Abstract
The human centromere protein B (CENP-B), one of the centromere components, specifically binds a 17 bp sequence (the CENP-B box), which appears in every other α-satellite repeat. In the present study, the crystal structure of the complex of the DNA-binding region (129 residues) of CENP-B and the CENP-B box DNA has been determined at 2.5 Å resolution. The DNA-binding region forms two helix–turn–helix domains, which are bound to adjacent major grooves of the DNA. The DNA is kinked at the two recognition helix contact sites, and the DNA region between the kinks is straight. Among the major groove protein-bound DNAs, this ‘kink–straight–kink’ bend contrasts with ordinary ‘round bends’ (gradual bending between two protein contact sites). The larger kink (43°) is induced by a novel mechanism, ‘phosphate bridging by an arginine-rich helix’: the recognition helix with an arginine cluster is inserted perpendicularly into the major groove and bridges the groove through direct interactions with the phosphate groups. The overall bending angle is 59°, which may be important for the centromere-specific chromatin structure.
Keywords: CENP-B/centromere/DNA bending/helix–turn–helix/protein–DNA complex
Introduction
The centromere is a region of the chromosome essential for its segregation during cell division, and has a special chromatin structure involving α-satellite DNA repeats and their associated proteins. In the human, the proteins associated with the centromere have been identified with specific autoantibodies for the centromere (Earnshaw and Rothfield, 1985). The human centromere proteins A, B and C (CENP-A, CENP-B and CENP-C, respectively) are such proteins, and they exhibit DNA-binding activities (Masumoto et al., 1989; Palmer et al., 1991; Saitoh et al., 1992; Sugimoto et al., 1994; Sullivan et al., 1994). These centromere-specific DNA-binding proteins are considered to be fundamental components in the formation of the special chromatin structure on the α-satellite DNA repeats of the centromere.
Among the centromere-specific DNA-binding proteins, CENP-B specifically binds a 17 bp sequence (the CENP-B box) (Earnshaw et al., 1989; Masumoto et al., 1989; Kipling et al., 1995), which appears in every other α-satellite repeat (Ikeno et al., 1994, 1998). The CENP-B box sequence positions nucleosomes adjacently (Yoda et al., 1998), and may therefore function as a cis-element for centromere-specific nucleosome assembly. In vivo analyses with cultured human cells revealed that the existence of the CENP-B box is essential for the formation of minichromosomes (Ikeno et al., 1998). On the other hand, CENP-B null mice appeared to be normal, except for lower body and testis weights, and uterine dysfunction (Hudson et al., 1998; Perez-Castro et al., 1998; Fowler et al., 2000), indicating the existence of functional homolog(s) of CENP-B. Actually, CENP-B-like proteins of unknown function have been identified in the human, such as the jerky-like protein (Toth et al., 1995) and the transposases encoded by the human Tigger1 and Tigger2 transposable elements (Smit and Riggs, 1996; Kipling and Warburton, 1997).
CENP-B is an 80 kDa protein that contains a DNA-binding region at the N-terminus (Yoda et al., 1992). In the present study, we determined the crystal structure of the DNA-binding region of CENP-B (CENP-B1–129) complexed with a 21 bp CENP-B box DNA at 2.5 Å resolution. This is the first structure of a centromere-specific protein–DNA complex.
Results and discussion
Structure determination
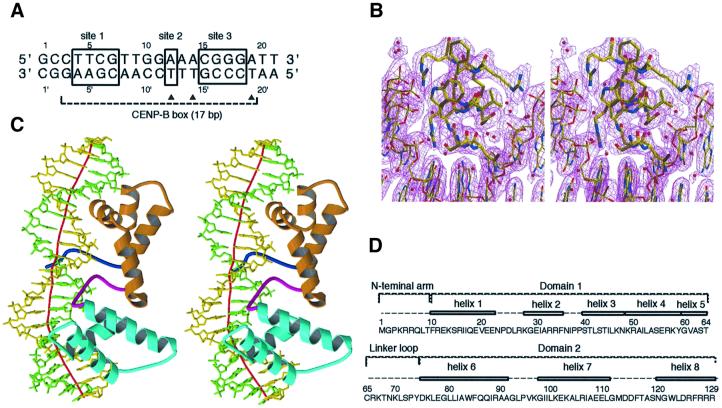
Recombinant CENP-B1–129 forms an inclusion body and, therefore, the protein was purified in the presence of 6 M urea. The complex of CENP-B1–129 and the CENP-B box DNA (21 bp; Figure 1A) was formed during the refolding process by dialysis against buffer without urea. The purified complex was concentrated up to 3–6 mg protein/ml, and the co-crystals were obtained by the hanging drop method. The crystals belong to the trigonal space group P3112, with unit cell constants of a = b = 83.4 Å, c = 139.0 Å, and contain one complex per asymmetric unit. A native diffraction data set was collected at 2.5 Å resolution at beamline BL44B2 in SPring-8, Harima. The structure was solved by multiple isomorphous replacement augmented with anomalous dispersion (MIRAS) (Table I), by the use of three isomorphous derivatives with duplex DNAs in which a thymidine was replaced by a 5-iodouridine at positions 12, 14 and 19, respectively (Figure 1A). A portion of the electron density map is shown in Figure 1B.
Fig. 1. The crystal structure of the CENP-B1–129 protein complexed with the 21mer DNA. (A) The 21mer DNA sequence with the CENP-B box. The three boxes, marked as sites 1, 2 and 3, indicate the essential bases for CENP-B binding to the CENP-B box DNA. Closed triangles indicate the thymine residues, which were each replaced by 5-iodouracil for phasing. (B) Stereo view of the (2|Fo| – |Fc|) electron density map for amino acid residues 119–129 and base pairs 15–20. (C) Overall structure of the CENP-B1–129·DNA complex. The N-terminal arm, domain 1, the linker loop and domain 2 are shown in blue, orange, magenta and cyan, respectively. The local DNA axis was calculated by the program CURVES (Lavery et al., 1988) and is shown in red. The structure was drawn by the programs MOLSCRIPT (Kraulis, 1991) and Raster3D (Merritt and Bacon, 1997). (D) The amino acid sequence and the secondary structure of CENP-B1–129.
Table I. Crystallographic statistics.
| Data set | Native | IdU12 | IdU14 | IdU19 |
|---|---|---|---|---|
| Data collection | ||||
| resolution (Å) | 2.5 | 2.5 | 2.5 | 3.5 |
| reflections | 417 349 | 619 774 | 397 619 | 58 051 |
| (unique) | (19 931) | (19 126) | (19 327) | (7012) |
| <I/σ(I)> | 13.9 | 14.5 | 8.1 | 2.2 |
| completeness (%) | 99.6 | 98.5 | 99.8 | 97.9 |
| Rmerge (%)a | 4.5 | 4.9 | 6.9 | 18.6 |
| Phasing | ||||
| phasing power | 1.34 | 0.96 | 0.80 | |
| Rcullis | 0.59 | 0.74 | 0.76 | |
| Refinement | ||||
| resolution (Å) | 30.0–2.5 | |||
| reflections | 19 313 | |||
| R-factor (%)b | 22.53 | |||
| Rfree (%)c | 26.53 | |||
| r.m.s.d. bond length (Å) | 0.006 | |||
| r.m.s.d. bond angles (°) | 1.08 |
aRmerge = Σ|I – <I>|/ΣI, where I is the intensity of an individual measurement and <I> is the mean intensity value.
bR-factor = Σ||Fo| – |Fc||/Σ|Fo|, where Fo and Fc are the observed and calculated structure factor, repectively.
cRfree is the same as the R-factor calculated using 5% of total reflections selected randomly.
Protein structure
The crystal structure of CENP-B1–129 is divided into four well-defined regions: the N-terminal arm (amino acid residues 1–9), domain 1 (residues 10–64), the linker loop (residues 65–74) and domain 2 (residues 75–129) (Figure 1C and D). Domains 1 and 2 of CENP-B1–129 have a helix–turn–helix motif and bind to adjacent major grooves of DNA. DNA-complexed structures of a tandem repeat of two helix–turn–helix domains have been determined for the DNA-binding domains of the human Myb protein (Ogata et al., 1994), the human Pax6 protein (Xu et al., 1999) and the yeast telomere binding protein RAP1 (König et al., 1996). Domain 1 of CENP-B1–129 has five α-helices (helices 1–5, Figure 1D). We previously reported the solution structure of a shorter CENP-B fragment consisting of residues 1–56 (Iwahara et al., 1998). Although the fragment was truncated in the middle of the helix 4 region, the solution structure of CENP-B1–56 can be superimposed onto the corresponding part of the present crystal structure of domain 1 in the DNA-bound CENP-B1–129. The presence of helices 4 and 5 in addition to the three α-helices of the canonical helix–turn–helix domain is characteristic of domain 1 of CENP-B; helix 1 is surrounded by helices 2–5 through many hydrophobic interactions. Domain 1 of CENP-B is structurally close to the DNA-binding domain of the Tc3A transposase (van Pouderoyen et al., 1997) (r.m.s.d. 1.44 Å), the N-sub domain of Pax6 (Xu et al., 1999) (r.m.s.d. 1.22 Å) and domain 1 of RAP1 (König et al., 1996) (r.m.s.d. 1.49 Å). Domain 2 of CENP-B has three α-helices (helices 6–8, Figure 1C and D), and the helix–turn–helix is formed by helices 7 and 8 and a turn region of seven amino acid residues. The C-subdomain of Pax6 and domain 2 of RAP1 show high similarity to domain 2 of CENP-B, with r.m.s.d. values of 1.97 and 2.18 Å, respectively (König et al., 1996; Xu et al., 1999). The CENP-B domains 1 and 2 themselves are similar to each other (r.m.s.d. 2.26 Å for the Cα atoms, except for the extra helices 4 and 5 of domain 1).
Base recognition and DNA kinks
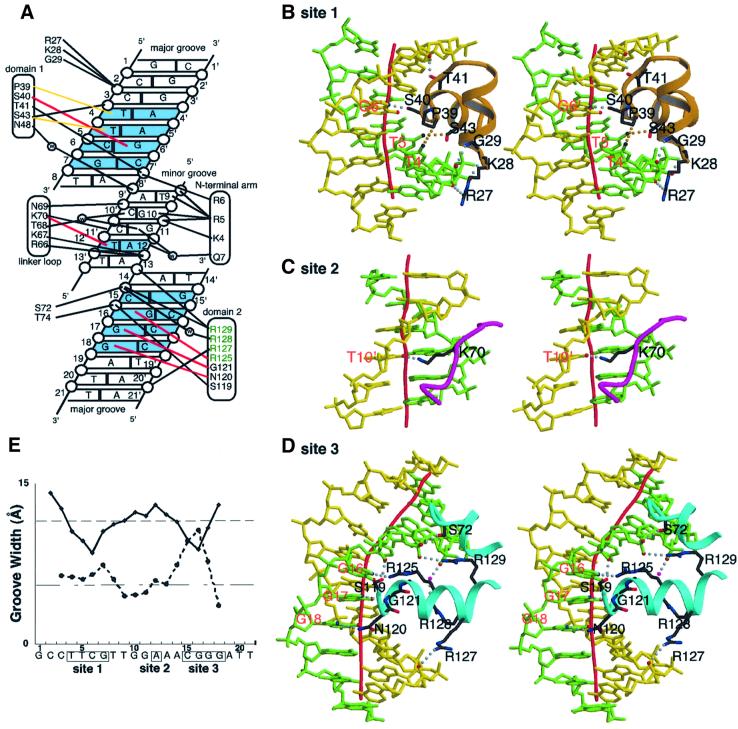
Nine out of the 17 bp of the CENP-B box are essential for the recognition by CENP-B (Masumoto et al., 1993). These essential base pairs are located in three distinct regions (sites 1, 2 and 3), which have the sequences T4–T5–C6–G7, A12, and C15–G16–G17–G18, respectively (Figure 1A). In the present complex structure, CENP-B1–129 makes direct contacts with these essential sites. Helix 3 of domain 1 lies along the major groove and recognizes the essential sequences in site 1 (Figure 2A and B). In site 1 (from T4:A4′ to G7:C7′), T4 and T5 of one strand are recognized by Pro39 and Ser43, respectively, through van der Waals interactions, and the N7 atom of G6′ of the other strand forms a hydrogen bond with the Oγ of Ser40 (Figure 2A and B). These interactions bend the DNA grooves at bp 6 of site 1, and induce a local DNA kink of 16° (Figures 2E and 3A). The G7:C7′ base pair, which is indispensable for the CENP-B binding (Masumoto et al., 1993), is not recognized directly by CENP-B in the crystal structure (Figure 2A). This base pair may be important for CENP-B to induce the kinked DNA structure, because replacement by another base pair, such as A:T, causes steric hindrance in the DNA structure (data not shown). In site 2 (A12:T12′), the O4 of T12′ is recognized by the Nζ atom of Lys70 of the linker loop through hydrogen bond formation in the minor groove (Figure 2A and C). The DNA around site 2 is only slightly distorted as compared with the B-form DNA. Arg5 of the N-terminal arm stacks with the base moiety of G10, and the side chain amino group of Arg5 forms a hydrogen bond with the O2 group of T9 in the same minor groove (Figure 2A).
Fig. 2. Protein–DNA interactions. (A) Schematic diagram summarizing the DNA contacts by CENP-B1–129. The essential base pairs in sites 1, 2 and 3 are colored blue. Water molecules are denoted as open circles labeled with W. Open circles represent phosphates. Hydrogen bonds and salt bridges with the backbone phosphate groups are indicated with thick black lines. Specific recognitions of bases in sites 1, 2 and 3 are shown in red (hydrogen bonds) and yellow (van der Waals interactions) lines. (B–D) Specific interactions at sites 1, 2 and 3, respectively (stereo view). DNA strands are shown in green and yellow. Dotted lines in gray represent hydrogen bonds, and those in orange represent van der Waals interactions. (E) Graphic representation of the widths of the major and minor grooves, calculated by CURVES (Lavery et al., 1988). The vertical axis indicates the groove width (Å) and the horizontal axis indicates the base number. The solid and dashed lines indicate the widths of the major and minor grooves, respectively.
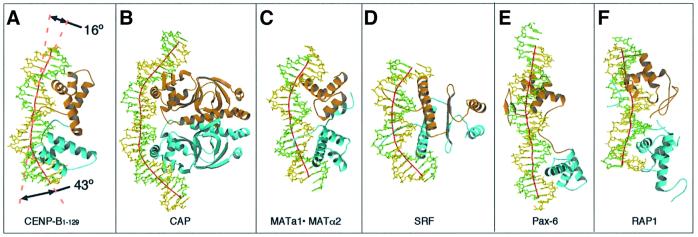
Fig. 3. Structural comparisons of the CENP-B·DNA complex (A) with the complexes of the E.coli CAP protein (B; Schultz et al., 1991), the yeast MATa1/MATα2 protein (C; Li et al., 1995), the human serum response factor (SRF) (D; Pellegrini et al., 1995), the human Pax6 protein (E; Xu et al., 1999) and the yeast RAP1 protein (F; König et al., 1996), respectively. The local DNA axis was calculated by the program CURVES (Lavery et al., 1988) and is shown as a red line. Arrows in (A) indicate the angles of local DNA kinks found in the CENP-B complex. All of the structures are represented in the orientation where the DNA molecules show the greatest bending.
Site 3, which consists of base pairs from C15:G15′ to G18:C18′ (Figure 1A), is kinked locally by CENP-B1–129 through a novel mechanism. In contrast to helix 3 of domain 1, the orientation of the recognition helix of domain 2 (helix 8) relative to the major groove is very different from the cases of canonical helix–turn–helix proteins. Actually, in site 3, helix 8 penetrates perpendicularly into the major groove around bp 16 (Figures 1D and 2D). Thus, N7 of G16, N7 of G17 and O6 of G18 are recognized by the side chain NH2 group of Arg125, the main chain NH group of Gly121 and the side chain NH2 group of Asn120, respectively, through hydrogen bonding (Figure 2A and D). In addition to the involvement of Arg125 in the base-specific recognition, helix 8 contains three more argine residues (Arg127, Arg128 and Arg129), all of which are involved in the DNA interaction (Figure 2A and D). The arginine cluster in the recognition helix is unique to the CENP-B domain 2 among helix– turn–helix proteins. Arg128 interacts with the 5′ phosphate group of C15 through a water molecule (Figure 2A). Remarkably, Arg127 and Arg129 bridge the major groove through direct interactions with the phosphate groups of A20′ and A14, respectively (Figure 2A and D). These bridges between the arginine residues of helix 8 and the backbone phosphates of the major groove drastically narrow the width of the major groove, expanding in turn that of the minor groove (Figure 2E and D), and induce the 43° local kink of the DNA at bp 16 in site 3 (Figure 3A). This kinking mechanism is unique, and is now named the ‘phosphate bridging by an arginine-rich helix’ (PBAH). The C15:G15′ base pair, which is essential to CENP-B binding (Masumoto et al., 1993), is not recognized directly by CENP-B. This is probably because C15:G15′ is sterically required as is G7:C7′ described above; the present kinked structure causes steric hindrance for the substitution of C15:G15′ by any other base pair (not shown).
A DNA kink by major groove protein binding has been found only for the CAP·DNA complex (Figure 3B, Schultz et al., 1991). The CAP protein forms a symmetric homodimer, and is bound with a DNA that is nicked at the protein contact sites. A sharp DNA kink (∼40°) was induced by the extensive interactions of the three α-helices with the bases and the phosphates on the major groove (Figure 4B).
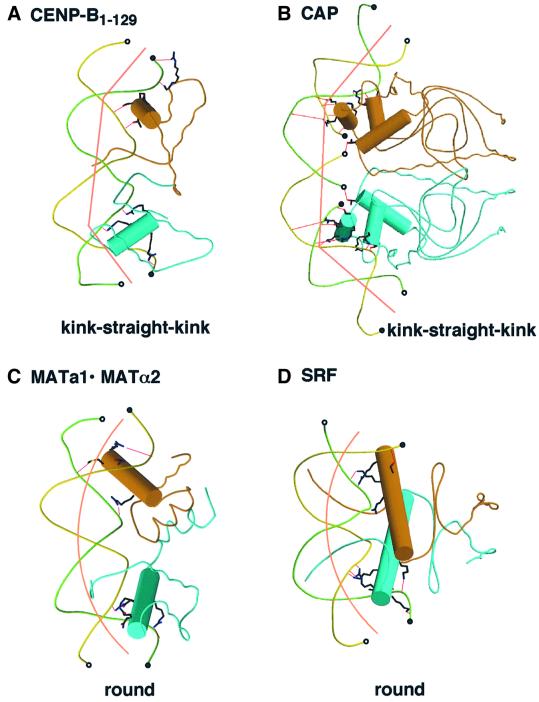
Fig. 4. Schematic drawings of DNA structures in the complexes with CENP-B (A), CAP (B), MATa1/MATα2 (C) and SRF (D). Cylinders indicate recognition helices for DNA binding. The 3′ and 5′ ends of DNA molecules are indicated by open and closed circles, respectively. The complex with CAP contains two molecules of double-stranded DNA, which has a four base overhang at the 3′ end, and the complexes with CENP-B, MATa1/MATα2 and SRF contain one molecule of double-stranded DNA.
Overall DNA bending
In the CENP-B·DNA complex, the kinks only occur at the two protein contact sites, and the DNA region between the two kinks is straight, resulting in a ‘kink–straight–kink’ bend (Figure 4A). In the CAP·DNA complex (Figure 4B), the DNA region between the two kinks is also straight, which might be due to the nicks in the DNA at the protein contact sites. In the CENP-B·DNA complex, the kinks at sites 1 and 3 are in the same plane, and the overall bending angle of 59° is therefore nearly equal to the sum of the two kink angles (Figure 3A). These kinks are not due to pseudocontinuous superhelix formation in the crystal (Li et al., 1995). One 21 bp DNA molecule contacts another symmetry-related DNA molecule in a manner twisted by 90° to form a straight pseudocontinuous DNA helix of 42 bp, but the two ends as well as the flank of the pseudocontinuous DNA helix have no crystal packing interactions with any other symmetry-related DNA molecules.
In contrast to the ‘kink–straight–kink’ bend of the CENP-B box DNA, other large DNA bends due to major groove protein binding occur gradually over the region between the two protein contact sites. These ‘round bends’ have thus far been observed mostly in systems that have extensive protein–protein interactions, such as dimer formation and domain–domain interactions. In the cases of helix–turn–helix proteins, the yeast MATa1·MATα2 heterodimer causes a round bend with a bending angle of 60° (Figures 3C and 4C) (Li et al., 1995). Similar round bends of DNA have been found for two non-helix–turn– helix proteins, the human serum response factor (SRF) and MCM1 (Pellegrini et al., 1995; Tan et al., 1998), which both belong to the MADS family (Figures 3D and 4D). These MADS family proteins form homodimers; coiled-coil α-helices formed by the well-conserved peptide segments (the MADS box) lie horizontally along the major groove of the specific DNA (Figure 3D). Therefore, the DNA bending mechanism of the CENP-B·DNA complex is totally different from that of the other DNA-bending proteins. On the other hand, the protein structures of the tandemly arranged helix–turn–helix domains of CENP-B are similar to those of the Pax6 and RAP1 DNA-binding regions (König et al., 1996; Xu et al., 1999). Two helix–turn–helix domains did not interact directly with each other, similar to the case of CENP-B in the complexes of the Pax6 and RAP1 DNA-binding regions with their cognate DNA sites. However, the DNA structures in the complexes are quite different; in the cases of Pax6 and RAP1, the DNAs were only slightly bent (Figure 3E and F).
Centromeric chromatin
It has been reported that CENP-B induced nucleosome positioning adjacent to the CENP-B box sequence (Yoda et al., 1998). The CENP-B box sequence exists in every other α-satellite repeat (171 bp), and CENP-B accommodates a pair of centromeric nucleosomes between two CENP-B boxes (Yoda et al., 1998). Furthermore, CENP-B bundles two distant CENP-B boxes through dimer formation (Yoda et al., 1998). On the other hand, it is proposed that the centromere nucleosome contains histones H2A, H2B, H4 and a centromere-specific histone H3 variant, CENP-A (Yoda et al., 2000). Recently, it was reported that depletion of CENP-A caused significant dispersions of CENP-B and CENP-C in mice (Howman et al., 2000), suggesting that CENP-A, CENP-B and CENP-C form the centromere cooperatively. In the present study, we found that binding of CENP-B to the CENP-B box sequence bends it by ∼60°. The kinked DNA structure induced by CENP-B should be taken into account when the mechanism of centromeric nucleosome condensation is investigated.
Materials and methods
Purification of CENP-B1–129
CENP-B 1–129 was overexpressed in Escherichia coli JM109 (DE3) cells under the control of the T7 promoter. The E.coli strains carrying the expression vector of CENP-B1–129 were grown at 37°C, and isopropyl-β-d-thiogalactopyranoside (IPTG; 1.5 mM) was added at an OD600 = 0.8 to induce the protein expression. After overnight cultivation at 37°C, the cells were harvested and were disrupted by sonication in buffer S [50 mM Tris–HCl pH 8.0, 5 mM EDTA, 100 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF)]. Then, the sample was centrifuged at 27 000 g for 20 min and CENP-B1–129 was recovered in the insoluble fraction. The precipitate was sonicated in buffer S and was centrifuged again. Proteins in the insoluble fraction were extracted under denaturing conditions with buffer A [6 M urea, 20 mM Tris–HCl pH 8.0, 10 mM dithiothreitol (DTT), 2 mM EDTA, 1 mM PMSF, 2.8 M NaCl] and, after centrifugation, the resulting pellet was discarded. The proteins including CENP-B1–129 were loaded onto a 30 ml butyl-Toyopearl M (TOHSO, Japan) column, and the resin was washed with 150 ml of buffer A. CENP-B1–129 was eluted with buffer A containing 0.1 M NaCl. The fractions were dialyzed against buffer C (6 M urea, 50 mM potassium phosphate pH 6.0, 5 mM DTT, 1 mM PMSF, 150 mM NaCl, 10% glycerol) and were loaded onto a 15 ml phosphocellulose column (Whatman P11). The resin was washed with 50 ml of buffer C, and CENP-B1–129 was eluted with a linear gradient of potassium phosphate pH 6.0, from 50 to 700 mM. The purified fractions were concentrated to 1–5 mg/ml by Centricon 10 ultrafiltration (Amicon).
DNA
The 21mer oligonucleotides containing the CENP-B box sequence were purchased from Oligo-Espec custom synthesis. These oligonucleotides were purified by Resource Q chromatography (Amersham Pharmacia Biotech) with a linear gradient of NaCl from 0.2 to 0.5 M. Three iodinated derivatives, in which the thymine is replaced by a 5-iodouracil at positions 12, 14 and 19 (IdU12, IdU14 and IdU19, respectively), were also prepared.
Purification of the CENP-B1–129·DNA complex
The purified CENP-B1–129 and both DNA strands were mixed in a 1:1 stoichiometry (molar ratio) under denaturing conditions. After an incubation for 120 min, the samples were dialyzed against buffer G (50 mM HEPES–KOH pH 7.5, 5 mM DTT, 150 mM NaCl, 10% glycerol). The CENP-B1–129·DNA complex was concentrated to 1–2 ml and was purified on a Hiload Superdex 75 (Pharmacia) gel filtration column in buffer G without glycerol.
Crystallization
The protein–DNA complex was concentrated by Centriprep-3 ultrafiltration (Amicon) to 0.2–0.4 mM (∼3–6 mg/ml). Co-crystals were grown at 20°C by the hanging drop vapor diffusion method. The drops contained 1 µl of the complex solution and 1 µl of the reservoir solution (100 mM sodium acetate, 50 mM sodium cacodylate pH 6.5 and 15% 2-methylpentane-2,4-diol). The drops were equilibrated against 0.5 ml of reservoir solution. Crystals grew in 1 day to a size of ∼0.3 × 0.3 × 0.5 mm3, and belong to the trigonal space group P3112 (a = b = 83.3 Å, c = 139.0 Å). Prior to data collection, the crystals were equilibrated in stabilization buffer (100 mM sodium acetate, 50 mM sodium cacodylate pH 6.5, 20% MPD) for 1 h. Then, the crystals were flash frozen in a stream of liquid nitrogen at 100 K. All data sets were collected from the beamline BL44B2 in SPring-8, Harima. The data were processed and scaled using the DENZO and SCALEPACK programs (Otwinowski and Minor, 1997).
Phasing and refinement
Phase calculations were carried out with the CCP4 program (Collaborative Computational Project Number 4, 1994). The iodine sites of IdU12, IdU14 and IdU19 were determined by the RSPS program (Collaborative Computational Project Number 4, 1994) from the isomorphous difference Patterson map. The initial phases from the IdU12 derivative were used to locate the positions of the iodine in the IdU14 and IdU19 derivatives by difference Fourier analysis, which were consistent with the Patterson search solutions. The heavy atom parameters were refined with the program MLPHALE (Collaborative Computational Project Number 4, 1994), and the resulting MIRAS map was of excellent quality. The map was improved further by density modification procedures, such as solvent flattening and phase extension, using the program DM (Collaborative Computational Project Number 4, 1994). An atomic model was fitted into the electron density map using the graphics program O (Jones et al., 1991). The positions of the iodines guided the assignment of the DNA residues in the electron density map. Crystallographic positional and simulated annealing refinements were carried out against the 2.5 Å data set using CNS (Brünger et al., 1998). The R-factor of the final model is 22.5% (Rfree = 26.5%) using data from 30 to 2.5 Å. Coordinates were deposited with the Protein Data Bank (RCSB id code: rcsb012438 and PDB ID code: 1HLV).
Acknowledgments
Acknowledgements
We thank Drs N.Kamiya and S.Adachi for the data collection at SPring-8, and also thank Drs M.Shirouzu and T.Terada for sharing beam time at SPring-8. This work was supported in part by the Bioarchitect Research Program (RIKEN) and also by a Grant-in-Aid from the Ministry of Education, Science, Sports and Culture, Japan.
References
- Brünger A.T et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Earnshaw W.C. and Rothfield,N. (1985) Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma, 91, 313–321. [DOI] [PubMed] [Google Scholar]
- Earnshaw W.C., Ratrie,H.,III and Stetten,G. (1989) Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma, 98, 1–12. [DOI] [PubMed] [Google Scholar]
- Fowler K.J., Hudson,D.F., Salamonsen,L.A., Edmondson,S.R., Earle,E., Sibson,M.C. and Choo,K.H.A. (2000) Uterine dysfunction and genetic modifiers in centromere protein B-deficient mice. Genome Res., 10, 30–41. [PMC free article] [PubMed] [Google Scholar]
- Howman E.V., Fowler,K.J., Newson,A.J., Redward,S., MacDonald,A.C., Kalitsis,P. and Choo,K.H.A. (2000) Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl Acad. Sci. USA, 97, 1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson D.F. et al. (1998) Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J. Cell Biol., 141, 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeno M., Masumoto,H. and Okazaki,T. (1994) Distribution of CENP-B boxes reflected in CREST centromere antigenic sites on long-range α-satellite DNA arrays of human chromosome 21. Hum. Mol. Genet., 3, 1245–1257. [DOI] [PubMed] [Google Scholar]
- Ikeno M., Grimes,B., Okazaki,T., Nakano,M., Saitoh,K., Hoshino,H., McGill,N., Cooke,H. and Masumoto,H. (1998) Construction of YAC-based mammalian artificial chromosomes. Nature Biotechnol., 16, 431–439. [DOI] [PubMed] [Google Scholar]
- Iwahara J., Kigawa,T., Kitagawa,K., Masumoto,H., Okazaki,T. and Yokoyama,S. (1998) A helix–turn–helix structure unit in human centromere protein B (CENP-B). EMBO J., 17, 827–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou,J-Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kipling D. and Warburton,P.E. (1997) Centromeres, CENP-B and Tigger too. Trends Genet., 13, 141–145. [DOI] [PubMed] [Google Scholar]
- Kipling D., Mitchell,A.P., Masumoto,H., Wilson,H.E., Nicol,L. and Cooke,H.J. (1995) CENP-B binds a novel centromeric sequence in the Asian mouse Mus caroli.Mol. Cell. Biol., 15, 4009–4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König P., Giraldo,R., Chapman,L. and Rhodes,D. (1996) The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell, 85, 125–136. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Lavery R. and Skelenar,H. (1988) Definiton of generalized helicoidal parameters and an axis of curvature for irregular nucleic acids. J. Biomol. Struct. Dyn., 6, 63–91. [DOI] [PubMed] [Google Scholar]
- Li T., Stark,M.R., Johnson,A.D. and Wolberger,C. (1995) Crystal structure of the MATa1/MATα2 homeodomain heterodimer bound to DNA. Science, 270, 262–269. [DOI] [PubMed] [Google Scholar]
- Masumoto H., Masukata,H., Muro,Y., Nozaki,N. and Okazaki,T. (1989) A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J. Cell Biol., 109, 1963–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H., Yoda,K., Ikeno,M., Kitagawa,K., Muro,Y. and Okazaki,T. (1993) Properties of CENP-B and its Target Sequence in a Satellite DNA. Springer-Verlag, Berlin.
- Merritt E.A. and Bacon,D.J. (1997) Raster3D: photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Ogata K., Morikawa,S., Nakamura,H., Sekikawa,A., Inoue,T., Kanai,H., Sarai,A., Ishii,S. and Nishimura,Y. (1994) Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell, 79, 639–648. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Palmer D.K., O’Day,K., Trong,H.L., Charbonneau,H. and Margolis,R.L. (1991) Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc. Natl Acad. Sci. USA, 88, 3734–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L., Tan,S. and Richmond,T.J. (1995) Structure of serum response factor core bound to DNA. Nature, 376, 490–497. [DOI] [PubMed] [Google Scholar]
- Perez-Castro A.V., Shamanski,F.L., Meneses,J.J., Lovato,T.L., Vogel,K.G., Moyzis,R.K. and Pedersen,R. (1998) Centromeric protein B null mice are viable with no apparent abnormalities. Dev. Biol., 201, 135–143 [DOI] [PubMed] [Google Scholar]
- Saitoh H., Tomkiel,J., Cooke,C.A., Patrie,H.,III, Maurer,M., Rothfield,N.F. and Earnshaw,W.C. (1992) CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell, 70, 115–125. [DOI] [PubMed] [Google Scholar]
- Schultz S.C., Shields,G.C. and Steitz,T.A. (1991) Crystal structure of a CAP–DNA complex: the DNA is bent by 90°. Science, 253, 1001–1007. [DOI] [PubMed] [Google Scholar]
- Smit A.F.A. and Riggs,A.D. (1996) Tiggers and DNA transposon fossils in the human genome. Proc. Natl Acad. Sci. USA, 93, 1443–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Yata,H., Muro,Y. and Himeno,M. (1994) Human centromere protein C (CENP-C) is a DNA-binding protein which possesses a novel DNA-binding motif. J. Biochem. (Tokyo), 116, 877–881. [DOI] [PubMed] [Google Scholar]
- Sullivan K.F., Hechenberger,M. and Masri,K. (1994) Human CENP-A contains a histone H3 related histome fold domain that is required for targeting to the centromere. J. Cell Biol., 127, 581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S. and Richmond,T.J. (1998) Crystal structure of the yeast MATα2/MCM1/DNA ternary complex. Nature, 391, 660–666. [DOI] [PubMed] [Google Scholar]
- Toth M., Grimsby,J., Buzsaki,G. and Donovan,G.P. (1995) Epileptic seizures caused by inactivation of a novel gene, jerky, related to centromere binding protein-B in transgenic mice. Nature Genet., 11, 71–75. [DOI] [PubMed] [Google Scholar]
- van Pouderoyen G., Ketting,R.F., Perrakis,A., Plasterk,R.H. and Sixma,T.K. (1997) Crystal structure of the specific DNA-binding domain of Tc3 transposase of C.elegans in complex with transposon DNA. EMBO J., 16, 6044–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.E, Rould,M.A., Xu,W., Epstein,J.A., Maas,R.L. and Pabo,C. (1999) Crystal structure of the human Pax6 paired domain–DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding. Genes Dev., 13, 1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda K., Kitagawa,K., Masumoto,H., Muro,Y. and Okazaki,T. (1992) A human centromere protein, CENP-B, has a DNA binding domain containing four potential α-helices at the NH2 terminus, which is separable from dimerizing activity. J. Cell Biol., 119, 1413–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda K., Ando,S., Okuda,A., Kikuchi,A. and Okazaki,T. (1998) In vitro assembly of the CENP-B/α-satellite DNA/core histone complex: CENP-B causes nucleosome positioning. Genes Cells, 3, 533–548. [DOI] [PubMed] [Google Scholar]
- Yoda K., Ando,S., Morishita,S., Houmura,K., Hashimoto,K., Takeyasu,K. and Okazaki,T. (2000) Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc. Natl Acad. Sci. USA, 97, 7266–7271. [DOI] [PMC free article] [PubMed] [Google Scholar]