Abstract
The aim of this study was to explore the molecular profile of proliferating rheumatoid arthritis synovial fibroblasts (RA-SF). Total RNA was extracted from two cultures of RA-SF (low-density [LD] proliferating cells and high-density [HD] nonproliferating cells) and suppression subtractive hybridization was performed to compare differential gene expression of these two cultures. Subtracted cDNA was subcloned, and nucleotide sequences were analyzed to identify each clone. Differential expression of distinct clones was confirmed by semiquantitative RT-PCR. The expression of certain genes in synovial tissues was examined by in situ hybridization. In both LD and HD cells, 44 clones were upregulated. Of the 88 total clones, 46 were identical to sequences that have previously been characterized. Twenty-nine clones were identical to cDNAs that have been identified, but with unknown functions so far, and 13 clones did not show any significant homology to sequences in GenBank (NCBI). Differential expression of distinct clones was confirmed by RT-PCR. In situ hybridization showed that certain genes, such as S100A4, NFAT5, unr and Fbx3, were also expressed predominantly in synovial tissues from patients with RA but not from normal individuals. The expression of distinct genes in proliferating RA-SF could also be found in RA synovium, suggesting that these molecules are involved in synovial activation in RA. Most importantly, the data indicate that the expression of certain genes in RA-SF depends on the stage of proliferation; therefore, the stage needs to be considered in any analysis of differential gene expression in SF.
Keywords: differential gene expression, molecular profile, proliferation, rheumatoid arthritis, synovial fibroblasts
Introduction
Recently, several lines of evidence indicated that the balance between apoptotic cell death and proliferation or differentiation appears to be modulated in the rheumatoid arthritis (RA) synovium [1-3]. In our laboratory, it was demonstrated that sentrin-1, which is identical to small ubiquitin-related modifier (SUMO)-1, is upregulated in cultured RA synovial fibroblasts (SF) as well as at sites of invasion [4]. Sentrin-1 is thought to modify the activity of Mdm2, which is also expressed in RA-SF [5]. Mdm2 is an E3 ubiquitin ligase for p53, and degrades p53 protein in a proteasome-dependent manner [6], suggesting that sentrin-1 is able to modulate the apoptotic pathway in RA synovial tissues. Moreover, we also reported that FLICE (Fas-associating protein with death domain [FADD]-like interleukin 1β converting enzyme [ICE]) inhibitory protein (FLIP), an inhibitor of caspase 8, is expressed predominantly at sites of bone and cartilage destruction in RA [7]. These results indicate that several anti-apoptotic mechanisms most likely play a critical role in maintaining the long-term activation of RA-SF and thereby contribute to matrix degradation.
In addition to the expression of these anti-apoptotic molecules, RA-SF show several features with regard to cell proliferation. Lafyatis et al.[8] reported that RA-SF reveal anchorage-independent cell growth under certain culture conditions, such as high serum concentration (20%) or platelet-derived growth factor (PDGF) supplementation. Moreover, it has been shown that PDGF and PDGF receptor [9], c-myc [10,11] and proliferating cell nuclear antigen (PCNA) [10], which are cell proliferation-associated molecules, are predominantly expressed in rheumatoid synovium.
On the basis of these observations, we hypothesized that proliferating RA-SF express a specific set of genes that promotes proliferation and/or inhibits the apoptotic process. Thus, the aim of this study was to investigate the expression profile of RA-SF during proliferation in comparison with that of nonproliferating RA-SF, and thereby to explore novel molecular mechanisms of synovial proliferation in RA.
Materials and methods
Preparation of synovial fibroblasts
Cultured RA-SF from one patient (SHK70) who fulfilled the 1987 revised criteria of the American College of Rheumatology for the disease were propagated from synovial tissue obtained during synovectomy, and were used for cDNA subtraction. Briefly, the tissue specimens were minced enzymatically, and the adherent cells were cultured in Dulbecco's modified Eagle's medium (Life Technologies AG, Basel, Switzerland) supplemented with 10% fetal calf serum. The cells were cultured until reaching confluence, then trypsinized and spread to new culture flasks in a 1:5 dilution. The cells were cultured for the sixth passage, and the cells in passage six were used for suppression subtractive hybridization. Two other RA-SF from different patients with RA (SHK78 and SHK80) were also used for RT-PCR.
RNA isolation and suppression subtractive hybridization
Total RNA was extracted with TRIZOL® reagent (Life Technologies AG, Basel, Switzerland) from two cultures, 4 days after seeding (LD; low-density cells) and 14 days after seeding (HD; high-density cells), respectively (see Supplementary material). From each culture, 1μg of total RNA was used for cDNA synthesis with the SMART™ PCR cDNA Synthesis Kit (BD Clontech, Basel, Switzerland), and suppression subtractive hybridization was performed with the PCR-Select™ cDNA Subtraction Kit (BD Clontech, Basel, Switzerland) according to the manufacturer's instructions. We performed two directional subtractions. In the forward (LD-specific) subtraction, we investigated the genes specifically expressed on day 4 in proliferating cells at low density. We also conducted reverse (HD-specific) subtraction to analyze the genes predominantly expressed on day 14 in nonproliferating cells at high density. Each subtracted cDNA was subcloned into PCR-Script Amp SK (+) cloning vector (Stratagene Europe, Amsterdam, The Netherlands). After plasmid DNA of each clone was purified using Concert™ Rapid Plasmid Purification Systems (Life Technologies AG, Basel, Switzerland), the nucleotide sequences were analyzed by commercial sequencing (Microsynth, Balgach, Switzerland), and BLAST searches (NCBI) were performed to identify the respective gene of each clone.
Semiquantitative RT-PCR
We designed primers specific for distinct genes, i.e. antizyme inhibitor (AZI), EGF-containing fibulin-like extracellular matrix protein 1 (EFEMP1), S100 calcium-binding protein A4 (S100A4), nuclear factor of activated T cells 5 (NFAT5), upstream of N-ras (unr), F-box only protein 3 (Fbx3), ribonuclease 4 (RNASE4), dipeptidylpeptidase IV (CD26), epithelial protein lost in neoplasm (EPLIN) and 13 kDa differentiation-associated protein (13kdap). Then, mRNA expression was analyzed by semiquantitative RT-PCR to confirm the differential expression of the subtracted cDNA. At first, template cDNA was diluted 1:3 and 1:9, and PCR with primers of housekeeping genes such as actin β and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was performed to compare the equal amounts of cDNA between two cultures in further experiments. Then, the efficiency of the amplification and the optimal cycles were confirmed for each set of primers. After this optimization, we compared the expression of each gene. All PCR reactions consisted of initial denaturation at 94°C for 10 min followed by 25 to 35 cycles at 94°C for 30 s, 58 to 60°C for 1 min, and 72°C for 90 s performed by using AmpliTaq Gold® DNA polymerase and GeneAmp PCR system 9700 (Applied Biosystems, Rotkreuz, Switzerland). PCR products were analyzed on 1.0% agarose gels.
In situ hybridization
The expression and distribution of distinct genes were examined in paraffin-embedded sections of synovial tissues from seven patients with RA and three normal individuals by in situ hybridization with digoxigenin-labeled RNA probes, which were generated by in vitro transcription with T3 or T7 RNA polymerases. The probes were detected with alkaline-phosphatase-conjugated anti-digoxigenin antibody F (ab), and visualized with 4-nitro blue tetrazolium chloride (NBT) and 5-bromo-4-chloro-3-indolyl-phosphate, 4-toluidine salt (BCIP) solutions (Roche Diagnostics, Rotkreuz, Switzerland). In each experiment, we counted the number of positive cells in the lining of synovial tissues, and the levels of mRNA expression were calculated as a percentage of positive cells.
Results
Identification of subtracted cDNA
We obtained 44 clones that were upregulated in LD cells and 44 clones that were upregulated in HD cells (Figs 1 and 2). Forty-six of the 88 clones were identical to sequences that have previously been identified and characterized. Twenty-nine clones were identical to cDNAs that have been identified, but with unknown functions so far, and 13 gene fragments did not show any significant homology to sequences in the GenBank (NCBI). In LD-specific clones, there were several extracellular matrix (ECM) proteins such as EFEMP1, fibrillin, versican, laminin and fibronectin. There were also some proliferation-related molecules such as AZI, Fbx3 and nucleosome assembly protein 1-like 1 (NAP1L1), or cell-activation-associated molecules such as S100A4 and NFAT5 (Fig. 1). On the other hand, in HD-specific clones, there were several molecules that induce protein synthesis, such as ribosomal proteins. We also found the expression of some differentiation markers such as 13kdap and CD26 and genes associated with inhibition of cell growth, such as EPLIN and RNASE4 (Fig. 2). We confirmed the efficiency of subtraction by PCR according to the manufacturer's instructions (data not shown).
Figure 1.
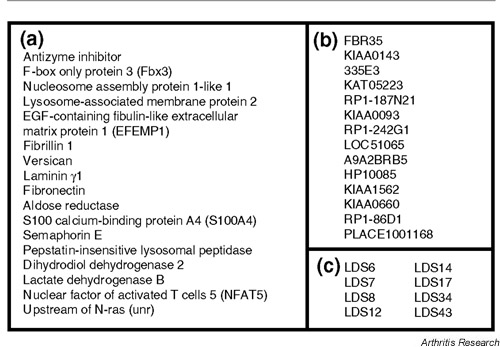
The list of clones obtained by forward (LD-specific) subtraction. We divided these clones into three categories. (a) This category consists of already identified and characterized genes. (b) This category includes the genes with sequences previously identified, but with unknown functions so far. The name of each clone is shown. (c) List of novel gene fragments that did not show any significant homology to sequences in GenBank (NCBI). We mention just the name of clones in our experiments.
Figure 2.
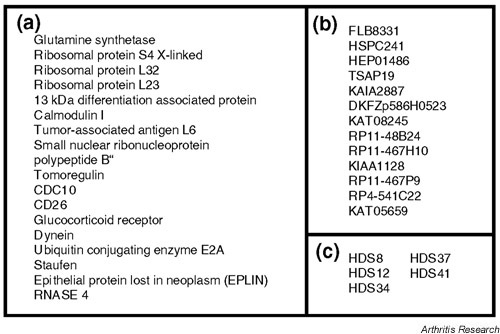
The list of clones obtained by reverse (HD-specific) subtraction. We divided these clones into the same three categories as described in Figure 1.
Confirmation of differential expression of distinct genes
The LD-specific genes such as EFEMP1, unr and Fbx3 (Fig. 3a), and the HD-specific genes such as CD26, 13kdap and RNASE4 (Fig. 3b) were upregulated in LD and HD cells, respectively. These results indicated that the subtracted cDNA represented the part of genes actually expressed in a culture condition-dependent manner. The results of actin-β and GAPDH indicated that we compared equal amounts of template cDNA in these experiments (Fig. 3c).
Figure 3.
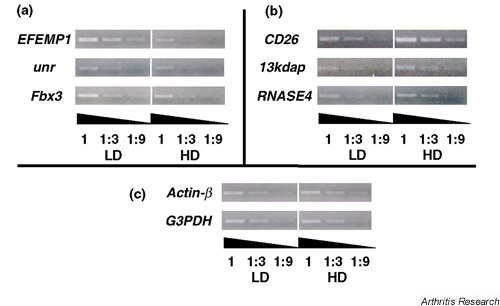
The differential expression of distinct genes. It was confirmed by semiquantitative RT-PCR. (a) LD-specific genes such as EFEMP1, unr and Fbx3, and (b) HD-specific genes such as CD26, 13kdap and RNASE4 were upregulated in LD and HD cells, respectively. (c) The results of actin-β and GAPDH indicated that we compared equal amounts of template cDNA in these experiments. We analyzed three independent RA-SF and show the representative results.
Expression of distinct genes in situ
We investigated the expression and distribution of distinct genes, namely S100A4, NFAT5, Fbx3 and unr, all of which were upregulated in LD cells, by in situ hybridization. S100A4 mRNA was expressed both in the lining and sublining layer of RA synovium (Fig. 4). The same expression pattern was shown by unr (see Supplementary material). We also detected the expression of NFAT5 mRNA in RA synovium (Fig. 5). Interestingly, NFAT5, as well as Fbx3, mRNA was also expressed at sites of bone destruction (Fig. 6 and see Supplementary material). These results indicate that NFAT5 and Fbx3 could be correlated not only with the proliferation but also with the activation and invasion of RA-SF in vivo. It is noteworthy that these four genes (i.e. S100A4, unr, NFAT5, Fbx3) were predominantly expressed in synovial tissues from patients with RA rather than from normal individuals (see Supplementary material), suggesting that these genes could contribute to the pathogenesis of RA.
Figure 4.
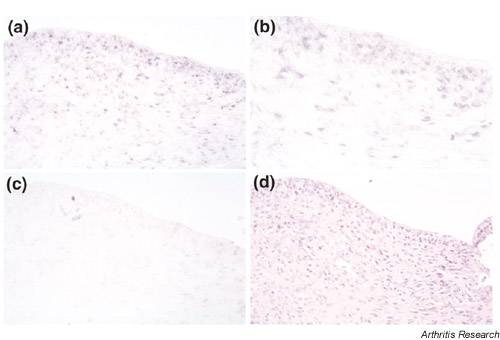
The expression of S100A4 mRNA in sections from patients with RA by in situ hybridization. This figure shows antisense (a,b) and sense (c) staining, respectively. We also performed HE staining (d) in serial sections. The original magnifications are 200× (a,c,d) and 400× (b), respectively.
Figure 5.
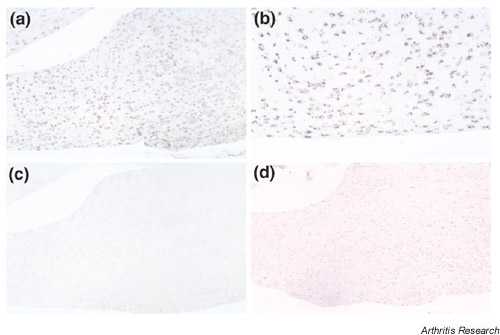
The expression of NFAT5 mRNA in sections from patients with RA by in situ hybridization. This figure shows antisense (a,b) and sense (c) staining, respectively. We also performed HE staining (d) in serial sections. The original magnifications are 100× (a,c,d) and 200× (b), respectively.
Figure 6.
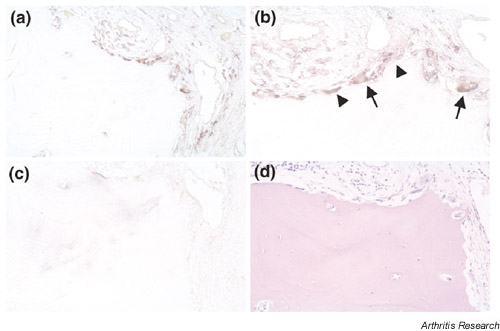
The expression of NFAT5 mRNA in sections from patients with RA by in situ hybridization. This figure shows antisense (a,b) and sense (c) staining, respectively. The positive signal was observed not only in fibroblast-like cells (arrowheads) but also in osteoclast-like cells (arrows). We also performed HE staining (d) in serial sections. The original magnifications are 200× (a,c,d) and 400× (b), respectively.
Discussion
We analyzed the expression profile of proliferating RA-SF in comparison with that of nonproliferating RA-SF, and we observed that distinct sets of genes, including several novel genes that have not been associated with RA, were expressed differentially in proliferating RA-SF by using a PCR-based suppression subtractive hybridization technique. It is useful for development of novel therapeutic approaches to identify the expression profiles in disease-associated cells or conditions [12,13], and there are some reports that identified genes specifically expressed in rheumatoid synovial cells by using a cDNA subtraction or a differential display method [14-17]. Since we used RA-SF from the same patient, at the same passage and cultured with the same medium, we could exclude genetic polymorphism between individuals, and could identify specific genes dependent on proliferation or cell density.
In LD-specific subtracted cDNA, we isolated several interesting molecules that play an important role in cell proliferation. For example, we found upregulation of several ECM proteins. ECM proteins not only serve just as a mechanical structure, but also play an important role in many biological functions, such as cell migration, proliferation and differentiation [18]. Indeed, it was reported that versican stimulates cell proliferation [19,20], and EFEMP1 stimulates DNA synthesis when microinjected into fibroblasts [21]. Fibronectin fragments play an important role in proliferation, adhesion and matrix metalloproteinase expression of RA-SF [22]. The expression of ECM proteins in proliferating RA-SF, therefore, could stimulate their proliferation and differentiation in an autocrine and/or paracrine manner.
The expression of proliferation-related molecules such as AZI, Fbx3 and NAP1L1 was also observed in LD cells. AZI induces polyamine synthesis, which is essential for cell growth [23,24], and it was reported that AZI is differentially expressed in tumor tissues rather than normal tissues [25]. Although the function of Fbx3 has not been identified so far, F-box proteins are important for controlling the cell cycle [26]. Simon et al.[27] demonstrated that NAP1L1 is highly expressed in proliferating cells. On the other hand, unr was also highly expressed in LD cells, and it was reported that unr interacts with apoptotic protease-activating factor-1 (Apaf-1) and could modulate a certain apoptotic pathway [28]. We can hypothesize, therefore, that these molecules are not only essential for cell survival and proliferation in general, but could also play an important role in the proliferation of RA-SF in vitro and in vivo.
In HD-specific subtracted cDNA, we also obtained several interesting molecules. Indeed, we found that CD26, EPLIN and RNASE4 were specifically expressed in non-proliferating RA-SF. CD26 was originally identified as an adenosine deaminase complexing protein and T-cell activation molecule, and it was reported that activated T cells as well as fibroblasts in rheumatoid synovium expressed this molecule [29]. Recently, it was shown that the malignant phenotype of melanoma cells is suppressed by the introduction of the exogenous CD26 gene [30]. EPLIN is downregulated or lost in cancer cell lines and inhibits cell proliferation when overexpressed [31,32]. On the other hand, RNASE 4 mediates the antiviral and antiproliferative roles of interferons and is induced during growth inhibition in murine cell lines [33]. Recently, Taniguchi et al. [34] showed that cyclin-dependent kinase inhibitor p16INK4a is induced in RA-SF by several growth-inhibited conditions, including irradiation, serum depletion and high cellular density. Surprisingly, they showed also that the gene transfer of p16 inhibits proliferation of synovial fibroblasts in vitro and suppresses rat adjuvant arthritis in vivo. In this context, EPLIN and RNASE4 genes might also be promising candidates for gene transfer as a novel approach to treating RA.
Conclusion
We demonstrated that proliferating RA-SF express a distinct set of genes clearly different from that of nonproliferating RA-SF. Moreover, a number of molecules correlated with cell proliferation and activation, including several novel genes, were isolated from RA-SF by using suppression subtractive hybridization. Cooperating with anti-apoptotic molecules such as sentrin-1 and FLIP, these molecules could be involved in synovial cell proliferation and activation and could contribute to matrix destruction at sites of invasion in RA. Last but not least, a consideration of cell density should be mandatory in any investigation examining cell proliferation or apoptosis in cell cultures.
Abbreviations
13kdap = 13 kDa differentiation-associated protein; Apaf-1 = apoptotic protease-activating factor-1; AZI = antizyme inhibitor; BCIP = 5-bromo-4-chloro-3-indolyl-phosphate, 4-toluidine salt; ECM = extracellular matrix; EFEMP1 = EGF-containing fibulin-like extracellular matrix protein 1; EPLIN = epithelial protein lost in neoplasm; Fbx3 = F-box only protein 3; FLICE = Fas-associating protein with death domain [FADD]-like interleukin 1β converting enzyme [ICE]; FLIP = FLICE inhibitory protein; HD = high-density; HE = hematoxylin and eosin; LD = low-density; NAP1L1 = nucleosome assembly protein 1-like 1; NBT = 4-nitro blue tetrazolium chloride; NCBI = National Center for Biotechnology Information; NFAT5 = nuclear factor of activated T cells 5; PCNA = proliferating cell nuclear antigen; PDGF = platelet-derived growth factor; RA = rheumatoid arthritis; RNASE4 = ribonuclease 4; RT-PCR = reverse transcriptase-polymerase chain reaction; S100A4 = S100 calcium-binding protein A4; SF = synovial fibroblasts; SUMO = small ubiquitin-related modifier; unr = upstream of N-ras.
Supplementary material
Supplementary Figure 1.
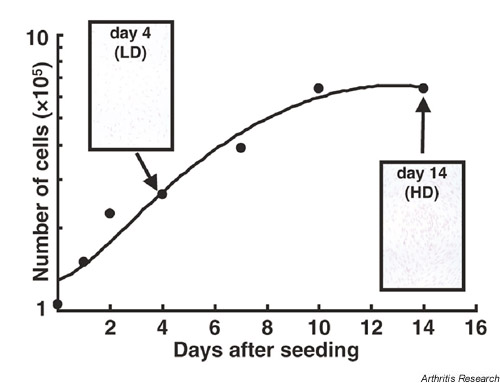
The strategy of our experiments. SF from one patient with RA (passage6) were cultured in 75 cm2 flasks until reaching confluence. Then, these cells were collected and seeded in 12-well culture plates (1.0 × 105 cells/well). At 1, 2, 4, 7, 10 and 14 days after seeding, the number of cells was counted by the trypan blue exclusion method. At 4 days, the cells at low density (LD) proliferated exponentially, and at 14 days, the cells at high density (HD) almost ceased to proliferate as a result of contact inhibition. The inserts show the HE staining of SF on chamber slides at 4 and 14 days after seeding, respectively.
Supplementary Figure 2.
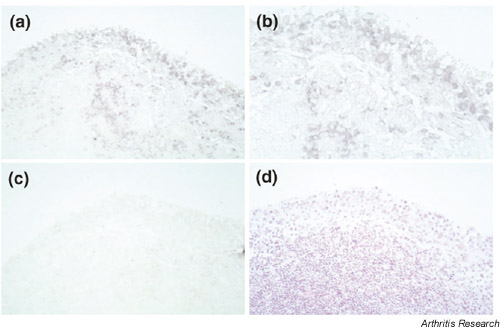
The expression of unr mRNA in sections from patients with RA as shown by in situ hybridization. This figure shows antisense (a,b) and sense (c) staining, respectively. We also performed HE staining (d) in serial sections. The original magnifications are 200× (a,c,d) and 400× (b), respectively.
Supplementary Figure 3.
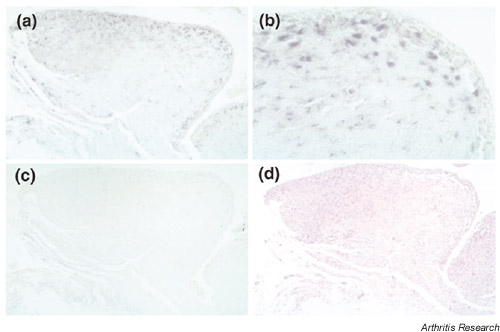
The expression of Fbx3 mRNA in sections from patients with RA as shown by in situ hybridization. This figure shows antisense (a,b) and sense (c) staining, respectively. We also performed HE staining (d) in serial sections. The original magnifications are 100× (a,c,d) and 400× (b), respectively.
Supplementary Figure 4.
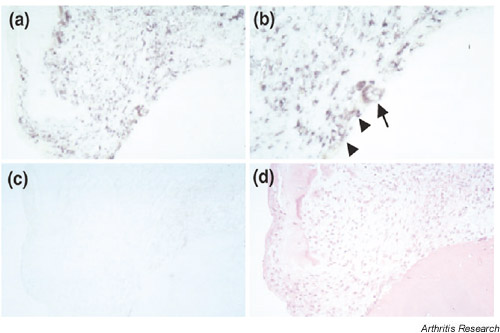
The expression of Fbx3 mRNA in sections from patients with RA by in situ hybridization. This figure shows antisense (a,b) and sense (c) staining, respectively. The positive signal was observed not only in fibroblast-like cells (arrowheads) but also in osteoclast-like cells (arrow). We also performed HE staining (d) in serial sections. The original magnifications are 200× (a,c,d) and 400× (b), respectively.
Supplementary Figure 5.
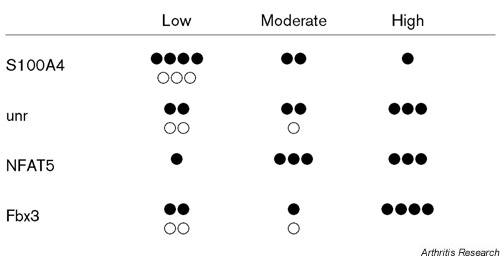
Summary of the expression of LD-specific genes in synovial tissues from patients with RA (●) and from normal individuals (○). In each experiment, we counted the number of positive cells in the lining of synovial tissues, and the levels of the mRNA expression were calculated as a percentage of positive cells. Then, we divided these data into three groups; namely, below 10% (Low), from 10 to 50% (Moderate) and more than 50% (High).
Acknowledgments
Acknowledgements
We would like to thank M Commazi, P Künzler and F Pataky for their technical assistance and G Pairo for the secretarial work. K Masuda is supported by the Japan Rheumatism Foundation, R Masuda by the Uehara Memorial Foundation and the other authors by their institutions.
References
- Firestein GS, Echeverri F, Yeo M, Zvaifler NJ, Green DR. Somatic mutations in the p53 tumor suppressor gene in rheumatoid arthritis synovium. Proc Natl Acad Sci USA. 1997;94:10895–10900. doi: 10.1073/pnas.94.20.10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Boyle DL, Shi Y, Green DR, Firestein GS. Dominant-negative p53 mutations in rheumatoid arthritis. Arthritis Rheum. 1999;42:1088–1092. doi: 10.1002/1529-0131(199906)42:6<1088::AID-ANR4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Muller-Ladner U, Gay RE, Nishioka K, Gay S. Ultra-structural demonstration of apoptosis, Fas and Bcl-2 expression of rheumatoid synovial fibroblasts. J Rheumatol. 1996;23:1345–1352. [PubMed] [Google Scholar]
- Franz JK, Pap T, Hummel KM, Nawrath M, Aicher WK, Shigeyama Y, Muller-Ladner U, Gay RE, Gay S. Expression of sentrin, a novel antiapoptotic molecule, at sites of synovial invasion in rheumatoid arthritis. Arthritis Rheum. 2000;43:599–607. doi: 10.1002/1529-0131(200003)43:3<599::AID-ANR17>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Muller-Ladner U, Nishioka K. p53 in rheumatoid arthritis: friend or foe? Arthritis Res. 2000;2:175–178. doi: 10.1186/ar84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschmann T, Fuchs SY, Lee CG, Pan ZQ, Ronai Z. SUMO-1 modification of Mdm2 prevents its self-ubiquitination and increases Mdm2 ability to ubiquitinate p53. Cell. 2000;101:753–762. doi: 10.1016/s0092-8674(00)80887-9. [DOI] [PubMed] [Google Scholar]
- Schedel J, Gay RE, Simmen B, Gay S. Flice-inhibitory protein (FLIP) expression at sites of cartilage and bone destruction in rheumatoid arthritis (RA) [abstract]. Arthritis Rheum. 2000;43:s160. doi: 10.1002/art.10309. [DOI] [PubMed] [Google Scholar]
- Lafyatis R, Remmers EF, Roberts AB, Yocum DE, Sporn MB, Wilder RL. Anchorage-independent growth of synoviocytes from arthritic and normal joints. Stimulation by exogenous platelet-derived growth factor and inhibition by transforming growth factor-beta and retinoids. J Clin Invest. 1989;83:1267–1276. doi: 10.1172/JCI114011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmers EF, Sano H, Wilder RL. Platelet-derived growth factors and heparin-binding (fibroblast) growth factors in the synovial tissue pathology of rheumatoid arthritis. Semin Arthritis Rheum. 1991;21:191–199. doi: 10.1016/0049-0172(91)90009-o. [DOI] [PubMed] [Google Scholar]
- Qu Z, Garcia CH, O'Rourke LM, Planck SR, Kohli M, Rosenbaum JT. Local proliferation of fibroblast-like synoviocytes contributes to synovial hyperplasia. Results of proliferating cell nuclear antigen/cyclin, c-myc, and nucleolar organizer region staining. Arthritis Rheum. 1994;37:212–220. doi: 10.1002/art.1780370210. [DOI] [PubMed] [Google Scholar]
- Trabandt A, Aicher WK, Gay RE, Sukhatme VP, Nilson-Hamilton M, Hamilton RT, McGhee JR, Fassbender HG, Gay S. Expression of the collagenolytic and Ras-induced cysteine proteinase cathepsin L and proliferation-associated oncogenes in synovial cells of MRL/I mice and patients with rheumatoid arthritis. Matrix. 1990;10:349–361. doi: 10.1016/s0934-8832(11)80142-3. [DOI] [PubMed] [Google Scholar]
- Liotta L, Petricoin E. Molecular profiling of human cancer. Nat Rev Genet. 2000;1:48–56. doi: 10.1038/35049567. [DOI] [PubMed] [Google Scholar]
- St Croix B, Rago C, Velculescu V, Traverso G, Romans KE, Mont-gomery E, Lal A, Riggins GJ, Lengauer C, Vogelstein B, Kinzler KW. Genes expressed in human tumor endothelium. Science. 2000;289:1197–1202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- Justen HP, Grunewald E, Totzke G, Gouni-Berthold I, Sachinidis A, Wessinghage D, Vetter H, Schulze-Osthoff K, Ko Y. Differential gene expression in synovium of rheumatoid arthritis and osteoarthritis. Mol Cell Biol Res Commun. 2000;3:165–172. doi: 10.1006/mcbr.2000.0211. [DOI] [PubMed] [Google Scholar]
- Mangasser-Stephan K, Dooley S, Welter C, Mutschler W, Hansel-mann RG. Identification of human semaphorin E gene expression in rheumatoid synovial cells by mRNA differential display. Biochem Biophys Res Commun. 1997;234:153–156. doi: 10.1006/bbrc.1997.6607. [DOI] [PubMed] [Google Scholar]
- Seki T, Selby J, Haupl T, Winchester R. Use of differential subtraction method to identify genes that characterize the phenotype of cultured rheumatoid arthritis synoviocytes. Arthritis Rheum. 1998;41:1356–1364. doi: 10.1002/1529-0131(199808)41:8<1356::AID-ART4>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Zanders ED, Goulden MG, Kennedy TC, Kempsell KE. Analysis of immune system gene expression in small rheumatoid arthritis biopsies using a combination of subtractive hybridization and high-density cDNA arrays. J Immunol Methods. 2000;233:131–140. doi: 10.1016/s0022-1759(99)00126-x. [DOI] [PubMed] [Google Scholar]
- Schuppan D, Somasundaram R, Dieterich W, Ehnis T, Bauer M. The extracellular matrix in cellular proliferation and differentiation. Ann NY Acad Sci. 1994;733:87–102. doi: 10.1111/j.1749-6632.1994.tb17259.x. [DOI] [PubMed] [Google Scholar]
- Yang BL, Zhang Y, Cao L, Yang BB. Cell adhesion and proliferation mediated through the G1 domain of versican. J Cell Biochem. 1999;72:210–220. doi: 10.1002/(SICI)1097-4644(19990201)72:2<210::AID-JCB5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cao L, Yang BL, Yang BB. The G3 domain of versican enhances cell proliferation via epidermial growth factor-like motifs. J Biol Chem. 1998;273:21342–21351. doi: 10.1074/jbc.273.33.21342. [DOI] [PubMed] [Google Scholar]
- Lecka-Czernik B, Lumpkin CK, Jr, Goldstein S. An overexpressed gene transcript in senescent and quiescent human fibroblasts encoding a novel protein in the epidermal growth factor-like repeat family stimulates DNA synthesis. Mol Cell Biol. 1995;15:120–128. doi: 10.1128/mcb.15.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barilla ML, Carsons SE. Fibronectin fragments and their role in inflammatory arthritis. Semin Arthritis Rheum. 2000;29:252–265. doi: 10.1016/s0049-0172(00)80012-8. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Murakami Y. Rapid and regulated degradation of ornithine decarboxylase. Biochem J. 1995;306:1–10. doi: 10.1042/bj3060001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J, Grahn B, Heby O. Antizyme inhibitor is rapidly induced in growth-stimulated mouse fibroblasts and releases ornithine decarboxylase from antizyme suppression. Biochem J. 2000;346 Pt 3:699–704. [PMC free article] [PubMed] [Google Scholar]
- Jung MH, Kim SC, Jeon GA, Kim SH, Kim Y, Choi KS, Park SI, Joe MK, Kimm K. Identification of differentially expressed genes in normal and tumor human gastric tissue. Genomics. 2000;69:281–286. doi: 10.1006/geno.2000.6338. [DOI] [PubMed] [Google Scholar]
- Winston JT, Koepp DM, Zhu C, Elledge SJ, Harper JW. A family of mammalian F-box proteins. Curr Biol. 1999;9:1180–1182. doi: 10.1016/S0960-9822(00)80021-4. [DOI] [PubMed] [Google Scholar]
- Simon HU, Mills GB, Kozlowski M, Hogg D, Branch D, Ishimi Y, Siminovitch KA. Molecular characterization of hNRP, a cDNA encoding a human nucleosome-assembly-protein-I-related gene product involved in the induction of cell proliferation. Biochem J. 1994;297:389–397. doi: 10.1042/bj2970389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SA, Brown EC, Coldwell MJ, Jackson RJ, Willis AE. Protein factor requirements of the Apaf-1 internal ribosome entry segment: roles of polypyrimidine tract binding protein and upstream of N-ras. Mol Cell Biol. 2001;21:3364–3374. doi: 10.1128/MCB.21.10.3364-3374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T, Williams YN, Baba H, Nosaka Y, Sugita T, Miyasaka N. Highly expressed dipeptidyl peptidase IV on fibroblast-like synoviocytes as well as activated helper T cells could be a target of RA therapy [abstract]. Arthritis Rheum. 2001;44:s88. [Google Scholar]
- Wesley UV, Albino AP, Tiwari S, Houghton AN. A role for dipeptidyl peptidase IV in suppressing the malignant phenotype of melanocytic cells. J Exp Med. 1999;190:311–322. doi: 10.1084/jem.190.3.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DD, Park NH, Denny CT, Nelson SF, Pe M. Characterization of transformation related genes in oral cancer cells. Oncogene. 1998;16:1921–1930. doi: 10.1038/sj/onc/1201715. [DOI] [PubMed] [Google Scholar]
- Maul RS, Chang DD. EPLIN, epithelial protein lost in neoplasm. Oncogene. 1999;18:7838–7841. doi: 10.1038/sj/onc/1203206. [DOI] [PubMed] [Google Scholar]
- Jacobsen H, Krause D, Friedman RM, Silverman RH. Induction of ppp(A2'p)nA-dependent RNase in murine JLS-V9R cells during growth inhibition. Proc Natl Acad Sci USA. 1983;80:4954–4958. doi: 10.1073/pnas.80.16.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K, Kohsaka H, Inoue N, Terada Y, Ito H, Hirokawa K, Miyasaka N. Induction of the p16INK4a senescence gene as a new therapeutic strategy for the treatment of rheumatoid arthritis. Nat Med. 1999;5:760–767. doi: 10.1038/10480. [DOI] [PubMed] [Google Scholar]


