Abstract
The generation of diacylglycerol (DAG) in response to receptor stimulation is a well-documented signalling mechanism that leads to activation of protein kinase C (PKC). Putative alternative effectors contain sequences that interact with DAGs, but the mechanisms of signal transduction are unknown. We have identified a Dictyostelium gene encoding a novel protein which contains a domain with high identity to the DAG-binding domain of PKC. It does not encode a PKC homologue as the conservation does not extend outside this region. We confirm that the proposed DAG-binding domain is sufficient to mediate interaction of a fusion protein with vesicles containing DAG. The protein also shows significant homology to mammalian phosphatidylinositol phosphate (PIP) kinases and we show that this domain has PIP kinase activity. The protein, PIPkinA, is enriched in the nucleus and abrogation of gene function by homologous recombination inhibits early developmental gene expression, blocking development at an early stage. Thus, we have identified a PIP kinase from Dictyostelium which is required for development, is a candidate effector for DAG and has the potential to synthesize nuclear PIP2.
Keywords: diacylglycerol/Dictyostelium/gene expression/nucleus/phosphatidylinositol phosphate kinase
Introduction
A wide range of signals lead to the transient or sustained generation of diacylglycerol (DAG) via activation of phospholipase activity (Nishizuka, 1992). The consequences of these signals for cell function are varied, but if they can be brought about by phorbol ester treatment they are assumed to involve protein kinase C (PKC) activation. In many cases, direct activation of PKC in response to increased DAG levels has been established through stabilization of membrane association of the enzyme after cell lysis. The DAG-binding domain of both novel and classical PKC family members is a cysteine- and histidine-rich motif (Ono et al., 1989). Two copies of this C6H2 motif are found in all PKC isoforms that interact with DAG, but a single copy has been shown to be sufficient for DAG interaction in vitro (Quest et al., 1994a). Other proteins have been isolated from mammalian cells which have domains homologous to those known to be responsible for DAG binding in PKC. These include n-chimaerin, a brain-specific protein isolated from human cells (Ahmed et al., 1990), the proto-oncogene c-vav (Katzav et al., 1989), and unc-13, which is encoded by a gene whose deletion causes uncoordinated movement in Caenorhabditis elegans (Maruyama and Brenner, 1991). The existence of these proteins raises the possibility that DAG can activate alternative signalling pathways in the cell and that some of the effects of DAG generation on cell function may be PKC independent.
In early development, Dictyostelium discoideum amoebae aggregate together in response to pulsatile emissions of cAMP from a signalling centre. In addition to its role as chemoattractant, extracellular cAMP acts at several stages in development to regulate gene expression (reviewed in Firtel, 1995). Extracellular cAMP binds to cell surface receptors, which stimulate multiple signalling pathways inside the cell. A phospholipase C (PLC) activity has been described which is present during early aggregation. The transient generation of inositol trisphosphate (IP3) is proposed to be due to activation of this PLC by a G protein-dependent mechanism (Bominaar et al., 1994). Hydrolysis of phosphatidylinositol bisphosphate (PIP2) by PLC would also lead to the transient generation of DAG. A role for DAG has been demonstrated in actin nucleation (Shariff and Luna, 1992), phagocytosis (Seastone et al., 1998, 1999) and regulation of gene expression (Ginsburg and Kimmel, 1989). However, the molecular details of the signalling pathways downstream of DAG generation have not been characterized. Several PKC-like activities have been identified in vegetative cells or during the early stages of development (e.g. Phillips et al., 1997), but the sequences encoding these proteins have not been determined.
We have identified a protein which contains a sequence tract that is highly homologous to the DAG-binding domain of mammalian PKCs and we show that the conserved domain can, indeed, interact with DAG. Primary structure analysis also identified a putative phosphatidylinositol phosphate (PIP) kinase domain and we demonstrate that this domain has PIP kinase activity when expressed heterologously. The protein, which we term PIPkinA, is enriched in the nucleus. Analysis of strains in which the gene encoding PIPkinA (Ddpik6) has been disrupted by homologous recombination reveals that this protein is required for early changes in developmental gene expression and aggregation.
Results
Isolation and sequence analysis of the gene encoding PIPkinA (Ddpik6)
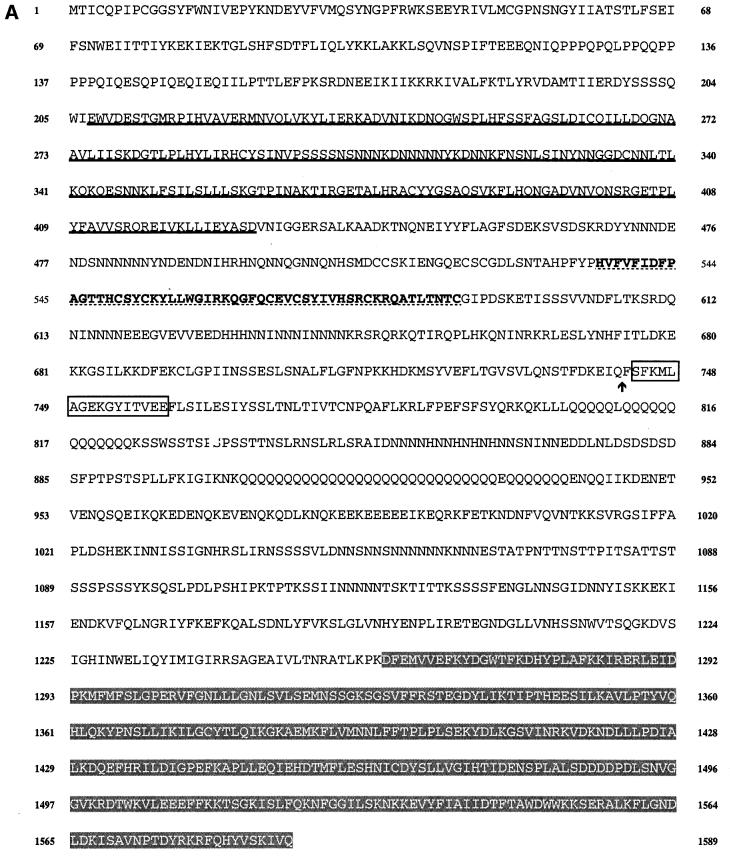
A synthetic oligonucleotide, based on the sequence of the DAG-binding domain of bovine PKCs, was used to screen a genomic library of Dictyostelium DNA and a set of overlapping clones was isolated. The entire gene sequence was determined (Figure 1A) and revealed an open reading frame of 4767 bp, which would give rise to a protein of 1589 amino acids with a predicted mol. wt of 182 kDa. Only one short intron is present in the gene, as indicated on Figure 1A. There are several long runs of repetitive AT-rich DNA within the coding sequence and several long tracts of AAC repeats. Similar AAC repeats have been found in the coding regions of many developmentally regulated genes (e.g. Agarwal et al., 1994; Schnitzler et al., 1994). As is normal for Dictyostelium genes, the non-coding DNA is extremely AT rich.
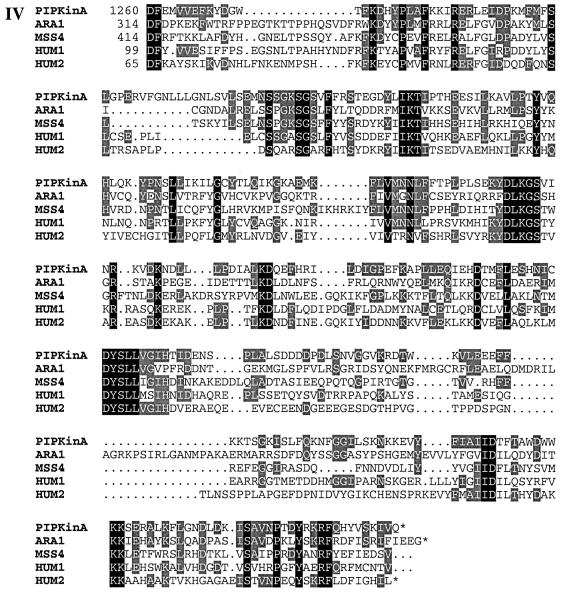
Fig. 1. (A) Sequence of the PIPkinA protein. The protein sequence of the PIPkinA open reading frame is shown. The areas of homology (as determined by BLAST sequence similarity searching) are indicated with the ankyrin-repeat domain underlined, the DAG-binding domain in bold with dotted underlining, the α-actinin homology domain boxed and the PIP kinase domain shaded. The position of the single small intron is marked with an arrow. The nucleotide sequence has been submitted to the DDBJ/EMBL/GenBank database under accession No. AF339903. (B) Sequence comparisons between PIPkinA and other proteins. Searching the EMBL database with the PIPkinA protein sequence revealed several areas of homology to other proteins which are shown here. (I) The homology between the DAG-binding domain of PIPkinA and the first copies of the repeat sequence from Drosophila PKC (Rosenthal et al., 1987) and bovine PKCγ (Coussens et al., 1986). The conserved His and Cys residues defining the motif are shaded and residues conserved in PIPkinA are underlined. N- and C-terminal deletion of the motif in PKCγ has revealed the N-terminal histidine residue to be essential, whereas the C-terminal cysteine can be deleted without loss of binding (Quest et al., 1994b). The spacing between these two residues and the central motif is different in PIPkinA with the insertion of extra amino acids, showing that the spacing is not critical for DAG association. (II) The sequence of the five ankyrin repeats found in PIPkinA with a consensus sequence (as defined by Bork, 1993) shown for comparison. Residues matching the consensus are underlined in each copy of the repeat. These 33 amino acid repeats have been found in a wide range of proteins including transcription factors such as SWI6 (Breeden and Nasmyth, 1987) and signalling proteins such as the Drosophila notch protein (Kidd et al., 1986). (III) The short stretch of sequence homology between PIPkinA and human α-actinin from skeletal muscle (SM) and non-muscle (NM) (Beggs et al., 1992). Residues conserved in PIPkinA are underlined. (IV) Alignment of the C-terminal region of PIPkinA with the PIPKs from Arabidopsis (ARA1) (Mikami et al., 1998), S.cerevisiae (MSS4) (Yoshida et al., 1994), human type 1α (HUM1) and human type IIα (HUM2) (Boronenkov and Anderson, 1995). Amino acids conserved in all five sequences are shaded in black and those conserved with PIPkinA are shaded in grey.
Sequence analysis revealed a stretch of DNA with high homology to the probe, and the putative protein sequence of this region has a high degree of identity to the DAG-binding domain of mammalian PKCs (Figure 1BI). The highest degree of homology is seen with a PKC homologue isolated from Drosophila (69% over the central portion) (Rosenthal et al., 1987), but there is significant homology to the mammalian sequences (62% with the first repeat from bovine PKCγ) (Coussens et al., 1986).
The PIPkinA open reading frame does not encode a PKC homologue as there is no significant homology to protein kinases (Hanks et al., 1988), and the sequence homology to mammalian PKCs does not extend outside the area required for DAG interaction. On searching a database of protein sequences, three other areas of interest became apparent. PIPkinA contains a region with a high degree of homology to human ankyrin. The homology lies in a region containing multiple repeats of the 33 amino acid ankyrin repeat, a motif present in a wide range of proteins (Sedgwick and Smerdon, 1999). In many cases, ankyrin repeats have been shown to be responsible for mediating protein–protein interactions. Dictyostelium PIPkinA has five copies of the ankyrin repeat (Figure 1BII). The third and fourth copies are interrupted by a 57 amino acid region containing several stretches of a single amino acid, most commonly asparagine. Similar homopolymeric stretches of amino acids have been reported in many Dictyostelium proteins, although their function is unknown (e.g. Agarwal et al., 1994; Schnitzler et al., 1994). The protein also contains a short region with high homology to a region of α-actinin (Figure 1BIII). The significance of the α-actinin homology region is not known.
The most extensive region of homology is between the C-terminal 330 amino acids and the phosphatidylinositol phosphate kinases (PIPKs) (Figure 1BIV). This homology includes the sequences that are conserved between all family members. PIPkinA shows greatest homology to human and yeast type I enzymes PIP5KIα (32%), Mss4 (31%), PIPKIIα (28%) and plant PIP kinases identified in Arabidopsis (30%) and Nicotiana (27%) (Yoshida et al., 1994; Boronenkov and Anderson, 1995; Mikami et al., 1998). Mammalian PIPKs of this family fall into two types. The type I enzymes phosphorylate PI 4-phosphate in the 5 position, whereas type II enzymes phosphorylate PI 5-phosphate in the 4 position (Rameh et al., 1997). PIPkinA shows a similar degree of homology to both classes. It has been shown that sequences at the C-terminus of PIPKs, called the activation loop, contribute to determining the substrate specificity of PIPKs (Kunz et al., 2000). This region of PIPkinA (residues 1540–1589) again shows a comparable degree of homology to both classes (36% with human type Iα and 38% with human type IIα) over this region and does not show distinguishing features which allow it to be placed in either class. The PIP kinase domain of PIPkinA shows a much lower degree of homology (20%) to the related family of PI 3-phosphate kinases such as Fab1p (Yamamoto et al., 1995; Cooke et al., 1998).
Pattern of expression of the gene encoding PIPkinA
The 5.1 kb PIPkinA transcript is expressed at an undetectable level in cells growing on a bacterial food source. It increases in abundance very rapidly upon starvation and is maintained at this level throughout the remainder of aggregation (Figure 2). The transcript continues to be present at a reduced level throughout the rest of development. As with many genes expressed during early development, the PIPkinA mRNA is present in axenically growing cells at an abundance characteristic of cells in early aggregation (data not shown).

Fig. 2. Developmental expression of PIPkinA. RNA was isolated from V12M2 Dictyostelium cells grown on bacteria and following starvation on agar plates to induce development. At the times indicated, total cellular RNA was extracted and the resulting northern blot probed with an RNA probe from the gene encoding PIPkinA. An identical transcript was detected using a 3′ proximal probe (data not shown).
Subcellular localization of PIPkinA
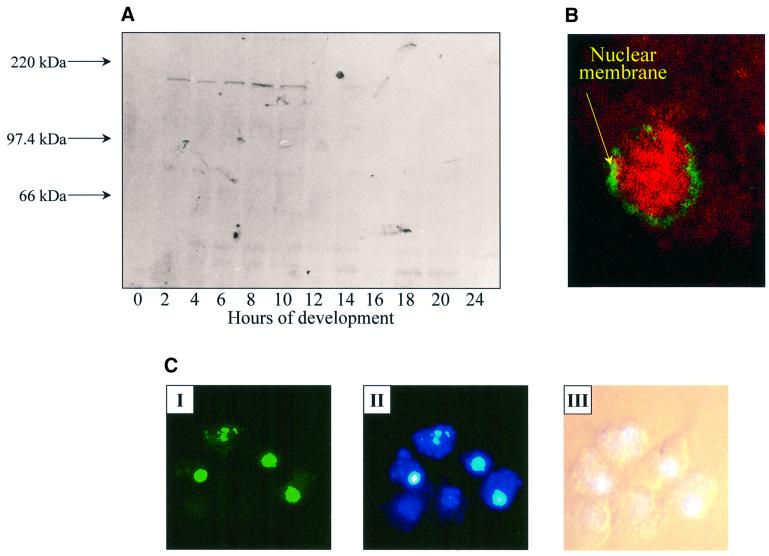
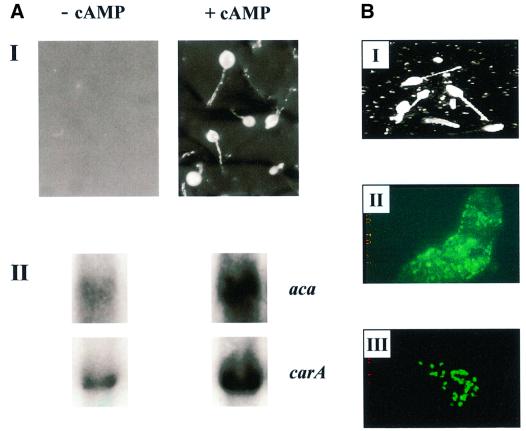
A polyclonal antiserum was raised against the ankyrin-repeat region of PIPkinA and western blot analysis confirmed the interaction of the antiserum with a single band of ∼180 kDa, with a time course of expression consistent with that seen by northern blot analysis (Figure 3A). Immunofluoresence revealed a nuclear enrichment of PIPkinA in growing cells (Figure 3B). The green staining indicates the position of the nuclear membrane, revealed using mab17, a monoclonal antibody that recognizes a component of the nuclear pore apparatus (Fukuzawa and Williams, 1997). Antibodies raised against the putative DAG-binding domain of PIPkinA also showed a nuclear enrichment (data not shown). Expres sion of a PIPkinA–green fluorescent protein (GFP) fusion protein, driven by a Dictyostelium promoter that expresses constitutively in early development, revealed that the fusion protein was enriched in nuclei of growing Dictyostelium amoebae, further confirming the nuclear localization of PIPkinA (Figure 3C). Cells expressing GFP alone showed no significant nuclear enrichment (data not shown). No change in the localization of the PIPkinA–GFP fusion protein was apparent in cells developed through to the aggregation stage of development (data not shown).
Fig. 3. Localization of PIPkinA protein. (A) Affinity-purified antisera raised against the ankyrin-repeat region of PIPkinA were used to investigate the time course of expression of the PIPkinA protein by western blotting of a developmental time course. Ax2 cells were developed on filters and harvested at the times shown. (B) The staining pattern observed when the affinity-purified antibody against the ankyrin-repeat domain (visualized using rhodamine) was used to stain vegetative amoebae. The green staining indicates the position of the nuclear membrane, revealed using a mouse monoclonal antibody (mab17) that recognizes a component of the nuclear pore apparatus (Fukuzawa and Williams, 1997). (C) A fusion protein containing the PIPkinA coding sequence with GFP fused to the C-terminus was expressed in Dictyostelium cells using a constitutive actin 15 promoter. The localization of the GFP fusion protein in nuclei was observed by fluorescence microscopy (I) and the enrichment in nuclei confirmedby staining with Hoechst 33258 (II). The bright field picture is shown for comparison (III).
PIPkinA is a DAG-binding protein
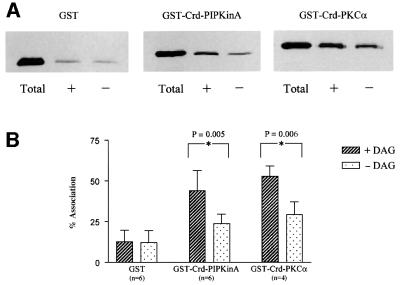
The ability of the cysteine-rich domain (Crd) to interact with DAG was determined by examining the association of a fusion protein containing the Crd with lipid vesicles containing DAG (Quest et al., 1994a). The sequence encoding the Crd (residues 528–591) was inserted downstream of the glutathione S-transferase (GST) gene so as to create a fusion protein. This protein was purified from bacterial lysates and its ability to associate with artificial lipid vesicles containing DAG was monitored by western blotting using antisera directed against the GST portion of the fusion protein. Association of the GST portion alone with the lipid vesicles was low and not significantly increased by the inclusion of DAG. In contrast, the Crd–GST fusion protein was able to associate with lipid vesicles and this association was significantly stimulated by the inclusion of DAG in the vesicles (Figure 4). Under identical conditions, a fusion protein containing GST fused to both of the Crds from bovine PKCα (residues 46–154) showed a comparable level of vesicle association to that of the PIPkinA Crd–GST fusion protein, both in the absence and presence of DAG. No increased association of the PIPkinA Crd was observed with vesicles containing a range of other lipids including PI(4,5)P2 (data not shown). It did not bind to the phorbol ester phorbol 12,13-dibutyrate (PDBu) in a phorbol binding assay, although, as a control, PDBu showed the expected high level of binding to the Crd of PKCα (data not shown).
Fig. 4. Association of GST fusion proteins with artificial membrane vesicles. (A) Lipid vesicles containing (+) or not containing (–) 5mol% DAG were incubated with GST alone or with fusion proteins incorporating the Crd from PIPkinA or bovine PKCα at room temperature for 30 min (Quest et al., 1994a). The vesicles were separated from the supernatant by centrifugation and the proportion of protein associated with the vesicles, compared with that originally added to the incubation, was assessed by western blotting using antisera directed against GST. Lanes marked ‘Total’ represent the amount of GST or fusion protein utilized in each assay. (B) Protein band intensities from western blots (as described above) were quantified by scanning densitometry. The term ‘% Association’ was defined as the intensity of a protein band recovered from a vesicular pellet expressed as a percentage relative to the intensity determined for the ‘Total’ amount of protein in each assay. All errors shown represent standard deviations of averaged values. The data was analysed using one-way analysis of variance (ANOVA) and showed significant increases in association in the presence of DAG for both GST–Crd–PIPkinA and GST–Crd–PKCα (p values indicated on the graph), but no significant increase for GST alone.
PIPkinA encodes PIP kinase activity
The C-terminal 369 amino acids of PIPkinA were fused in-frame to GST and the lipid kinase activity of the purified protein confirmed by the generation of PIP2 from PIP (Figure 5). The addition of phosphatidic acid did not stimulate the kinase activity, unlike mammalian type I enzymes (data not shown).
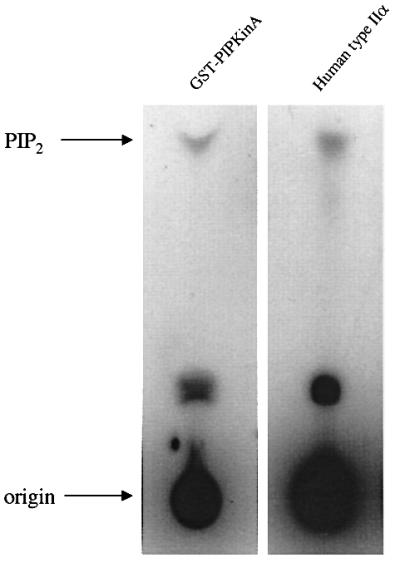
Fig. 5. PIP kinase activity of the C-terminal domain of PIPkinA.GST alone or a fusion protein of GST and the C-terminal domain of PIPkinA was purified by elution from glutathione–Sepharose 4B beads, and incubated with PIP in the presence of [γ-32P]ATP for 30 min at 22°C. Recombinant human type IIα PIP kinase was used as a control. Lipids were extracted into chloroform/methanol and the generation of radiolabelled PIP2 was assessed by TLC.
PIPkinA protein plays a role in early development
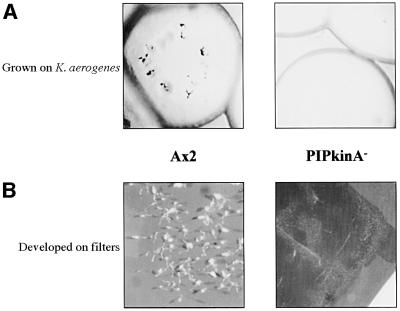
The function of the PIPkinA protein was investigated by abrogating gene function by homologous recombination. Gene disruption was confirmed by Southern blot analysis and no transcript from the gene encoding PIPkinA was detectable by RT–PCR (data not shown). Two independently isolated disruptant strains showed identical phenotypes. PIPkinA– cells show a marked defect in their ability to develop. The null strain does not aggregate when allowed to develop after growth on a bacterial lawn. When plated for development on filters, most of the cells failed to form aggregates even after 48 h (Figure 6). However, at the edges of the filters, small aggregates occasionally formed and went on to produce small, but apparently normal, fruiting bodies.
Fig. 6. Phenotype of developing PIPkinA– cells. (A) Ax2 and PIPkinA– cells grown and developed in association with Klebsiella aerogenes. Individual clones of cells grow by feeding on the bacterial lawn, and as they clear the lawn, parental cells initiate development in the centre of cleared area. PIPkinA– cells clear the lawn as normal but no aggregates form. (B) Ax2 and PIPkinA– cells developed on filters for 48 h. Mature fruiting bodies are visible in the parental Ax2 cells, whereas the PIPkinA– cells fail to aggregate and develop.
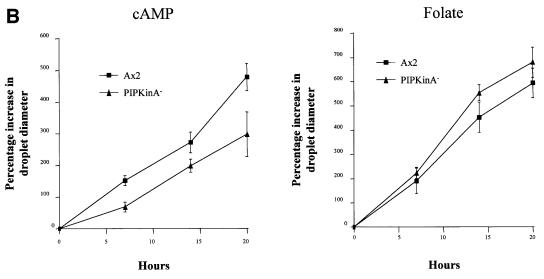
The defect in aggregation could be due to a defect in the ability of the cells to undergo chemotaxis or to generate cAMP. Northern analysis revealed defects in induction of early developmental genes in PIPkinA– cells (Figure 7A). There was a slight increase in the expression of the cAMP receptor cAR1, but not the further increase which is associated with the onset of cAMP pulses. There are also significantly lower levels of the transcripts encoding the extracellular phosphodiesterase (PDE) and its inhibitor (PDI), and the adenylyl cyclase required to generate the relay signal (ACA). Consistent with the primary defect being in gene expression, PIPkinA– cells were able to undergo chemotaxis when plated on buffered agar containing either folic acid or cAMP (Figure 7B). This spreading drop assay suggests that the chemotactic response is still intact; the reduced response to cAMP is most likely explained by the low levels of expression of the genes required, such as the cAMP receptor cAR1.
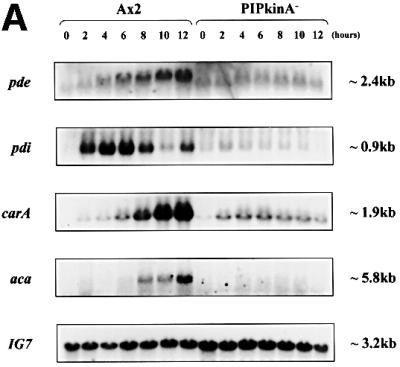
Fig. 7. Aggregation defect in PIPKinA– cells. (A) Northern blot analysis of early developmental gene expression in PIPkinA– cells. Ax2 and PIPkinA– cells were allowed to develop on filters and RNA was extracted at 2 h intervals. The resulting northern blot was probed for the expression of a range of genes normally induced during early development, and also IG7, a constitutively expressed gene, to control for equal RNA loading. aca, adenylyl cyclase; pde, phosphodiesterase; pdi, phosphodiesterase inhibitor; carA, the cAMP receptor cAR1. (B) Chemotaxis assay. The ability of parental Ax2 and PIPkinA– cells to undergo chemotaxis was investigated using the spreading spot assay as described in Tillinghast and Newell (1987). One microlitre droplets of cells were placed on the surface of non-nutrient buffered agar containing either cAMP or folate at a concentration of 10 µM. At the times indicated, the percentage increase in droplet diameter was calculated as described in Varnum and Soll (1981). All errors shown are standard deviations of averaged values following measurements from five droplets of cells.
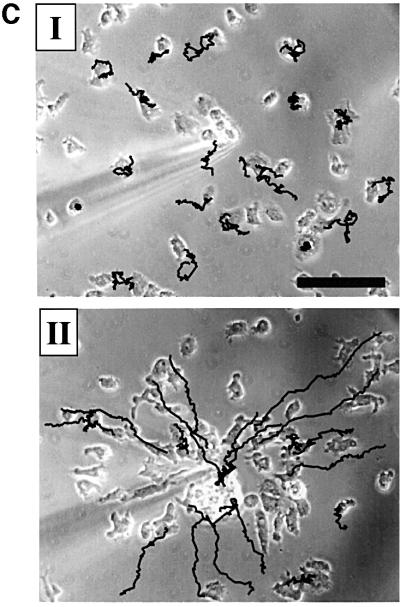
The aggregation defect could be rescued by exogenously pulsing the cells with cAMP for 5 h in shaking suspension prior to plating on filters, or by mixing experiments (Figure 8). Pulsing the cells with cAMP was able to induce expression of cAR1 and a slight increase in expression of ACA was also apparent on a long exposure of the northern blot (Figure 8A). The ability of PIPkinA– cells to respond normally when provided with exogenous pulses was confirmed using mixtures containing PIPkinA– cells marked by expression of GFP driven by a constitutive promoter, and parental Ax2 cells. The mixtures were allowed to develop and the mutant cells were able to respond normally to the signals generated by the parental cells and were, therefore, included in the aggregates. As few as 25% of parental cells were sufficient to rescue the aggregation of the majority of PIPkinA– cells. The mutant cells were evenly distributed throughout the slug and the final fruiting body, and were able to contribute to stalk and spore cell formation (Figure 8B). The slightly reduced chemotaxis to cAMP seen in the spreading drop assay is most likely due to the reduced expression of the developmentally regulated genes such as the cAMP receptor cAR1. This was confirmed by comparing the movement of pulsed and non-pulsed PIPkinA– cells towards a source of cAMP. PIPkinA– cells which had been developed in shaking suspension with cAMP pulses to induce gene expression showed increased polarity and movement towards the cAMP source compared with cells developed without exogenous pulses (Figure 8C).
Fig. 8. PIPkinA– cells can be rescued by exogenous cAMP pulsing. (A) PIPkinA– cells were developed for 5 h in shaking suspension, with or without the addition of pulses of cAMP (100 nM every 5 min). The cells were then plated on filters for development. Cells were harvested after 5 h of pulsing, and the levels of the transcripts encoding cAR1 and ACA were compared by northern blotting. Prolonged exposure of the blot was required to detect the low level of the ACA transcript. The blot was re-probed with IG7 to confirm equal loading (data not shown). (B) Parental Ax2 cells (25%) were mixed with 75% PIPkinA– cells, marked by the expression of GFP from a constitutive promoter. The cells were plated for development on filters and the final fruiting bodies photographed after 24 h (I). The distribution of GFP-labelled PIPkinA– cells in the slug (II) was determined by fluorescence microscopy, as was the production of labelled spore cells (III). (C) PIPkinA– cells were developed in shaking suspension for 6 h without (I) and with (II) cAMP pulsing. The images show the chemotactic response towards a micropipette containing 10–4 M cAMP after 30 min. The cells that have not been pulsed show a poor chemotactic response. The cells that have been pulsed for 6 h show a strong chemotactic response. The black lines indicate the cell movement tracks over the preceding 9 min of the sequence. Scale bar: 50 µm.
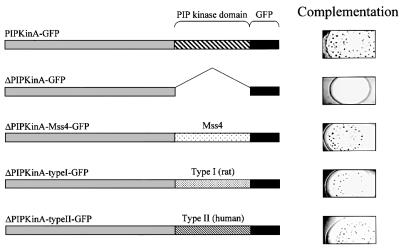
Complementation of the aggregation defect in PIPkinA– cells
The aggregation defect in PIPkinA– cells could be rescued by expression of a PIPkinA–GFP fusion protein driven by a constitutive promoter (Figure 9). Complementation of the aggregation defect was dependent on the presence of the PIP kinase domain, as expression of a fusion protein lacking this domain failed to complement the defect. In order to determine whether PIPkinA can be classified as a type I or type II enzyme, constructs were made to express chimeric proteins in which the PIP kinase domain of PIPkinA was replaced with either type I or type II sequences. When the PIP kinase domain of PIPkinA was replaced with the equivalent sequences from Saccharomyces cerevisiae Mss4 (type I), rat type Iβ and human type IIβ, all of these chimeric proteins were able to rescue the aggregation defect (Figure 9). This confirms that PIPkinA is a PIPK, but suggests that it cannot readily be assigned into one of the previously identified classes.
Fig. 9. Complementation of the aggregation defect in PIPkinA– cells. The constitutive actin 15 promoter was used to drive expression of a PIPkinA–GFP fusion protein in PIPkinA– cells. Constructs in which the PIP kinase domain was removed from the fusion protein (ΔPIPkinA–GFP), and then subsequently replaced with the equivalent domain from Mss4 (ΔPIPkinA–Mss4–GFP), rat type Iβ (ΔPIPkinA–typeI–GFP) and human type IIβ (ΔPIPkinA–typeII–GFP), were generated as described in Materials and methods. These fusion proteins were expressed in PIPkinA– cells and the ability of the resulting cells to form aggregates, following growth on bacteria, assessed. All the fusion proteins were expressed at comparable levels (data not shown).
Discussion
In early Dictyostelium development, the signalling pathways stimulated by pulses of cAMP lead to many changes in cell function. These include the production of more cAMP via the activation of adenylyl cyclase to relay the signal, cytoskeletal reorganization to allow chemotaxis towards the initial source of cAMP, and changes in gene expression. A PLC activity has been described during this early phase (Bominaar et al., 1994) that leads to the generation of DAG, and recent evidence suggests that it is activated by an extracellular factor used by cells to sense their density (conditioned medium factor; CMF) via a heterotrimeric G protein-dependent pathway (Brazill et al., 1998). A PLC gene has been isolated from Dictyostelium and is a member of the PLCδ family (Drayer and van Haastert, 1992).
We have identified a protein which contains a DAG-binding domain (Crd) and demonstrated that the Crd is sufficient to mediate interaction of a GST fusion protein with artificial lipid vesicles containing DAG. The degree of interaction is comparable with that mediated by the two copies of this motif found in bovine PKCα when expressed under the same conditions, and is of a similar level to the published association of a single repeat from PKCγ (Quest et al., 1994a). Unlike the equivalent domain in PKCα and n-chimaerin, the DAG-binding domain of PIPkinA does not interact with the phorbol ester PDBu. This is not surprising since not all known isoforms of PKC are activated by phorbol esters, and this is especially true for the PKCs found in lower eukaryotes (Ogita et al., 1990).
Despite the presence of a C6H2 motif, the gene clearly does not encode a PKC homologue because it contains none of the conserved sequences present in all known protein kinases. This raised the possibility that PIPkinA transmits a DAG signal via an alternative route. Clues as to PIPkinA function lie in the presence of other conserved protein motifs. The presence of ankyrin repeats in PIPkinA, for example, suggests that they may interact with specific proteins in the cell and this interaction may mediate or localize signal transduction in response to DAG generation.
There is a high degree of homology between the C-terminal 330 amino acids of PIPkinA and the PIP kinases of other eukaryotes, including the conservation of motifs conserved between all other family members which are known to play a role in catalysis (reviewed in Anderson et al., 1999). The homology between PIPkinA and both type I and type II PIPKs is comparable, including that seen between the activation domains that contribute to substrate selection (Kunz et al., 2000). The lower degree of sequence homology with the PIPKs responsible for phosphorylation of PI 3-P (such as Fab1) makes it much less likely that PIPkinA is a member of this family. We have shown that a GST fusion protein incorporating the C-terminal 369 residues from PIPkinA possesses PIPK activity. The substrate used contains a mixture of lipids, including both PI 4-P and PI 5-P, and so does not distinguish between type I and type II enzyme activity. Attempts made to determine the specificity using pure synthetic PI 4-P and PI 5-P as substrates have so far been unsuccessful (K.Guo, C.Pears and K.Hinchliffe, unpublished observations). Either these are not the correct substrates or an activator present in the impure preparations is missing from the pure synthetic lipids. Strikingly, fusion proteins containing the PIP kinase domain of PIPkinA replaced with the equivalent sequences from both mammalian type I and type II PIPKs were able to complement the aggregation defect associated with disruption of the gene encoding PIPkinA. This is unlike the situation in yeast, where the lethality associated with disruption of the type I MSS4 gene can only be rescued by expression of type I and not type II enzymes (Kunz et al., 2000). The similar degree of sequence homology with both type I and type II enzymes and the in vivo complementation results suggest that PIPkinA cannot be easily classified as a type I or type II enzyme.
The function of the PIPkinA protein has been investigated by generating strains in which the gene has been disrupted. Loss of PIPkinA function is not lethal to Dictyostelium cells, unlike loss of the homologous Mss4 in yeast (Yoshida et al., 1994). This suggests either that another PIPK is present during growth or that PIP2 is not essential for growth of Dictyostelium. Analysis of the data from the Dictyostelium genome sequencing project reveals the presence of at least two more potential PIPK genes (C.Pears, unpublished observations). Early development is impaired in PIPkinA– cells which fail to form aggregates on filters. In other organisms, type I PIPKs have also been shown to play a role in various cellular processes (Anderson et al., 1999), including remodelling the actin cytoskeleton and in secretion and vesicular transport. We have no evidence that changes in the actin cytoskeleton play a significant part in the aggregation defect of PIPkinA– cells. PIPkinA– cells can undergo chemotaxis, and providing exogenous pulses of cAMP greatly increases both the polarization of the cells and their ability to move towards a source of cAMP. PIPkinA– cells can also participate normally in aggregation either when pulsed with exogenous cAMP or when mixed with parental Ax2 cells. In the resulting chimeric fruiting bodies, the null cells show an ability to make both spore and stalk cells. This is consistent with a role for PIPkinA in early development, but suggests no direct role in cell fate decisions.
Analysis of the expression of early developmental genes in PIPkinA– cells reveals loss of expression of genes induced by cAMP pulses (e.g. cAR1) and those dependent on the activity of cAMP-dependent protein kinase (e.g. PDI) (Schulkes and Schaap, 1995). As the aggregation defect can be rescued by pulsing the cells with cAMP, the main defect must be the inability to generate pulses, and once pulses can be made, the other genes required can be induced sufficiently to allow the completion of development. Cell–cell signalling can be conveniently assayed by the appearance of optical density waves, which directly reflect underlying cAMP waves through the aggregation field (reviewed in Patel et al., 2000). When PIPkinA– cells are plated for development, very few of these dark field waves are apparent and they always initiate from the edge of the filters. If the cells are pulsed before plating, many more centres are apparent and they are scattered all over the filter (D.Dormann and C.J.Weijer, unpublished observations). Expression of cAR1 is rescued by pulsing the null cells with exogenous cAMP. Interestingly we can detect very little expression of ACA, the adenylyl cyclase involved in generating cAMP for events in early aggregation, although a slight increase in ACA expression levels was induced by pulsing. The induction of ACA is one of the earliest events in development. This suggests that PIPkinA may act in a signalling pathway very early during development and would be consistent with it playing a part in signals transduced by CMF, the protein that stimulates PLC activity. PIPkinA– cells showed activation of cAMP-induced signalling pathways such as activation of the MAPkinase ERK2 (data not shown), which was only slightly reduced compared with parental cells. This, combined with the localization of PIPkinA, suggests that the defect is likely to be at the level of gene expression in the nucleus rather than at the plasma membrane.
DAG, PIP2 and both type I and type II PIPKs have been found in the nuclei of mammalian cells and, although their role there is unknown, there is suggestive evidence that they may be important for the regulation of gene expression (Boronenkov et al., 1998; D’Santos et al., 1998). The constitutive presence of Dictyostelium PIPkinA in the nucleus also suggests a function in the generation of a nuclear pool of PIP2 required for the gene expression changes during early development. The presence of a DAG-binding domain and the ankyrin-repeat motif in PIPkinA suggest potential mechanisms for the regulation of enzyme activity and localization, and will facilitate further genetic analysis to delineate possible molecular links between signal transduction, nuclear phosphatidylinositol metabolism and gene expression.
Materials and methods
Isolation of a gene encoding PIPkinA (Ddpik6) and sequence determination
The gene encoding PIPkinA was isolated from a genomic library, using as a probe an oligonucleotide of 77 residues, designed to encode the N-terminal cysteine and histidine-rich repeat from bovine PKCα, but using Dictyostelium codon preferences. The sequence of the gene was determined from an overlapping set of genomic clones and from cDNAs isolated from a bacteriophage λ library prepared using mRNA from growing cells. The presence of an intron was predicted by the extreme AT content of non-coding Dictyostelium DNA and confirmed by sequencing the mRNA directly using PCR products primed with oligonucleotides flanking the predicted intron. We have called the gene Ddpik6, following the nomenclature of Zhou et al. (1995), who isolated five Dictyostelium inositol lipid kinase genes.
Generation of a GST–Crd fusion protein and assessment of membrane association
PCR was used to amplify the DNA sequence between nucleotide residues 1584 and 1773 of the Ddpik6 coding sequence to include the putative DAG-binding region (Crd). The PCR product was inserted into the expression vector pGEX-KG (Pharmacia) such that a fusion protein between the GST protein and the required portion of PIPkinA was expressed in Escherichia coli. A similar strategy was used to create a fusion protein containing amino acid residues 46–154 of bovine PKCα, containing two Crd repeats, as a positive control for DAG association. The GST fusion proteins, and GST alone, were purified by affinity chromatography using standard protocols.
The ability of the fusion proteins to associate with artificial lipid vesicles was determined as described in Quest et al. (1994a). Liposomes (100 µg/ml) of phosphatidylserine and phosphatidylcholine (ratio 1:1) were incubated with GST or GST fusion proteins (1 µg) in 50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 1 mM dithiothreitol, 5 µg/ml butylated hydroxytoluene, 0.1 mM diethylene triamine penta-acetic acid and 1 mM EGTA. 1,2-dioleoyl glycerol (C18:1, [cis]-9) (Sigma) was added to 5 mol% of the lipid. After incubation at room temperature for 30 min, the liposomes were pelleted by centrifugation at 100 000 r.p.m. in a Beckman TL-100 centrifuge for 15 min at 4°C. The pellet and supernatant were separated, and the fusion proteins associated with the vesicles analysed by western blotting using anti-GST antisera (Sigma) and visualized by enhanced chemiluminescence (Amersham Pharmacia Biotech.).
Generation of GST–PIPkin fusion protein and PIP kinase assay
The putative PIP kinase domain, amino acid residues 1220–1589 of the open reading frame, was amplified by PCR and fused in-frame to GST. After expression in bacteria, the fusion protein (GST–PIPK) was purified using glutathione–Sepharose 4B beads following the manufacturer’s instructions (Amersham Pharmacia Biotech.).
The kinase reaction was carried out in 50 µl reactions performed for 30 min at room temperature in a final concentration of 50 mM Tris–HCl pH 7.4, 2 mM EGTA, 20 mM MgCl2, 50 µM PIP, 0.4% Triton X-100, 50 µM [γ-32P]ATP (10 µCi) and 10 µg of purified GST or GST–PIPK protein, in the presence or absence of phosphatidic acid (100 µM). The reaction was stopped by the addition of 200 µl of 1 N HCl. Phospholipids were extracted with chloroform:methanol:(12 N) HCl (200:100:1), separated by thin-layer chromatography in chloroform:methanol: H2O:(15 N) NH4OH (90:90:20:7) and detected by autoradiography.
Generation of antibodies
Polyclonal antisera were raised in rabbits against purified GST fusion proteins containing the PIPKinA ankyrin-repeat motif (amino acid residues 207–429). Depletion of antisera against GST and subsequent purification on a fusion protein affinity column were carried out as described in Maeda and Firtel (1997).
Growth and development of Dictyostelium
Dictyostelium strain Ax2 was grown in HL-5 medium at 22°C (Watts and Ashworth, 1970). For development, exponentially growing cells were harvested and washed three times in KK2 (16.5 mM KH2PO4, 3.8 mM K2HPO4 pH 6.2) and developed on Millipore nitrocellulose filters supported on a buffered pad. Chemotaxis assays were carried out as described in Tillinghast and Newell (1987) and the ratio of the diameter of the spot on agar containing the chemoattractant to the initial diameter plotted against time (Varnum and Soll, 1981). For pulsing experiments, cells were resuspended in KK2 at 1 × 107/ml, shaken at 110 r.p.m. for 2 h and then pulsed with 100 nM cAMP at 5 min intervals for 5 h.
Isolation of RNA and northern blot analysis
Total cellular RNA was isolated and northern blot analysis carried out as described in Huang et al. (1997). A riboprobe was used to detect expression of mRNA encoding PIPkinA. The RNA probe was transcribed using T7 polymerase from a subclone of a central AccI fragment (nucleotides 635–1768) from the Ddpik6 gene. Expression of early developmental genes was detected using randomly primed 32P-labelled cDNA.
Generation of PIPkinA– cells
A 3.76 kb fragment of the PIPkinA coding region (nucleotide residues 31–3796) was inserted into the vector pGEM-7Zf(+). A cassette encoding the blasticidin resistance gene downstream of the actin 15 promoter was inserted into the NheI site at position 2243 within the coding region of PIPkinA, relative to the start site of the open reading frame (labelled +1). The resulting plasmid was electroporated into Ax2 cells and blasticidin-resistant clones screened for homologous recombinants by PCR using primers flanking the insertion site. Two single clones with a disrupted Ddpik6 gene were identified in two separate electroporation experiments, and these clones showed indistinguishable phenotypes. PIPkinA– cells were grown in medium supplemented with 5 µg/ml blasticidin.
Complementation experiments
A vector to express a PIPkinA–GFP fusion protein in Dictyostelium was created using a restriction fragment stretching from residues 31 to 4767 of Ddpik6, which contains the entire coding sequence apart from the first 10 amino acids. This was inserted into the pAct15-GFP vector (kindly provided by W.-T.Chang) between the constitutive actin 15 promoter (including the actin 15 start codon) and the sequences coding for GFP. A construct lacking the PIP kinase domain of PIPkinA (ΔPIPkinA) was generated by inserting a DNA fragment from 31–3796 into the same vector to create pKG3001. The PIP kinase domain was replaced with the homologous domains from other PIPKs by PCR amplification of the relevant fragment and insertion between the deleted PIPkinA sequences and GFP in pKG3001. The sequences encoded the C-terminal 333 amino acid residues of Mss4 from S.cerevisiae (Yoshida et al., 1994), 320 residues of rat type Iβ and 340 amino acid residues of human type IIβ (Ciruela et al., 2000). The resulting plasmids were all sequenced to confirm that all the fusions were in-frame and that no mutations were present following PCR amplification. The plasmids created were transformed into the PIPkinA– strain and selected with 20 µg/ml G418.
Acknowledgments
Acknowledgements
We are particularly indebted to Kath Hinchliffe for her help with lipid kinase assays as well as providing recombinant type I and type II enzymes and cDNA clones. We would like to thank H.Mahubani, J.Kirk, E.Coghill and A.Salako for technical assistance at various stages of the project, and Julian Gross and Nigel Goode for critical reading of the manuscript. We are grateful to M.Hall and T.Beck for supplying the MSS4 gene. This work was supported by Wellcome Trust Programme Grant 039899/Z to J.G.W., the BBSRC and Wellcome Grant 039430 to C.P. R.N. is the recipient of a BBSRC studentship.
References
- Agarwal A., Sloger,M., Oyama,M. and Blumberg,D. (1994) Analysis of a novel cAMP inducible prespore gene in Dictyostelium: evidence for different patterns of cAMP regulation. Differentiation, 57, 151–162. [DOI] [PubMed] [Google Scholar]
- Ahmed S., Kozma,R., Monfries,C., Hall,C., Lim,H., Smith,P. and Lim,L. (1990) Human brain n-chimaerin cDNA encodes a novel phorbol ester receptor. Biochem. J., 272, 767–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R., Boronenkov,I., Doughman,S., Kunz,J. and Loijens,J. (1999) Phosphatidylinositol phosphate kinases, a multifaceted family of signaling enzymes. J. Biol. Chem., 274, 9907–9910. [DOI] [PubMed] [Google Scholar]
- Beggs A., Byers,T., Knoll,J., Boyce,F., Bruns,G. and Kunkel,L. (1992) Cloning and characterisation of two human skeletal muscle α-actinin genes located on chromosomes 1 and 11. J. Biol. Chem., 267, 9281–9288. [PubMed] [Google Scholar]
- Bominaar A.A., Kesbeke,F. and Van Haastert,H.P. (1994) Phospholipase C in Dictyostelium. Cyclic AMP surface receptor and G-protein-regulated activity in vitro. Biochem. J., 297, 181–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P. (1993) Hundreds of ankyrin-like repeats in functionally diverse proteins: mobile modules that cross phyla horizontally. Proteins, 17, 363–374. [DOI] [PubMed] [Google Scholar]
- Boronenkov I. and Anderson,R. (1995) The sequence of phosphatidylinositol-4-phosphate, 5-kinase defines a novel family of lipid kinases. J. Biol. Chem., 270, 2881–2884. [DOI] [PubMed] [Google Scholar]
- Boronenkov I., Loijens,J. and Anderson,R. (1998) Phosphoinositide signalling pathways in nuclei are associated with nuclear speckles containing pre-mRNA processing factors. Mol. Biol. Cell, 9, 3547–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazill D., Lindsey,D., Bishop,J. and Gomer,R. (1998) Cell density sensing mediated by a G protein coupled receptor activating phospholipase C. J. Biol. Chem., 273, 8161–8168. [DOI] [PubMed] [Google Scholar]
- Breeden L. and Nasmyth,K. (1987) Similarity between cell cycle genes of budding yeast and fission yeast and the notch gene of Drosophila. Nature, 329, 651–654. [DOI] [PubMed] [Google Scholar]
- Ciruela A., Hinchliffe,K., Divecha,N. and Irvine,R. (2000) Nuclear targeting of the β isoform of type II phosphatidylinositol phosphate kinase by its α-helix 7. Biochem. J., 346, 3587–3591. [PMC free article] [PubMed] [Google Scholar]
- Cooke F., Dove,S., McEwen,R., Painter,G., Holmes,A., Hall,M., Michell,R. and Parker,P. (1998) The stress-activated phosphatidyl inositol 3-phosphate 5-kinase Fab1p is essential for vacuole function in S.cerevisiae. Curr. Biol., 8, 1219–1222. [DOI] [PubMed] [Google Scholar]
- Coussens L., Parker,P., Rhee,L., Yang-Feng,T., Chen,E., Waterfield,M., Francke,U. and Ullrich,A. (1986) Multiple, distinct forms of bovine and human protein kinase C suggest diversity in cellular signalling pathways. Science, 233, 859–865. [DOI] [PubMed] [Google Scholar]
- Drayer A.L. and van Haastert,P.J. (1992) Molecular cloning and expression of a phosphoinositide-specific phospholipase C of Dictyostelium discoideum. J. Biol. Chem., 267, 18387–18392. [PubMed] [Google Scholar]
- D’Santos C., Clarke,J. and Divecha,N. (1998) Phospholipid signalling in the nucleus. Biochim. Biophys. Acta, 1436, 201–232. [DOI] [PubMed] [Google Scholar]
- Firtel R. (1995) Integration of signalling information in controlling cell fate decisions in Dictyostelium. Genes Dev., 9, 1427–1444. [DOI] [PubMed] [Google Scholar]
- Fukuzawa M. and Williams,J. (1997) cudA: a Dictyostelium gene with pleiotropic effects on cell differentiation and slug behaviour. Development, 124, 2719–2728. [DOI] [PubMed] [Google Scholar]
- Ginsburg G. and Kimmel,A.R. (1989) Inositol trisphosphate and diacylglycerol can differentially modulate gene expression in Dictyostelium. Proc. Natl Acad. Sci. USA, 86, 9332–9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks S., Quinn,A. and Hunter,T. (1988) The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science, 241, 42–52. [DOI] [PubMed] [Google Scholar]
- Huang H., Takagawa,D., Weeks,G. and Pears,C. (1997) Cells at the centre of Dictyostelium aggregates become spores. Dev. Biol., 192, 564–571. [DOI] [PubMed] [Google Scholar]
- Katzav S., Martin-Zanca,D. and Barbacid,M. (1989) vav, a novel human oncogene derived from a locus ubiquitously expressed in hemapoietic cells. EMBO J., 8, 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd S., Kelley,M. and Young,M. (1986) Sequence of the notch locus of Drosophila melanogaster: relationship of the encoded protein to mammalian clotting and growth factors. Mol. Cell. Biol., 6, 3094–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J., Wilson,M., Kisseleva,M., Jurley,J., Majerus,P. and Anderson,R. (2000) The activation loop of phosphatidylinositol phosphate kinases determines signaling specificity. Mol. Cell, 5, 1–11. [DOI] [PubMed] [Google Scholar]
- Maeda M. and Firtel,R. (1997) Activation of the MAPK ERK2 by the chemoattractant folic acid in Dictyostelium. J. Biol. Chem., 272, 23690–23695. [DOI] [PubMed] [Google Scholar]
- Maruyama I. and Brenner,S. (1991) A phorbol ester/diacylglycerol-binding protein encoded by the unc-13 gene of C.elegans. Proc. Natl Acad. Sci. USA, 88, 5729–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami K., Katagiri,T., Iuchi,S., Yamaguchi-Shinozaki,K. and Shinozaki,K. (1998) A gene encoding phosphatidyinositol-4-phosphate 5-kinase is induced by water stress and abscisic acid in Arabidopsis thaliana. Plant J., 15, 563–568. [DOI] [PubMed] [Google Scholar]
- Nishizuka Y. (1992) Intracellular signalling by hydrolysis of phospholipids and activation of protein kinase C. Science, 258, 607–614. [DOI] [PubMed] [Google Scholar]
- Ogita K. et al. (1990) Protein kinase C in Saccharomyces cerevisiae: comparison with the mammalian enzyme. Proc. Natl Acad. Sci. USA, 87, 5011–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y., Fujii,T., Igarishi,K., Kuno,T., Tanaka,C. and Nishizuka,Y. (1989) Phorbol ester binding to protein kinase C requires a cysteine-rich zinc-finger-like sequence. Proc. Natl Acad. Sci. USA, 86, 4868–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel H., Guo,K., Parent,C., Gross,J., Devreotes,P. and Weijer,C. (2000) A temperature sensitive adenylyl cyclase mutant of Dictyostelium. EMBO J., 19, 2247–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips P., Thio,M. and Pears,C. (1997) A protein kinase C-like activity involved in the chemotactic response of Dictyostelium discoideum. Biochim. Biophys. Acta, 1349, 72–80. [DOI] [PubMed] [Google Scholar]
- Quest A., Bardes,E. and Bell,R. (1994a) A phorbol ester binding domain of PKCγ. High affinity binding to a GST/cys2 fusion protein. J. Biol. Chem., 269, 2953–2960. [PubMed] [Google Scholar]
- Quest A., Bardes,E. and Bell,R. (1994b) A phorbol ester binding domain of PKCγ: deletion analysis of the cys2 domain defines a minimal 43 amino acid peptide. J. Biol. Chem., 269, 2961–2970. [PubMed] [Google Scholar]
- Rameh L., Tolias,K., Duckworth,B. and Cantley,L. (1997) A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature, 390, 192–196. [DOI] [PubMed] [Google Scholar]
- Rosenthal A., Rhee,L., Yadegari,R., Paro,R., Ullrich,A. and Goeddel,D. (1987) Structure and nucleotide sequence of a Drosophila melanogaster protein kinase C gene. EMBO J., 6, 433–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitzler G., Fischer,W. and Firtel,R. (1994) Cloning and characterization of the G-box binding factor, an essential component of the developmental switch between early and late development in Dictyostelium. Genes Dev., 8, 502–514. [DOI] [PubMed] [Google Scholar]
- Schulkes C. and Schaap,P. (1995) cAMP-dependent protein kinase activity is essential for preaggregative gene expression in Dictyostelium. FEBS Lett., 368, 381–384. [DOI] [PubMed] [Google Scholar]
- Seastone D., Lee,E., Bush,J., Knecht,D. and Cardelli,J. (1998) Overexpression of a novel rho family GTPase, RacC, induces unusual actin-based structures and positively affects phagocytosis in Dictyostelium discoideum. Mol. Biol. Cell, 9, 2891–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seastone D., Zhang,L., Buczynski,G., Rebstein,P., Weeks,G., Speigelman,G. and Cardelli,J. (1999) The small Mr Ras-like GTPase Rap1 and the phospholipase C pathway act to regulate phagocytosis in Dictyostelium discoideum. Mol. Biol. Cell, 10, 393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick S. and Smerdon,S. (1999) The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem. Sci., 24, 311–316. [DOI] [PubMed] [Google Scholar]
- Shariff A. and Luna,E.J. (1992) Diacylglycerol-stimulated formation of actin nucleation sites at plasma membranes. Science, 256, 245–247. [DOI] [PubMed] [Google Scholar]
- Tillinghast H.S. and Newell,P.C. (1987) Chemotaxis towards pteridines during development of Dictyostelium. J. Cell Sci., 87, 45–53. [DOI] [PubMed] [Google Scholar]
- Varnum B. and Soll,D. (1981) Chemoresponsiveness to cAMP and folic acid during growth, development and dedifferentiation in Dictyostelium. Differentiation, 18, 151–160. [DOI] [PubMed] [Google Scholar]
- Watts D. and Ashworth,J. (1970) Growth of myxamoebae of the cellular slime mould Dictyostelium discoideum in axenic culture. Biochem. J., 119, 171–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., DeWald,D., Bronenkov,I., Anderson,R., Emr,S. and Koshland,D. (1995) Novel PI(4)P 5-kinase homologue, Fab1p, essential for normal vacuole function and morphology in yeast. Mol. Biol. Cell, 6, 525–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Ohya,Y., Nakano,A. and Anraku,Y. (1994) Genetic interactions among genes involved in the STT4-PKC1 pathway of S. cerevisiae. Mol. Gen. Genet., 242, 631–640. [DOI] [PubMed] [Google Scholar]
- Zhou K., Takegawa,K., Emr,S. and Firtel,R. (1995) A phosphatidylinositol (PI) kinase gene family in Dictyostelium discoideum: biological roles of putative mammalian p110 and yeast Vps34p PI 3-kinase homologues during growth and development. Mol. Cell. Biol., 15, 5645–5656. [DOI] [PMC free article] [PubMed] [Google Scholar]