, which is operated by the Department of Energy, Office of Basic Energy Sciences. This work was supported in part by National Institutes of Health grants GM856775, DK20503 and DK02794.
References
- Battersby A.R. (1978a) The discovery of nature’s biosynthetic pathways. Experientia, 34, 1–13. [DOI] [PubMed] [Google Scholar]
- Battersby A.R. (1978b) Ideas and experiments in biosynthesis. Ciba Found. Symp., 53, 25–51. [DOI] [PubMed] [Google Scholar]
- Battersby A.R. and Leeper,F.J. (1990) Biosynthesis of the pigments of life: Mechanistic studies on the conversion of porphobilinogen to uroporphyrinogen III. Chem. Rev., 90, 1261–1274. [Google Scholar]
- Battersby A., Fookes,C., McDonald,E. and Meegan,M. (1978) Bio synthesis of type-III porphyrins: proof of intact enzymatic conversion of the head-to-tail bilane into uro’gen III by intramolecular arrangement. J. Chem. Soc. Chem. Commun., 185–186. [Google Scholar]
- Carson M. (1991) Ribbons 2.0. J. Appl. Crystallogr., 24, 968–961. [Google Scholar]
- Cassidy M.A., Crockett,N., Leeper,F.J. and Battersby,A.R. (1991) Synthetic studies on the proposed spiro intermediate for biosynthesis of the natural porphyrins: the stereochemical probe. J. Chem. Soc. Chem. Commun., 6, 384–386. [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cowtan K.D. and Zhang,K.Y. (1999) Density modification for macromolecular phase improvement. Prog. Biophys. Mol. Biol., 72, 245–270. [DOI] [PubMed] [Google Scholar]
- de la Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy-atom parameter refinement in the MIR and MAD methods. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Desnick R.J., Glass,I.A., Xu,W., Solis,C. and Astrin,K.H. (1998) Molecular genetics of congenital erythropoietic porphyria. Semin. Liver Dis., 18, 77–84. [DOI] [PubMed] [Google Scholar]
- Drennan C.L., Huang,S., Drummond,J.T., Matthews,R.G. and Ludwig,M.L. (1994) How a protein binds B12: A 3.0 Å X-ray structure of B12-binding domains of methionine synthase. Science, 266, 1669–1674. [DOI] [PubMed] [Google Scholar]
- Engh R.A. and Huber,R. (1991) Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. A, 47, 392–400. [Google Scholar]
- Higgins D.G. and Sharp,P.M. (1989) Fast and sensitive multiple sequence alignments on a microcomputer. Comput. Appl. Biosci., 5, 151–153. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1997) Dali/FSSP classification of three-dimensional protein folds. Nucleic Acids Res., 25, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch W. and Sander,C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- Kappas A., Sassa,S., Galbraith,R.A. and Nordmann,Y. (1995) The porphyrias. In Scriver,C.R., Beaudet,A.L., Sly,W.S. and Valle.D. (eds), The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill, New York, NY, pp. 2103–2160.
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Leeper F.J. (1994) The evidence for a spirocyclic intermediate in the formation of uroporphyrinogen III by cosynthase. Ciba Found. Symp., 180, 111–123. [DOI] [PubMed] [Google Scholar]
- Mathewson J. and Corwin,A. (1961) Biosynthesis of pyrrolepigments: a mechanism for porphybilinogen polymerization. J. Am. Chem. Commun., 1313–1315. [Google Scholar]
- Murshudov G.N., Vagin,A.A., Lebedev,A., Wilson,K.S. and Dodson,E.J. (1999) Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. D, 55, 247–255. [DOI] [PubMed] [Google Scholar]
- Murzin A.G., Brenner,S.E., Hubbard,T. and Chothia,C. (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. (1993) Oscillation data reduction program. In Sawyer,L., Isaacs,N. and Bailey,S. (eds), Data Collection and Processing. SERC Daresbury Laboratory, Warrington, UK, pp. 56–62.
- Pichon C., Atshaves,B.P., Stolowich,N.J. and Scott,A.I. (1994a) Evidence for an intermediate in the enzymatic formation of uroporphyrinogen III. Bioorg. Med. Chem., 2, 153–168. [DOI] [PubMed] [Google Scholar]
- Pichon C., Atshaves,B.P., Xue,T., Stolowich,N.J. and Scott,A.I. (1994b) Studies on uro’gen III synthase with modified bilanes. Bioorg. Med. Chem., 4, 1105–1110. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V., Finch,J.T., Graziano,V., Lee,P.L. and Sweet,R.M. (1993) Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature, 362, 219–223. [DOI] [PubMed] [Google Scholar]
- Rogounovitch T., Takamura,N., Hombrados,I., Morel,C., Tanaka,T., Kameyoshi,Y., Shimizu-Yoshida,Y., de Verneuil,H. and Yamashita,S. (2000) Congenital erythropoietic porphyria: a novel homozygous mutation in a Japanese patient. J. Invest. Dermatol., 115, 1156. [DOI] [PubMed] [Google Scholar]
- Saval A.H. and Tirado,A.M. (1999) Congenital erythropoietic porphyria affecting two brothers. Br. J. Dermatol., 141, 547–550. [DOI] [PubMed] [Google Scholar]
- Scott A.I. (1993) How nature synthesizes vitamin B12—a survey of the last four billion years. Angewandte Chemie, 32, 1223–1376. [Google Scholar]
- Stark W.M., Hart,G.J. and Battersby,A.R. (1986) Synthetic studies on the proposed spiro intermediate for biosynthesis of the natural porphyrins: inhibition of cosynthase. J. Chem. Soc. Chem. Commun., 465. [Google Scholar]
- Takamura N., Hombrados,I., Tanigawa,K., Namba,H., Nagayama,Y., de Verneuil,H. and Yamashita,S. (1997) Novel point mutation in the uroporphyrinogen III synthase gene causes congenital erythropoietic porphyria of a Japanese family. Am. J. Med. Genet., 70, 299–302. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S.F., Bishop,D.F. and Desnick,R.J. (1987) Purification and properties of uroporphyrinogen III synthase from human erythrocytes. J. Biol. Chem., 262, 1268–1273. [PubMed] [Google Scholar]
- Tsai S.F., Bishop,D.F. and Desnick,R.J. (1988) Human uroporphy rinogen III synthase: molecular cloning, nucleotide sequence, and expression of a full-length cDNA. Proc. Natl Acad. Sci. USA, 85, 7049–7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner C.A., Yoo,H.W., Roberts,A.G. and Desnick,R.J. (1992) Congenital erythropoietic porphyria: identification and expression of exonic mutations in the uroporphyrinogen III synthase gene. J. Clin. Invest., 89, 693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]