Abstract
Processes vital to life such as respiration and photosynthesis critically depend on the availability of tetrapyrroles including hemes and chlorophylls. tRNA-dependent catalysis generally is associated with protein biosynthesis. An exception is the reduction of glutamyl-tRNA to glutamate-1-semialdehyde by the enzyme glutamyl-tRNA reductase. This reaction is the indispensable initiating step of tetrapyrrole biosynthesis in plants and most prokaryotes. The crystal structure of glutamyl-tRNA reductase from the archaeon Methanopyrus kandleri in complex with the substrate-like inhibitor glutamycin at 1.9 Å resolution reveals an extended yet planar V-shaped dimer. The well defined interactions of the inhibitor with the active site support a thioester-mediated reduction process. Modeling the glutamyl-tRNA onto each monomer reveals an extensive protein–tRNA interface. We furthermore propose a model whereby the large void of glutamyl-tRNA reductase is occupied by glutamate-1-semialdehyde-1,2-mutase, the subsequent enzyme of this pathway, allowing for the efficient synthesis of 5-aminolevulinic acid, the common precursor of all tetrapyrroles.
Keywords: crystal structure/glutamyl-tRNA reductase/metabolic channeling/tetrapyrrole biosynthesis/tRNA
Introduction
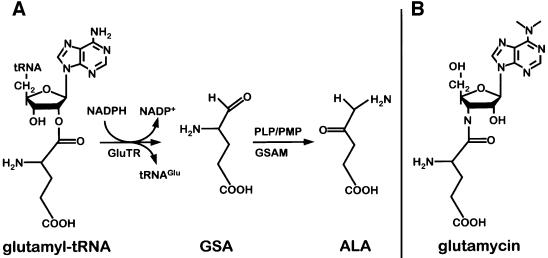
The function of tRNA is associated primarily with ribosomal protein biosynthesis (Al-Karadaghi et al., 2000). Central to this process is the tRNA-mediated activation of amino acids catalyzed by aminoacyl-tRNA synthetases. In protein biosynthesis, this activation is the energetic prerequisite for peptide bond formation at the ribosome. Intriguingly, tRNA-activated glutamate is also utilized to initiate a second major metabolic pathway, tetrapyrrole biosynthesis in plants, archaea and the majority of bacteria (Jahn et al., 1992). Here the common precursor of all tetrapyrroles, 5-aminolevulinic acid (ALA), is generated from glutamyl-tRNA. In a first step, the activated glutamate is reduced to glutamate-1-semialdehyde (GSA) by the NADPH-dependent enzyme glutamyl-tRNA reductase (GluTR). In this reaction, a cysteine residue nucleophilically attacks the aminoacyl bond of glutamyl-tRNA leading to a highly reactive thioester intermediate (Moser et al., 1999). Subsequently, the thioester is reduced via hydride transfer from NADPH, leading to the formation of GSA. In a second step, GSA is transaminated by glutamate-1-semialdehyde aminomutase (GSAM), leading to the formation of ALA (Smith et al., 1992) (Figure 1).
Fig. 1. Pathway of tRNA-dependent formation of 5-aminolevulinic acid (ALA) and structure of the substrate-like inhibitor glutamycin. (A) Glutamyl-tRNA reductase (GluTR) reduces tRNA-bound glutamate to glutamate-1-semialdehyde (GSA). Glutamate-1-semialdehyde aminomutase (GSAM) transaminates GSA to ALA. (B) The inhibitor glutamycin is an analog of the 3′-terminal nucleotide of acylated tRNAGlu.
Here we present the first crystal structure of a GluTR, in complex with the competitive substrate-like inhibitor glutamycin. The structure was solved by multiple isomorphous replacement (MIR) (Table I) and refined at a resolution of 1.95 Å (Table II).
Table I. MIR phasing statistics.
| Native | TlCl | K2IrCl6 | K2HgI4 | |
|---|---|---|---|---|
| Soak (mM) | – | 0.6 | 3 | 2 |
| Soaking time (days) | – | 2 | 2 | 3 |
| Resolution (Å) | 1.9 | 2.5 | 2.6 | 3.0 |
| Completeness (%) | 91.1 | 100.0 | 99.7 | 99.6 |
| (last shell) | 69.1 | 99.7 | 99.6 | 99.3 |
| Redundancy | 2.5 | 5.4 | 5.2 | 5.7 |
| Average I/σ | 12.0 | 15.9 | 9.8 | 9.2 |
| Rsym (%)a | 7.3 | 8.3 | 7.4 | 8.5 |
| (last shell) | 32.4 | 30.5 | 24.4 | 21.0 |
| Sites | – | 1 | 1 | 1 |
| Rcullis (%)b | – | 0.80 | 0.67 | 0.73 |
| Phasing power | – | 1.53 | 1.60 | 1.19 |
| Figure of merit | 0.79 |
aRsym = Σj|Ij – <I>|Σj Ij with j = (hkl, i), where Ij is the intensity for the reflection j, and <I> is the mean intensity for multiply measured reflections.
bRcullis = Σj||FPH – FP| – FH|/Σj FH with j = (centric hkl), where FH is the calculated heavy atom structure factor.
Table II. Refinement statistics.
| Resolution (Å) | 1.95 |
|---|---|
| No. of atoms: protein, inhibitor, water | 3143, 29, 374 |
| R-factor (%)a | 19.7 |
| Rfree (%)b | 26.0 |
| R.m.s.d. bond lengths (Å) | 0.03 |
| R.m.s.d. bond angles (°) | 3.0 |
aR-factor = Σ|Fo – Fc|/ΣFo, where Fo and Fc are the observed and calculated structure factors, respectively.
bRfree is the cross-validated R-factor computed for a test set of 5% of the reflections, which were omitted during refinement (Brünger, 1992).
Results and discussion
Description of the structure
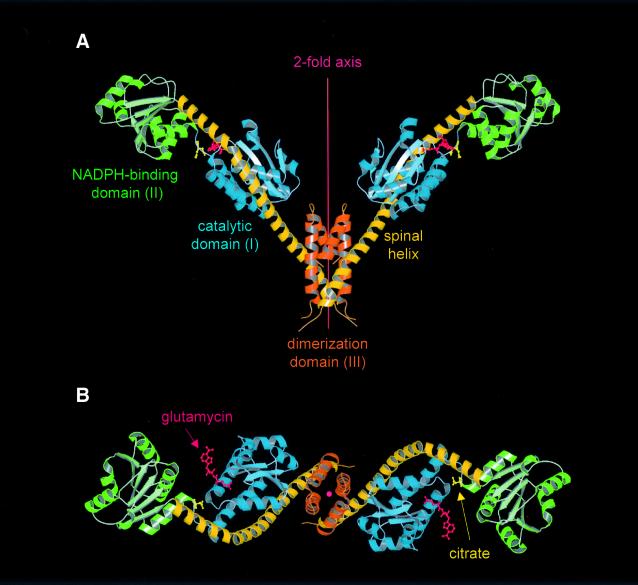
GluTR is a dimeric protein with an unusual V-shaped fold (Figure 2). The monomers, each constituting one leg of the V, are ∼125 Å in length, 45 × 45 Å2 in cross-section and together span a maximum width of ∼165 Å. Each monomer consists of three distinct domains linearly arranged along a common axis and connected by a curved ‘spinal’ α-helix of 110 Å in length. Residues 1–145 constitute the N-terminal domain (domain I) consisting of two subdomains (Figure 3A). Residues 1–77 form a central antiparallel four-stranded β-sheet decorated on one side by three α-helices. The topology, though modified to βαββααβ, most closely resembles the common βαββαβ-motif of, amongst others, RNA-binding proteins (Lindahl et al., 1994; Schluenzen et al., 2000). Residues 78–141 form three antiparallel α-helices packed against the central β-sheet. Together with the central part of the spinal α-helix, they constitute a distorted antiparallel four-helix bundle. A short linker (residues 142–148) connects domain I to domain II (residues 149–285). Domain II has a classical nucleotide-binding fold (Carugo and Argos, 1997) composed of a central six-stranded parallel β-sheet, characteristic βαβ motifs and a conserved glycine-rich loop (Figure 3B). Domain II is directly followed by the spinal α-helix (residues 286–352) comprising 18 α-helical turns with a visible kink induced by Pro305. C-terminally, the spinal α-helix becomes part of a three-helix dimerization domain (domain III), which forms an unusual six-helix bundle with the neighboring GluTR monomer around a crystallographic 2-fold axis (Figure 3C). Residues 355–360, connecting the spinal helix to the second helix of domain III, are not visible in the electron density map. As a result, it is unclear whether α-helices from one subunit are arranged alongside each other or whether α-helices from alternate subunits interdigitate.
Fig. 2. Structure of the GluTR dimer viewed (A) perpendicular to and (B) along the 2-fold axis. Monomers consist of three structural domains: (I) an N-terminal catalytic domain (blue); (II) an NAPDH-binding domain (green); and (III) a C-terminal dimerization domain (orange)—connected by an extended 18-turn ‘spinal’ α-helix (dark-yellow). Glutamycin (red) binds at the catalytic domain. At the deep end of the large crevice between domains I and II a citrate anion (yellow) is bound. Figures 2, 3 and 5 were generated using MOLSCRIPT (Kraulis, 1991) rendered with RASTER3D (Merrit and Murphy, 1994).
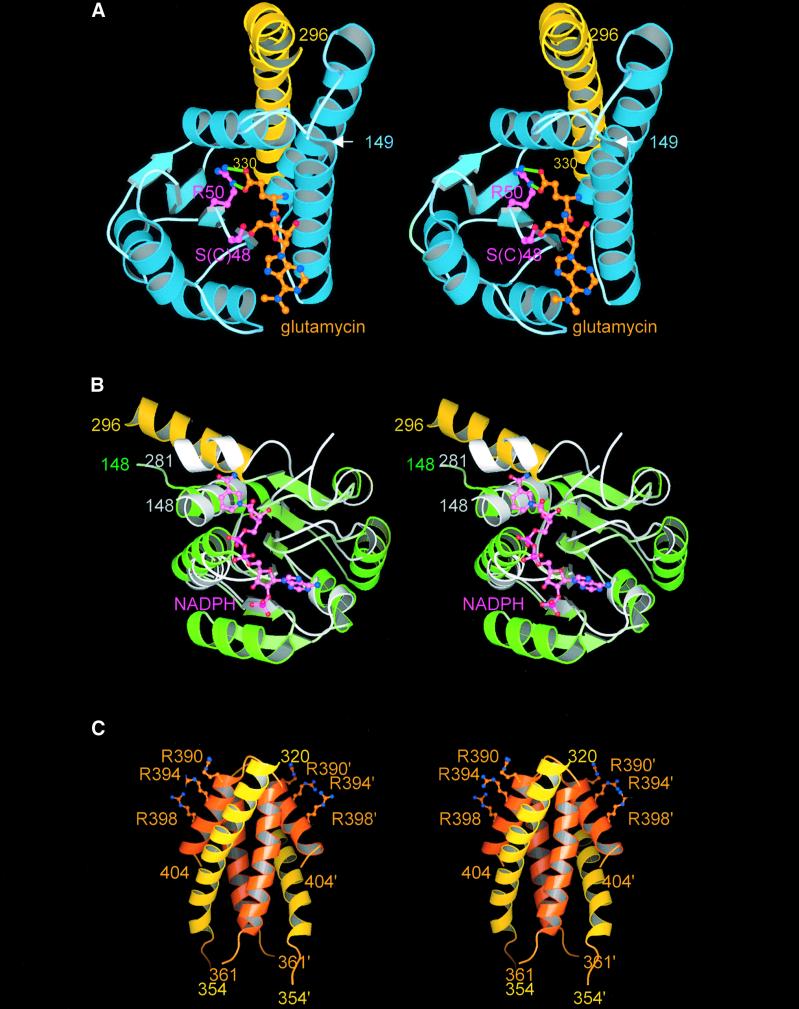
Fig. 3. Stereo images of the three domains of GluTR. Coloring of the domains and the spinal helix are as in Figure 2. (A) Catalytic domain with bound glutamycin (orange) viewed towards the glutamate recognition pocket. The salt bridge-like interactions between the glutamate moiety of glutamycin and Arg50 (pink) are indicated by green lines. The catalytic side chain of Ser/Cys48 is also shown. (B) NADPH-binding domain of GluTR superimposed on the NADPH-binding domain of tetrahydrofolate dehydrogenase/cyclohydrolase (light gray). NADPH bound to this domain is shown in pink. (C) Dimerization domain. Residues of the second monomer are primed. Also shown are the side chains of Arg390, Arg394 and Arg398 that we postulate may interact with the anticodon region of glutamyl-tRNA.
Active site and catalytic mechanism
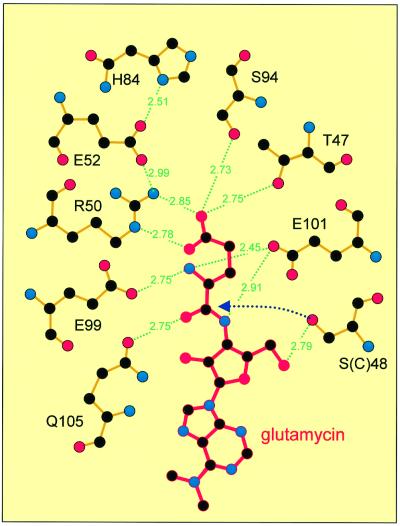
The present crystal structure of GluTR is a complex with the inhibitor glutamycin (Moser et al., 1999), a synthetic glutamate analog of the aminonucleoside antibiotic compound puromycin (Figure 1). Glutamycin is bound within a deep pocket in domain I of GluTR specifically recognized by an array of strictly conserved residues (Figures 3A and 4). The principal interaction at the bottom of the pocket is the bidentate salt bridge between the carboxylate group of glutamycin and Arg50, allowing for the differentiation of glutamate as opposed to glutamine. Other hydrogen bonds to the glutamycin carboxylate involve the side chains of Ser94 and Thr47. The α-amino-group of glutamycin is recognized by Glu99, Glu101 and Gln105, and the amide group and ribose O3 atom again by Glu101. Arg50, which is largely buried, is held in position by an intricate hydrogen-bonding network of likewise highly conserved residues including His84, which has been shown to be essential for catalytic activity (Moser et al., 1999). Analogously, in aspartyl-tRNA synthetase (Eiler et al., 1999) and presumably glutamyl-tRNA synthetase (Sekine et al., 2001), the substrate carboxylate group is also recognized specifically via a conserved arginine at the bottom of the amino acid recognition pocket.
Fig. 4. Glutamycin (red) bound within the glutamate recognition pocket of GluTR (orange bonds). Primarily the glutamate moiety of glutamycin is recognized through a network of specific hydrogen bonds; the ribose moiety is recognized to a lesser extent. The initial step of GluTR catalysis, the nucleophilic attack of the carbonyl carbon by Cys48 (here Ser48), is indicated (dotted arrow). Adapted from LIGPLOT (Wallace et al., 1995).
The proposed catalytic mechanism for the enzyme (Moser et al., 1999) is in good agreement with the crystal structure (Figure 4). The activated α-carboxylate of glutamyl-tRNA is nucleophilically attacked by a conserved cysteine (Cys48), leading to a covalent thioacyl intermediate and release of tRNAGlu. In the present structure, Cys48 (as well as other cysteines) had to be substituted by serine to prevent non-specific intermolecular disulfide bond formation during crystallization. The mutation C48S, apart from preventing oligomerization, leads to the complete inactivation of the enzyme (Moser et al., 1999). The side chain of Ser48, exposed at the rim of the binding pocket, is in close proximity (3.9 Å) to the α-carbonyl carbon atom of glutamycin. In a second step, the highly reactive thioacyl intermediate is then reduced to GSA by an SN2-like hydride transfer from NADPH. The precise position of the nicotinamide moiety in the active complex cannot be derived from the present structure as the NADPH-binding site in domain II is too remote from the glutamate recognition pocket (see below).
Domain II closely resembles the NADPH-binding domain of human tetrahydrofolate dehydrogenase/cyclohydrolase (Holm and Sander, 1996; Allaire et al., 1998), the r.m.s. deviation for main chain atoms being 2.3 Å for 104 common residues with structurally well conserved binding pockets for the adenine moiety (Figure 3B). In GluTR, however, the NADPH-binding site is empty despite attempts to co-crystallize GluTR with NADPH or smaller analogs. It appears that the second α-helix (αB, residues 176–190) and the glycine-rich loop (connecting β1–αB, residues 174–176) are laterally shifted in GluTR, leading to a slight compression of the diphosphate recognition pocket (between the glycine-rich loop and loop β4–αE, residues 234–236) (Carugo and Argos, 1997), thus preventing NADPH binding. Based on superpositions with homologous structures, we nevertheless have derived a theoretical position for NADPH (Figure 3B). The distance between the nicotinamide moiety and the glutamate-binding pocket is, however, ∼21 Å. The NADPH-binding pocket obliquely faces the large active site crevice between domains I and II, suggesting that domain II is required to tilt substantially relative to the remaining structure in order to position NADPH for substrate reduction. Possibly this tipping of domain II induces the opening of the NADPH-binding pocket and occurs in concert with glutamyl-tRNA binding. The present open structure of GluTR may therefore be described as a ‘pre-active’ state.
The large crevice separating domains I and II is delimited at its deep end by the N-terminal third of the spinal α-helix and the loop connecting domains I and II. Within this crevice, we have identified a well-defined citrate anion specifically contacting residues of both domains I (Gln100) and II (Ser152), as well as the connecting loop (Val148) and the spinal α-helix (Arg300) (Figure 2). The citrate proved to be mandatory for the successful crystallization of the enzyme, presumably by stabilizing the present ‘pre-active’ GluTR conformation. Interestingly, Arg300, which binds the citrate anion with a total of three different hydrogen bonds, is conserved in all plant and Archaea GluTR sequences, but not, however, in all bacteria (see http://www.sanger.ac.uk/srs6bin/cgi-bin/wgetz?-e+[PFAM-ID:glutr]). Its side chain extends through a curve in the interdomain loop, presenting its positive charge at the bottom of the crevice. It is plausible that the citrate anion may partly mimic the acceptor stem of the glutamyl-tRNAGlu substrate, either as a counter ion for a tRNA backbone phosphate or for specific base recognition.
Comparison of GluTR and its theoretical model
Using multiple sequence alignments, secondary structure prediction, homology modeling and biochemical evidence, Brody et al. (1999) have proposed a theoretical model of GluTR. Subunit B of succinyl-CoA synthase served as an overall template. In principle, the model correctly identifies the domains of GluTR, including the glutamyl-thioester region (part of domain I), the NADPH-binding region (domain II) and the α-helical, glutamyl-tRNA-binding region (domain III).
Of the three domains, only the NADPH-binding domain, however, resembles its true three-dimensional structure. Clearly, the large collection of related domains available from known protein structures favors a structural comparison. Intricate details of glutamate recognition and the overall glutamyl-tRNA surface are, of course, beyond the scope of such a model. However, aspects including the dimerization domain were also not identified by the theoretical model.
Glutamyl-tRNA recognition
Does the present GluTR structure allow for the prediction of binding of the natural substrate glutamyl-tRNA? Based on the well defined location of glutamycin in the present structure, we manually positioned the acceptor stem bearing both the 5′ and 3′ ends of Thermus thermophilus tRNAGlu (Sekine et al., 2001) into the active site of GluTR and rotated the tRNA around the RNA double-helical axis of the acceptor stem. This procedure favors a single orientation for the tRNAGlu in which substantial surface complementarity between the tRNAGlu and the enzyme is achieved without incurring steric clashes (Figure 5A). In this orientation, the 3′ strand of the amino acid acceptor stem (ribonucleotides 68–70) interacts with domain I (helix A), while nucleotides 11–13 and 24–25 of the dihydrouridine hairpin fit into two neighboring shallow depressions on the surface of domain I formed by α-helices B and C. Also the anticodon bases 34–36 of the anticodon hairpin are in close proximity to α-helical domain III, thus allowing in principle for the specific recognition of the anticodon. This mode of tRNA recognition is reminiscent of the discriminating GluRS, where the anticodon of glutamyl-tRNA is recognized by an all α-helical domain (Sekine et al., 2001). Organisms lacking glutaminyl-tRNA synthetase (GlnRS) (Schön et al., 1988) mischarge glutaminyl-tRNA with glutamate and convert this to glutamine using an amidotransferase. In these organisms, GluTR needs to discriminate between glutamyl-tRNAGln and glutamyl-tRNAGlu. Anticodon recognition critically supports such discrimination. Our proposed model is corroborated by studies of barley GluTR where tRNAGlu recognition has been shown to involve nucleotides in the anticodon stem of the tRNAGlu (Willows et al., 1995). In addition, the mutual extended dimensions of GluTR and the tRNA substrate, and the proposed anti codon recognition suggest that the fidelity of tRNAGlu recognition is ensured by a multitude of individual interactions.
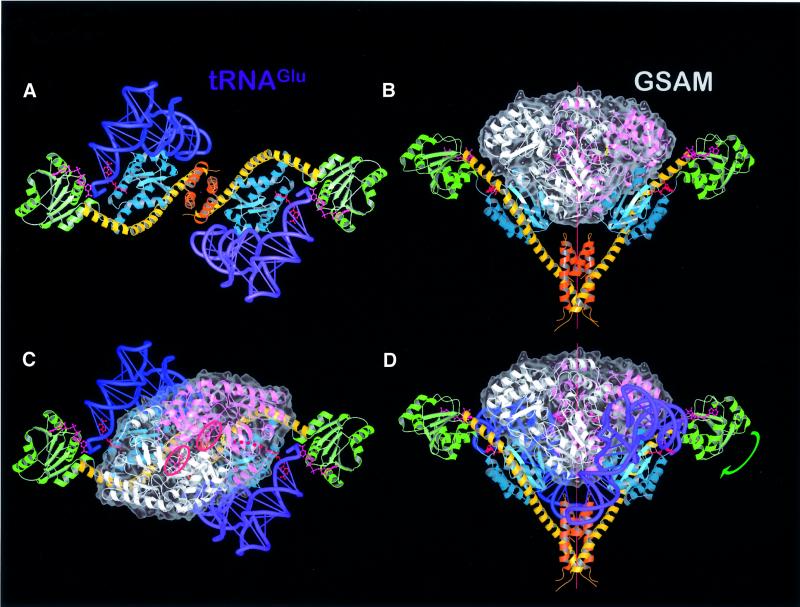
Fig. 5. Model complexes. (A) Model of GluTR–2tRNAGlu viewed along the 2-fold axis. A GluTR dimer can independently bind two tRNAs (blue and violet). (B) The open V-shape of GluTR provides sufficient space to allow the following enzyme of the tetrapyrrole biosynthesis pathway, GSAM (white and pink), to bind. The translucent surface of GSAM (gray) indicates significant surface complementarity between both protein dimers, which share a 2-fold symmetry axis. (C and D) Combining complexes (A) and (B) gives rise to a proposed ternary complex GluTR–GSAM–2tRNAGlu without steric clashes between individual constituents. The NADPH-binding domain would need to tip and rotate around the end of the spinal helix [green arrow in (D)], to close the active center and initiate the reaction. The proposed substrate path of GSA from the active site of GluTR to that of GSAM is indicated as a dotted red line in (C). The active sites of the GSAM monomers are emphasized by two red rings marking the PLP cofactors.
In our present model of the enzyme–substrate complex, tRNAGlu and domain II of GluTR interact only marginally. In fact, glutamyl-tRNA and domain I together create a large shallow surface that could accommodate domain II. To achieve this, the latter would need to swing around the N-terminal tip of the spinal α-helix. Simulating this movement in the model (arrow in Figure 5D) places NADPH in close proximity to the glutamate-binding pocket. Similarly, a slight relaxation of the elbow angle of tRNAGlu would significantly improve the fit around domain I. Such deviations from free tRNA structures have been observed before (Nissen et al., 1995, 1999; Schmitt et al., 1998). In particular, the flexible, single-stranded 3′ end of tRNAGlu as taken from the complex with T.thermophilus glutamyl-tRNA synthetase (Sekine et al., 2001) is required to adopt a tight turn conformation reminiscent of glutaminyl-tRNA bound to GlnRS (Rath et al., 1998). This conformational change has been incorporated into the model as seen in Figure 5A.
Model of a ternary complex between GluTR, glutamyl-tRNA and GSAM
In animals, fungi and some bacteria, ALA is synthesized in a one-step condensation of succinyl-CoA and glycine by ALA synthase (Kikuchi et al., 1958; May et al., 1995). The tRNA-dependent ALA formation in plants and most bacteria, in contrast, requires the concerted action of the two enzymes GluTR and GSAM (Figure 1). As yet, no biochemical evidence has indicated a direct interaction between both enzymes despite being metabolically linked by the reactive and therefore transient aldehyde GSA. A close spatial proximity between GluTR and GSAM, therefore, would ensure the efficient formation of ALA by preventing leakage of the aldehyde from this synthetic pathway. The V-shaped structure of the GluTR suggests an attractive solution to this metabolic problem. Placing GluTR alongside the similarly dimeric structure of GSAM from Synechococcus sp. (Hennig et al., 1997) immediately suggests that the open space delimited by the GluTR monomers is strikingly similar to the volume occupied by GSAM. Docking both enzymes along their 2-fold axes leads to a model complex (Figure 5B) with a striking degree of surface complementarity. In the case of GluTR, domain I and the spinal α-helix of each monomer are involved in inter-protein contacts. To improve the mutual fit further requires a small tilting of GSAM, which translates into a slight widening of the GluTR grip by moving the catalytic domains outward relative to the dimerization domain. Concomitantly, this movement of domain I relative to domain III brings the tRNA anticodon loop of tRNAGlu into closer contact with domain III of GluTR, which, as proposed, may have a role in anticodon recognition (see above). In addition, widening the GluTR embrace allows GSAM to approach the flexible loop connecting the two C-terminal α-helices in domain III.
As described, both tRNAGlu and GSAM may be docked separately onto GluTR, each in a single plausible position. Though docked separately, the model of the ternary complex of GluTR, tRNAGlu and GSAM does not lead to steric clashes between GSAM and tRNAGlu (Figure 5C and D). Instead, GSAM and tRNAGlu appear to share a substantial binding surface, indicating that the proposed ternary complex is stabilized by mutual recognition between each pair of the three components. The common binding interface for tRNAGlu therefore appears to be prerequisite for tRNAGlu recognition and binding.
Residues on the surface of both GluTR and GSAM are not particularly strongly conserved. Similarly, both tRNA sequences and nucleotide modifications vary from one species to another. Therefore, local complementarity via salt bridges or hydrophobic interactions is most probably species dependent.
Metabolic channeling between GluTR and GSAM
The most striking result of the GluTR–tRNAGlu–GSAM model complex is, however, that the putative active site entrance of each GSAM monomer (Hennig et al., 1997) is positioned opposite a partly opened depression in domain I of GluTR. This depression, defined largely by Thr10, His11, Glu13, His84 and Glu93, is separated from the glutamate recognition pocket only by Arg50. The proposed complex thus implies that the GluTR product GSA could leave the enzyme via this ‘back door’ of the glutamate recognition pocket and directly channel to the active site of GSAM, a distance of ∼26 Å, without coming into contact with the aqueous environment (red dotted line in Figure 5C; Figure 6). The proposed channeling consequently would ensure the catalytic efficiency of this pathway. Despite extended surface complementarity between GluTR and GSAM, residues contributing to this interface are not conserved. Instead species-specific interactions may have evolved. As the two protein structures used in the present modeling originate from distantly related organisms, predictions of individual pairs of interacting residues is not possible.
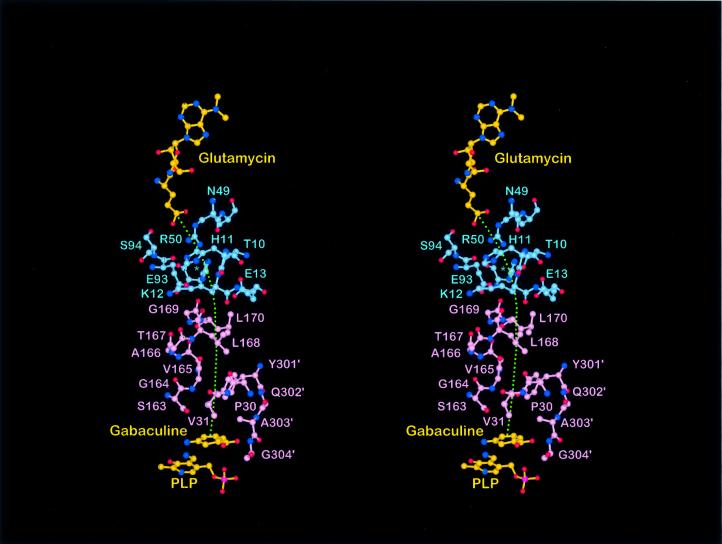
Fig. 6. In the proposed complex GluTR–GSAM (blue/pink residues), the respective active sites are separated by ∼26 Å. GSA, the product of GluTR and the substrate of GSAM, would channel from GluTR to GSAM (green, dotted line). In GluTR, there is an opening immediately behind the central active site residue Arg50 surrounded by the loop residues Thr10–Glu13, Glu93–Ser94 and His84 blue asterisk). In the putative complex, this comes to lie opposite a partly disordered loop 159–172 (Ser163–Leu170 shown) proposed to be the active site entrance for GSAM (Hennig et al., 1997) and to be essential for catalysis (Contestabile et al., 2000). Gabaculine, an inhibitor of GSAM, indicates the position GSA presumably would occupy to initiate its conversion to ALA.
GSAM is known to be an allosteric dimeric enzyme (Hennig et al., 1997; Contestabile et al., 2000) potentially dictating the overall reaction rate of the complex. As indicated by the disordered lid covering the active site of one monomer (Hennig et al., 1997), GSAM oscillates between two states where at any particular time one monomer is in a contracted, active state, while the other is in a relaxed state, allowing the product ALA to leave the complex. In oscillating between the two states, GSAM would, in our model, induce corresponding changes in the GluTR monomers, possibly optimizing the tRNAGlu recognition surface. Once glutamyl-tRNA had bound, GSA would be produced and channeled to GSAM. The complex could then switch to the second state, inducing the conversion of GSA to ALA.
Conclusions and outlook
The unusual V-shaped structure of GluTR suggests that the enzyme is capable of forming multiple and extended inter actions not only with its natural substrate glutamyl-tRNA but also simultaneously with the subsequent enzyme of tetrapyrrole biosynthesis, GSAM. The proposed ternary complex provides the basis for the analysis of the complex protein–protein as well as the protein–RNA interactions required for the initiation of tetrapyrrole biosynthesis in plants and most bacteria. In addition, it may represent a unique model system to study metabolic channeling between two essential enzymes at both the functional and structural level. As this pathway is ubiquitous throughout the biosphere, merely excluding animals and some pro karyotes, the proposed complex furthermore may allow the rational design of novel herbicides and antibiotics.
Materials and methods
Protein crystallization and data collection
Details of recombinant production and purification of GluTR have been published elsewhere (Moser et al., 1999). GluTR was crystallized by hanging drop vapor diffusion at 4°C with a protein concentration of 9.6 mg/ml in 0.2 M NaCl, 2 mM MgCl2, 100 mM HEPES pH 7.5, 0.2 M sodium citrate and 30% (v/v) methylpentenediole as precipitant. Lens-shaped crystals grew to a size of 0.3 × 0.1 × 0.1 mm3 within 2 weeks.
Phasing, model building and refinement
Crystals contain one molecule per asymmetric unit and belong to space group P21212 with a = 78.3 Å, b = 98.7 Å and c = 68.6 Å. All data were collected at 100 K using an MSC-Rigaku R-AXIS IV++ imaging plate detector on a Rigaku RU-H3R X-ray generator equipped with Osmic Confocal MaxFlux™ Optics. Data were processed with Denzo/Scalepack (Otwinowski and Minor, 1997) and truncate (CCP4, 1994). ‘Native’ data were obtained from crystals of a mutant, where cysteines at positions 6, 42, 48, 90 and 393 were replaced by serine. Heavy atom derivative soaks were performed using the triple mutant C42S/C48S/C90S. Phasing was initiated with difference Patterson search using SOLVE (Terwilliger and Berendzen, 1996) and performed with SHARP (de la Fortelle and Bricogne, 1997), making use of multiple isomorphous and anomalous scattering (MIRAS) phase probabilities. The subsequent phase-modified map (using SOLOMON; Abrahams and Leslie, 1996) was readily traced with O (Jones et al., 1991). Refinement was performed using CNS (Brünger et al., 1998) followed by Refmacs (Murshudov et al., 1997). PROCHECK (Laskowski et al., 1993) was used for structure validation. The current model of GluTR contains residues 1–404 but excludes residues 355–360 which are disordered. Eighty five percent of the residues were in the most favored regions of the Ramachandran plot. Outliers cumulate in two poorly defined loop regions located in domain III (residues A349, R352, L353, Q363, L384, P385 and D386). Other outliers of possible functional relevance include D18, R23, R59, R390 and A391. Coordinates and structure factors of GluTR have been deposited at the Protein Data Base (entry code 1gpj).
Acknowledgments
Acknowledgements
We thank Dr S.Sekine and Professor S.Yokohama (University of Tokyo) for providing us with the coordinates of glutamyl-tRNA. We thank Professors Brian Matthews (University of Oregon) and Dieter Söll (Yale) for helpful discussions and comments on the manuscript. Synchrotron beam time on beamlines BW6 and BW7a (DESY and MPI, Hamburg) and financial support by the Deutsche Forschungsgemeinschaft (Bonn, Germany) are also gratefully acknowledged.
References
- Abrahams J.P. and Leslie,A.G.W. (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D, 52, 30–42. [DOI] [PubMed] [Google Scholar]
- Al-Karadaghi S., Kristensen,O. and Liljas,A. (2000) A decade of progress in understanding the structural basis of protein synthesis. Prog. Biophys. Mol. Biol., 73, 167–193. [DOI] [PubMed] [Google Scholar]
- Allaire M., Li,Y., MacKenzie,R.E. and Cygler,M. (1998) The 3-D structure of a folate-dependent dehydrogenase/cyclohydrolase bifunctional enzyme at 1.5 Å resolution. Structure, 6, 173–182. [DOI] [PubMed] [Google Scholar]
- Brody S.S., Gough,S.P. and Kannangara,C.G. (1999) Predicted structure and fold recognition for the glutamyl tRNA reductase family of proteins. Proteins: Struct. Funct. Genet., 37, 485–493. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. (1992) Free R-value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature, 355, 472–475. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system (CNS): a new software system for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Carugo O. and Argos,P. (1997) NADP-dependent enzymes. I: conserved stereochemistry of cofactor binding. Proteins: Struct. Genet., 28, 10–28. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Contestabile R., Angelaccio,S., Maytum,R., Bossa,F. and John,R.A. (2000) The contribution of a conformationally mobile, active site loop to the reaction catalyzed by glutamate semialdehyde aminomutase. J. Biol. Chem., 275, 3879–3886. [DOI] [PubMed] [Google Scholar]
- de la Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy-atom parameter refinement for the multiple isomorphous replacement and multiwavelenth anomalous diffraction methods. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Eiler S., Dock-Bregeon,A., Moulinier,L., Thierry,J.C. and Moras,D. (1999) Synthesis of aspartyl-tRNA(Asp) in Escherichia coli—a snapshot of the second step. EMBO J., 18, 6532–6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig M., Grimm,B., Contestabile,R., John,R.A. and Jansonius,J.N. (1997) Crystal structure of glutamate-1-semialdehyde aminomutase: an α2-dimeric vitamin B6-dependent enzyme with asymmetry in structure and active site reactivity. Proc. Natl Acad. Sci. USA, 94, 4866–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1996) Alignment of three-dimensional protein structures: network server for database searching. Methods Enzymol., 266, 653–662. [DOI] [PubMed] [Google Scholar]
- Jahn D., Verkamp,E. and Söll,D. (1992) Glutamyl-transfer RNA: a precursor of heme and chlorophyll biosynthesis. Trends Biochem. Sci., 17, 215–218. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kikuchi G., Kumar,A., Talmage,P. and Shemin,D. (1958) The enzymatic synthesis of δ-aminolevulinic acid. J. Biol. Chem., 233, 1214–1219. [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots or protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lindahl M. et al. (1994) Crystal structure of the ribosomal protein S6 from Thermus thermophilus. EMBO J., 13, 1249–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May B.K, Dogra,S.C., Sadlon,T.J., Bhasker,C.R., Cox,T.C. and Bottomley,S.S. (1995) Molecular regulation of heme biosynthesis in higher vertebrates. Prog. Nucleic Acid Res. Mol. Biol., 51, 1–51. [DOI] [PubMed] [Google Scholar]
- Merrit E.A. and Murphy,M.E.P. (1994) Raster3D version 2.0: a program for photorealistic molecular graphics. Acta Crystallogr. D, 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Moser J., Lorenz,S., Hubschwerlen,C., Rompf,A. and Jahn,D. (1999) Methanopyrus kandleri glutamyl-tRNA-reductase. J. Biol. Chem., 274, 30679–30685. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin,A.A. and Dodson,E.H. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D, 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Nissen P., Kjeldgaard,M., Thirup,S., Polekhina,G., Reshetnikkova,L., Clark,B.F. and Nyborg,J. (1995) Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu and a GTP analog. Science, 270, 1464–1472. [DOI] [PubMed] [Google Scholar]
- Nissen P., Thirup,S., Kjeldgaard,M. and Nyborg,J. (1999) The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Struct. Fold. Des., 7, 143–156. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Rath V.L., Silvian,L.F., Beijer,B., Sproat,B.S. and Steitz,T.A. (1998) How glutaminyl-tRNA synthetase selects glutamine. Structure, 6, 439–449. [DOI] [PubMed] [Google Scholar]
- Schluenzen F. et al. (2000) Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell, 102, 615–623. [DOI] [PubMed] [Google Scholar]
- Schmitt E., Panvert,M., Blanquet,S. and Mechulam,Y. (1998) Crystal structure of methionyl-tRNAfMet transformylase complexed with the initiator formyl-methionyl-tRNAfMet. EMBO J., 17, 6819–6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schön A., Kannangara,C.G., Gough,S. and Söll,D. (1988) Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature, 331, 187–190. [DOI] [PubMed] [Google Scholar]
- Sekine S., Nureki,O., Shimada,A., Vassylyev,D.G. and Yokoyama,S. (2001) Structural basis for anticodon recognition by discriminating glutamyl-tRNA synthetase. Nature Struct. Biol., 8, 189–191. [DOI] [PubMed] [Google Scholar]
- Smith M.A., Kannangara,C.G. and Grimm,B. (1992) Glutamate 1-semialdehyde aminotransferase: anomalous enantiomeric reaction and enzyme mechanism. Biochemistry, 31, 11249–11254. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1996) Correlated phasing in multiple isomorphous replacement. Acta Crystallogr. D, 52, 749–757. [DOI] [PubMed] [Google Scholar]
- Wallace A.C., Laskowski,R.A. and Thornton,J.M. (1995) LIGPLOT: a program to generate schematic diagrams of protein–ligand interactions. Protein Eng., 8, 127–134. [DOI] [PubMed] [Google Scholar]
- Willows R.D., Kannangara,C.G. and Pontoppidan,B. (1995) Nucleotides of tRNA (Glu) involved in recognition by barley chloroplast glutamyl-tRNA synthetase and glutamyl-tRNA-reductase. Biochim. Biophys. Acta, 1263, 228–234. [DOI] [PubMed] [Google Scholar]