Abstract
Duplexes of 21–23 nucleotide (nt) RNAs are the sequence-specific mediators of RNA interference (RNAi) and post-transcriptional gene silencing (PTGS). Synthetic, short interfering RNAs (siRNAs) were examined in Drosophila melanogaster embryo lysate for their requirements regarding length, structure, chemical composition and sequence in order to mediate efficient RNAi. Duplexes of 21 nt siRNAs with 2 nt 3′ overhangs were the most efficient triggers of sequence-specific mRNA degradation. Substitution of one or both siRNA strands by 2′-deoxy or 2′-O-methyl oligonucleotides abolished RNAi, although multiple 2′-deoxynucleotide substitutions at the 3′ end of siRNAs were tolerated. The target recognition process is highly sequence specific, but not all positions of a siRNA contribute equally to target recognition; mismatches in the centre of the siRNA duplex prevent target RNA cleavage. The position of the cleavage site in the target RNA is defined by the 5′ end of the guide siRNA rather than its 3′ end. These results provide a rational basis for the design of siRNAs in future gene targeting experiments.
Keywords: PTGS/RNA interference/small interfering RNA
Introduction
Post-transcriptional gene silencing (PTGS) mediated by double-stranded (ds) RNA represents an evolutionarily conserved cellular defence mechanism for controlling the expression of alien genes in protists, filamentous fungi, plants and animals (Fire, 1999; Bass, 2000; Cogoni and Macino, 2000; Carthew, 2001; Hammond et al., 2001b; Sharp, 2001; Tuschl, 2001; Voinnet, 2001; Waterhouse et al., 2001). It is believed that random integration of alien genes (such as transposons) or viral infection causes production of dsRNA, which activates sequence-specific degradation of homologous single-stranded mRNA or viral genomic RNA, thereby preventing expression or replication of the foreign genetic material. The dsRNA is used as the guide RNA in this sequence-specific RNA degradation process. In some cases, dsRNA may also be involved in amplification of the silencing signal important for systemic spread (Palauqui et al., 1997; Voinnet et al., 1998) or long-term maintenance of silencing (Dalmay et al., 2000; Mourrain et al., 2000; Smardon et al., 2000). In animals, the dsRNA-triggered silencing effect is referred to as RNA interference (RNAi; Fire et al., 1998).
One important feature of the mechanism of RNAi is the processing of long dsRNAs into duplexes of 21–25 nucleotide (nt) RNAs. These short RNA products were first detected in plant tissues that exhibited transgene- or virus-induced PTGS (Hamilton and Baulcombe, 1999), but were also found later in fly embryos and worms injected with long dsRNAs (Parrish et al., 2000; Yang et al., 2000) or in extracts from Drosophila melanogaster Schneider-2 (S2) cells that were transfected with dsRNA (Hammond et al., 2000). The processing reaction of long dsRNAs to 21–23 nt RNAs was first recapitulated in vitro, in extracts prepared from D.melanogaster embryos (Zamore et al., 2000) and later in extracts from S2 cells (Bernstein et al., 2001). In the embryo lysate, it was observed that the target mRNA was cleaved in ∼21 nt intervals (Zamore et al., 2000) and that synthetic 21 and 22 nt RNA duplexes added to the lysate were able to guide efficient sequence-specific mRNA degradation, while duplexes of 30 bp dsRNA were inactive (Elbashir et al., 2001b). The 21 nt RNA products were therefore named small interfering RNAs or silencing RNAs (siRNAs).
A ribonuclease III enzyme, dicer, is required for processing of long dsRNA into siRNA duplexes (Bernstein et al., 2001). It was recently shown that dicer has an additional cellular function and is also required for excision of 21 and 22 nt small temporal RNAs (stRNAs) from ∼70 nt stable stem–loop precursors (Grishok et al., 2001; Hutvágner et al., 2001). These tiny expressed RNA molecules are important regulators of developmental timing and control the translation of downstream regulatory genes (Ambros, 2000; Moss, 2000; Pasquinelli et al., 2000). stRNAs are different from siRNAs in that the target mRNA is not degraded during silencing (Wightman et al., 1993; Olsen and Ambros, 1999) and they are single stranded (Reinhart et al., 2000), while siRNAs are believed to be double stranded (Elbashir et al., 2001b; Hutvágner et al., 2001).
In RNAi, a siRNA-containing endonuclease complex cleaves a single-stranded target RNA in the middle of the region complementary to the 21 nt guide siRNA of the siRNA duplex (Elbashir et al., 2001b). This cleavage site is one helical turn displaced from the cleavage site that produced the siRNA from long dsRNA, suggesting dramatic conformational and/or compositional changes after processing of long dsRNA to 21 nt siRNA duplexes. The target RNA cleavage products are rapidly degraded because they either lack the stabilizing cap or poly(A) tail. A protein component of the ∼500 kDa endonuclease or RNA-induced silencing complex (RISC) was recently identified and is a member of the argonaute family of proteins (Hammond et al., 2001a); however, it is currently unclear whether dicer is required for RISC activity.
It is also unknown whether RISC contains single- or double-stranded siRNAs. By analogy to stRNA excision, it may be envisaged that only one of the strands of a siRNA duplex is incorporated into a catalytic siRNP, but because of the symmetry of the siRNA duplex, two approximately equal populations of sense and antisense strand-containing catalytic siRNPs are produced. Synthetic siRNA duplexes cleaved sense as well as antisense target RNAs in the middle of the region covered by the siRNA duplex in D.melanogaster lysate (Elbashir et al., 2001b). However, longer dsRNAs did not produce symmetric sense and antisense target RNA cleavage sites in embryo lysate (Elbashir et al., 2001b), suggesting that the direction of processing of long dsRNA defined which of the strands of the resulting siRNA duplex could be used for guiding target degradation. Some protein, involved in the production of the 21 nt siRNA duplexes, may be deposited on the siRNA duplex to mark the strand that is going to be used for guiding target RNA cleavage.
Despite the lack of profound mechanistic understanding, RNAi has rapidly developed into an important tool for reverse genetics and has been widely applied in Caenorhabditis elegans (Fraser et al., 2000; Gönczy et al., 2000; Piano et al., 2000; Maeda et al., 2001), as well as in insects (see references in Lam and Thummel, 2000) and insect cell lines (Clemens et al., 2000; Hammond et al., 2000; Ui-Tei et al., 2000). RNAi has also been shown to occur in a variety of vertebrates by targeting of mRNAs important for embryonic development. In differentiated mammalian cells, dsRNAs with >30 bp generally activate the interferon response, which leads to a global shut-off in protein synthesis as well as non-specific mRNA degradation (Stark et al., 1998). This unspecific response to long dsRNAs can be bypassed using 21 nt siRNA duplexes, resulting in specific knock-down of the expression of the targeted gene (Elbashir et al., 2001a; Hutvágner et al., 2001), providing a new method for analysis of mammalian gene function in cultured cells.
Here we describe the results of a systematic analysis of the length, secondary structure, sugar backbone and sequence specificity of siRNA duplexes for RNAi, using the established D.melanogaster embryo in vitro system. The most potent siRNA duplexes are 21 nt long, comprising a 19 nt base-paired sequence with 2 nt 3′-overhanging ends. The 5′ end of the target-complementary siRNA strand (guide siRNA) sets the ruler to define the position of target RNA cleavage. Furthermore, we find that target recognition is extremely specific, as even single nucleotide mismatches between the siRNA duplex and the target mRNA abolish interference. These results provide a rational basis for the design of siRNAs for future gene targeting experiments.
Results
Variation of the 3′ overhang in duplexes of 21 nt siRNAs
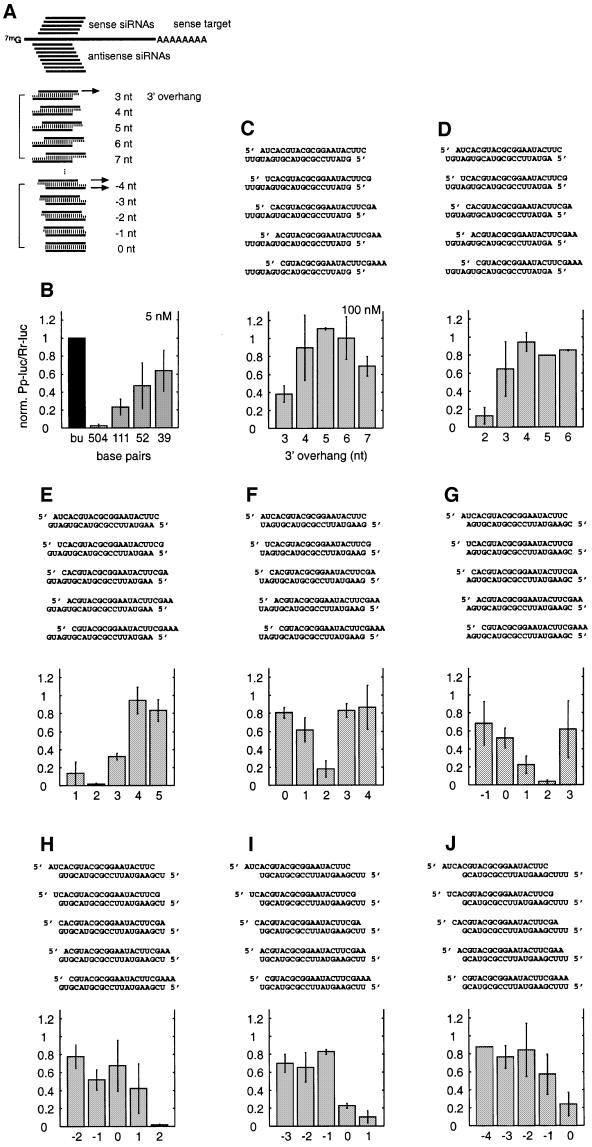
We reported previously that two or three unpaired nucleotides at the 3′ end of siRNA duplexes were more efficient in target RNA degradation than blunt-ended duplexes (Elbashir et al., 2001b). To perform a more comprehensive analysis of the function of the terminal nucleotides, we synthesized five 21 nt sense siRNAs, each displaced by one nucleotide relative to the target RNA, and eight 21 nt antisense siRNAs, each displaced by one nucleotide relative to the target (Figure 1A). By combining these sense and antisense siRNAs, a series of eight siRNA duplexes with symmetric overhanging ends were generated spanning a range from 7 nt 3′ overhang to 4 nt 5′ overhang. The interference was measured using the dual luciferase assay system (Tuschl et al., 1999; Zamore et al., 2000). siRNA duplexes were directed against firefly luciferase mRNA and sea pansy luciferase mRNA was used as internal control. The luminescence ratio of target to control luciferase activity was determined in the presence of siRNA duplex and was normalized to that observed in its absence. For comparison, the interference ratios of long dsRNAs (39–504 bp) are shown in Figure 1B (Elbashir et al., 2001b). The interference ratios were determined at concentrations of 5 nM for long dsRNAs (Figure 1A) and at 100 nM for siRNA duplexes (Figure 1C–J). The 100 nM concentration of siRNAs was chosen because complete processing of 5 nM 504 bp dsRNA would result in 120 nM total siRNA duplexes.
Fig. 1. Variation of the 3′ overhang of duplexes of 21 nt siRNAs. (A) Outline of the experimental strategy. The capped and polyadenylated sense target mRNA is depicted and the relative positions of sense and antisense siRNAs are shown. Eight series of duplexes according to the eight different antisense strands were prepared. The siRNA sequences and the number of overhanging nucleotides were changed in 1 nt steps. (B) Normalized relative luminescence of target luciferase (Photinus pyralis, Pp-luc) to control luciferase (Renilla reniformis, Rr-luc) in D.melanogaster embryo lysate in the presence of 5 nM blunt-ended dsRNAs (Elbashir et al., 2001b). The luminescence ratios determined in the presence of dsRNA were normalized to the ratio obtained for a buffer control (bu; black bar). Normalized ratios less than 1 indicate specific interference. (C–J) Normalized interference ratios for eight series of 21 nt siRNA duplexes. The sequences of siRNA duplexes are depicted above the bar graphs. Each part shows the interference ratio for a set of duplexes formed with a given antisense guide siRNA and five different sense siRNAs. The number of overhanging nucleotides (3′ overhang, positive numbers; 5′ overhang, negative numbers) is indicated on the x-axis. Data points were averaged from at least three independent experiments. Error bars represent standard deviations.
The ability of 21 nt siRNA duplexes to mediate RNAi is dependent on the number of overhanging nucleotides or base pairs formed. Duplexes with 4–6 3′-overhanging nucleotides were unable to mediate RNAi (Figure 1C–F), as were duplexes with two or more 5′-overhanging nucleotides (Figure 1G–J). The duplexes with 2 nt 3′ overhangs were most efficient in mediating RNA interference, although the efficiency of silencing was also sequence dependent and up to 12-fold differences were observed for different siRNA duplexes with 2 nt 3′ overhangs (compare Figure 1D–H). Duplexes with blunted ends, 1 nt 5′ overhang or 1–3 nt 3′ overhangs were sometimes functional and sometimes completely inactive. The small silencing effect observed for the siRNA duplex with 7 nt 3′ overhang (Figure 1C) may be due to an antisense effect of the long 3′ overhang rather than to RNAi. Comparison of the efficiency of RNAi between long dsRNAs (Figure 1B) and the most effective 21 nt siRNA duplexes (Figure 1E, G and H) indicates that a single siRNA duplex at 100 nM concentration can be as effective as 5 nM 504 bp dsRNA.
Length variation of the sense siRNA paired to an invariant 21 nt antisense siRNA
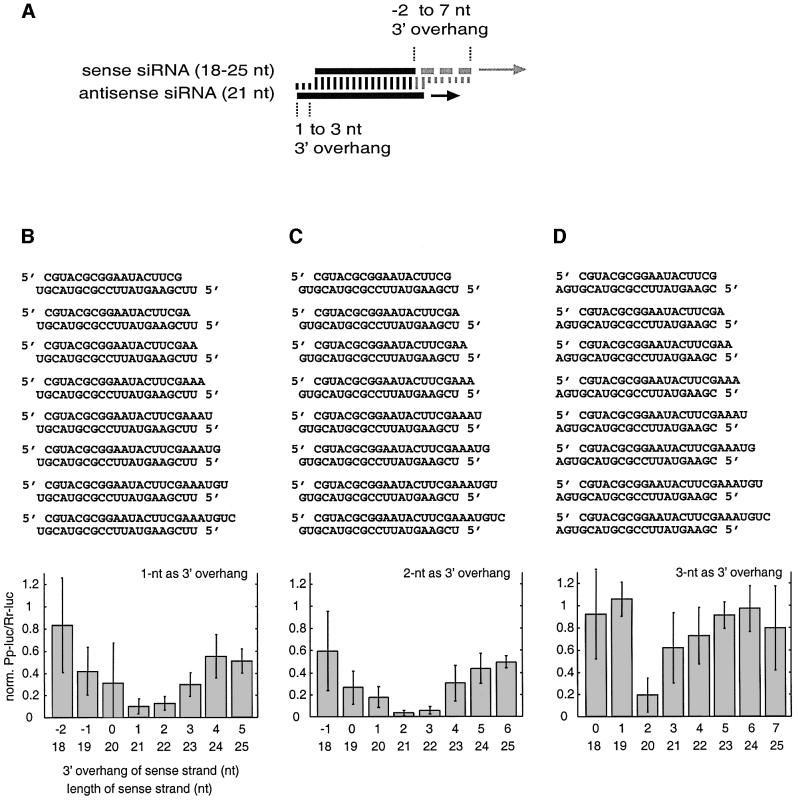
In order to investigate the effect of the length of siRNAs on RNAi, we prepared three series of siRNA duplexes, combining three 21 nt antisense strands with eight 18–25 nt sense strands. The 3′ overhang of the antisense siRNA was fixed to 1, 2 or 3 nt in each siRNA duplex series, while the sense siRNA was varied at its 3′ end (Figure 2A). Independently of the length of the sense siRNA, we found that duplexes with 2 nt 3′ overhang of antisense siRNA (Figure 2C) were more active than those with 1 or 3 nt 3′ overhang (Figure 2B and D). In the first series, with 1 nt 3′ overhang of antisense siRNA, duplexes with 21 and 22 nt sense siRNAs, carrying a 1 and 2 nt 3′ overhang of sense siRNA, respectively, were most active. Duplexes with 19–25 nt sense siRNAs were also able to mediate RNA, but to a lesser extent. Similarly, in the second series, with 2 nt overhang of antisense siRNA, the 21 nt siRNA duplex with 2 nt 3′ overhang was most active and any other combination with the 18–25 nt sense siRNAs was active to a significant degree. In the last series, with 3 nt antisense siRNA 3′ overhang, only the duplex with a 20 nt sense siRNA and 2 nt sense 3′ overhang was able to reduce target RNA expression. Together, these results indicate that the length of the siRNA as well as the length of the 3′ overhang are important, and that duplexes of 21 nt siRNAs with 2 nt 3′ overhang are optimal for RNAi.
Fig. 2. Variation of the length of the sense strand of siRNA duplexes. (A) Representation of the experiment. Three 21 nt antisense strands were paired with eight sense siRNAs. The siRNAs were changed in length at their 3′ end. The 3′ overhang of the antisense siRNA was 1 nt (B), 2 nt (C) or 3 nt (D), while the sense siRNA overhang was varied for each series. The sequences of the siRNA duplexes and the corresponding interference ratios are indicated.
Length variation of siRNA duplexes with a constant 2 nt 3′ overhang
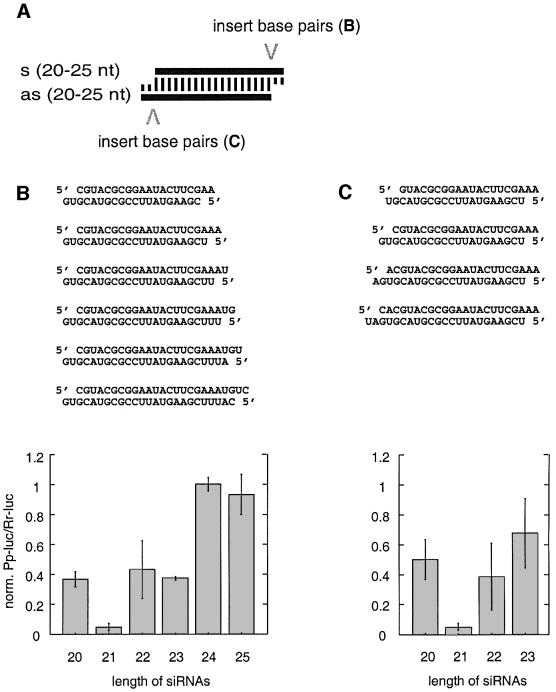
We then examined the effect of simultaneously changing the length of both siRNA strands by maintaining symmetrical 2 nt 3′ overhangs (Figure 3A). Two series of siRNA duplexes were prepared, including the 21 nt siRNA duplex of Figure 1H as reference. The length of the duplexes was varied between 20 and 25 bp by extending the base-paired segment at the 3′ end of the sense siRNA (Figure 3B) or at the 3′ end of the antisense siRNA (Figure 3C).
Fig. 3. Variation of the length of siRNA duplexes with preserved 2 nt 3′ overhangs. (A) Graphic representation of the experiment. The 21 nt siRNA duplex is identical in sequence to the one shown in Figures 1H and 2C. The siRNA duplexes were extended to the 3′ side of the sense siRNA (B) or the 5′ side of the sense siRNA (C). The siRNA duplex sequences and the respective interference ratios are indicated.
Duplexes of 20–23 bp caused specific repression of target luciferase activity, but the 21 nt siRNA duplex was at least 8-fold more efficient than any of the other duplexes. siRNA duplexes of 24 and 25 nt did not result in any detectable interference. Sequence-specific effects were minor as variations on both ends of the duplex produced similar effects.
2′-deoxy- and 2′-O-methyl-modified siRNA duplexes
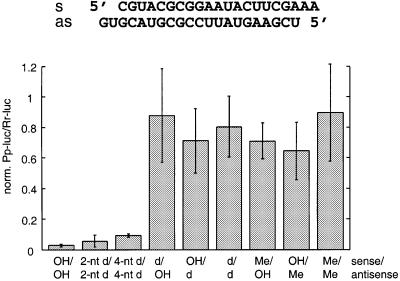
To assess the importance of the siRNA ribose residues for RNAi, duplexes with 21 nt siRNAs and 2 nt 3′ overhangs with 2′-deoxy- or 2′-O-methyl-modified strands were examined (Figure 4). Substitution of the 2 nt 3′ overhangs by 2′-deoxynucleotides had no effect and even the replacement of two additional ribonucleotides by 2′-deoxyribonucleotides adjacent to the overhangs in the paired region produced significantly active siRNAs. Thus, 8 out of 42 nt of a siRNA duplex were replaced by DNA residues without loss of activity. Complete substitution of one or both siRNA strands by 2′-deoxy residues, however, abolished RNAi, as did complete substitution by 2′-O-methyl residues.
Fig. 4. Substitution of the 2′-hydroxyl groups of the siRNA ribose residues. The 2′-hydroxyl groups (OH) in the strands of siRNA duplexes were replaced by 2′-deoxy (d) or 2′-O-methyl (Me). 2 and 4 nt 2′-deoxy substitutions at the 3′ ends are indicated as 2- and 4-nt d, respectively. Uridine residues were replaced by 2′-deoxythymidine.
Definition of target RNA cleavage sites
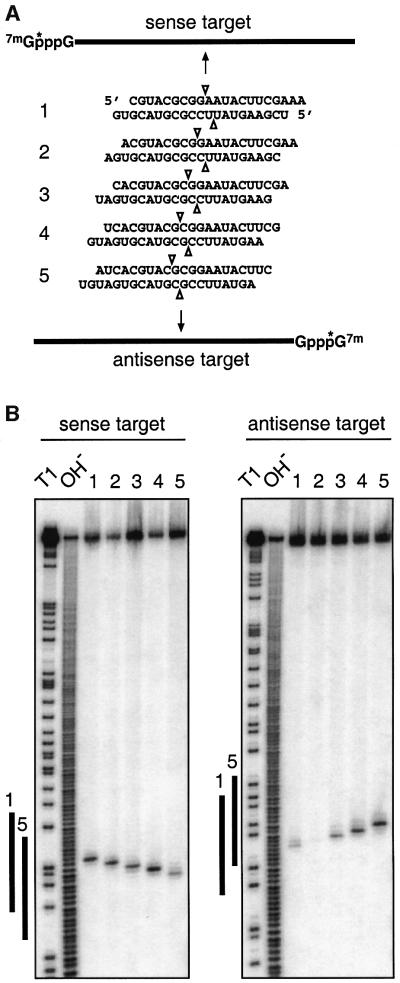
Target RNA cleavage positions were previously determined for 22 nt siRNA duplexes and for a 21 and 22 nt duplex (Elbashir et al., 2001b). The position of target RNA cleavage was located in the centre of the region covered by the siRNA duplex, 11 or 12 nt downstream of the first nucleotide that was complementary to the 21 or 22 nt siRNA guide sequence. Five distinct 21 nt siRNA duplexes with 2 nt 3′ overhang (Figure 5A) were incubated with 5′ cap-labelled sense or antisense target RNA in D.melanogaster lysate (Tuschl et al., 1999; Zamore et al., 2000). The 5′ cleavage products were resolved on sequencing gels (Figure 5B). The amount of sense target RNA cleaved correlated with the efficiency of siRNA duplexes determined in the translation-based assay, and siRNA duplexes 1, 2 and 4 (Figures 5B, 1E, G and H) cleaved target RNA faster than duplexes 3 and 5 (Figures 5B, 1D and F). Notably, the sum of radioactivity of the 5′ cleavage product and the input target RNA were not constant over time and the 5′ cleavage products did not accumulate. Presumably, the cleavage products, once released from the siRNA–endonuclease complex, were rapidly degraded due to the lack of either the poly(A) tail or the 5′ cap.
Fig. 5. Mapping of sense and antisense target RNA cleavage by 21 nt siRNA duplexes with 2 nt 3′ overhangs. (A) Representation of 32P (asterisk) cap-labelled sense and antisense target RNAs and siRNA duplexes. The position of sense and antisense target RNA cleavage is indicated by triangles on top and below the siRNA duplexes, respectively. (B) Mapping of target RNA cleavage sites. After 2 h incubation of 10 nM target RNA with 100 nM siRNA duplex in D.melanogaster embryo lysate, the 5′ cap-labelled substrate and the 5′ cleavage products were resolved on 6% sequencing gels. Length markers were generated by partial RNase T1 digestion (T1) and partial alkaline hydrolysis (OH) of the target RNAs. The bold lines to the left of the images indicate the region covered by the siRNA strands 1 and 5 of the same orientation as the target.
The cleavage sites for both sense and antisense target RNAs were located in the middle of the region spanned by the siRNA duplexes. The cleavage sites for each target produced by the five different duplexes varied by 1 nt according to the 1 nt displacement of the duplexes along the target sequences. The targets were cleaved precisely 11 nt downstream of the target position complementary to the 3′-most nucleotide of the sequence-complementary guide siRNA (Figure 5).
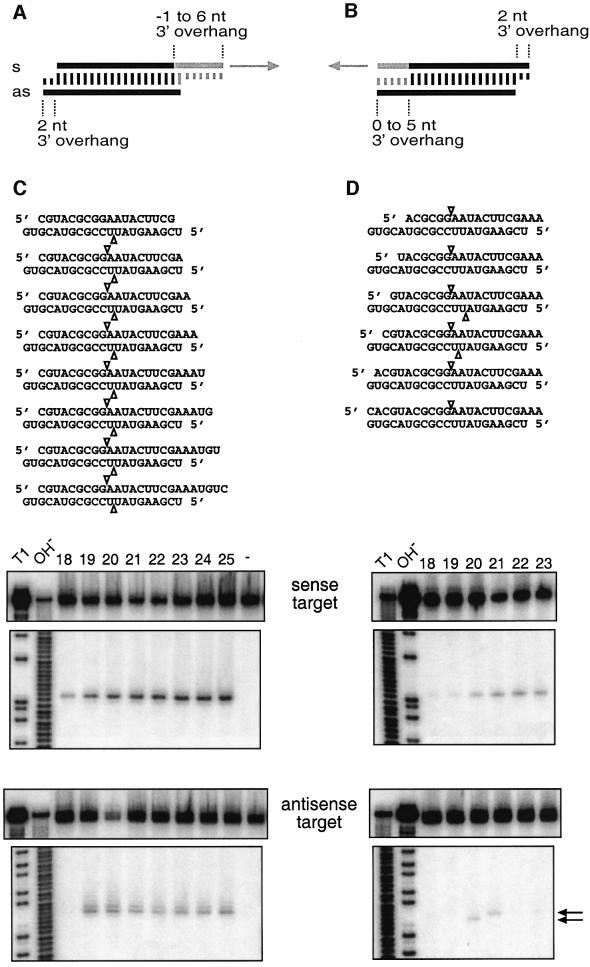
In order to determine whether the 5′ or the 3′ end of the guide siRNA sets the ruler for target RNA cleavage, we devised the experimental strategy outlined in Figure 6A and B. A 21 nt antisense siRNA, which was kept invariant for this study, was paired with sense siRNAs that were modified in length at either of their 5′ or 3′ ends. The position of sense and antisense target RNA cleavage was determined as described above. Changes in the 3′ end of the sense siRNA, monitored for 1 nt 5′ overhang to 6 nt 3′ overhang, did not affect either the position of sense nor antisense target RNA cleavage (Figure 6C). Changes in the 5′ end of the sense siRNA did not affect the sense target RNA cleavage (Figure 6D, top), as expected, because the antisense siRNA was unchanged. The residual amount of uncleaved sense target RNA (Figure 6C and D top) correlated with the efficiency of siRNA duplexes determined in translation-based assays (Figure 2C and data not shown), but did not correlate with the amount of detected cleavage product. Accumulation of cleavage products was more pronounced for the longer and less efficient siRNA duplexes, suggesting that product release may have become rate limiting. Because the antisense siRNA was kept unchanged while the sense siRNA was varied, an alteration in product release implies a role of the sense siRNA strand in the target RNA degradation process.
Fig. 6. The 5′ end of a guide siRNA defines the position of target RNA cleavage. (A and B) Representation of the experimental strategy. The antisense siRNA was the same in all siRNA duplexes, but the sense strand was varied between 18 and 25 nt by changing the 3′ end (A) or 18 and 23 nt by changing the 5′ end (B). The position of sense and antisense target RNA cleavage is indicated by triangles on top and below the siRNA duplexes, respectively. (C and D) Analysis of target RNA cleavage using cap-labelled sense (top) or antisense (bottom) target RNAs. The residual amount of targeted substrate and the cap-labelled 5′ cleavage products are shown. The sequences of the siRNA duplexes are indicated and the length of the sense siRNA strands is marked on top. The control lane, marked with a dash in (C), shows target RNA incubated in absence of siRNAs. Markers were as described in Figure 5. The arrows in (D), bottom, indicate the target RNA cleavage sites that differ by 1 nt.
Changes in the 5′ end of the sense siRNA, in contrast to its 3′ end, strongly affected antisense target RNA cleavage (Figure 6D, bottom). The antisense target was only cleaved when the sense siRNA was 20 or 21 nt in size. The position of cleavage differed by 1 nt, suggesting that the 5′ end of the target-recognizing siRNA sets the ruler for target RNA cleavage. This position is located between nucleotide 10 and 11 when counting in an upstream direction from the target nucleotide paired to the 5′-most nucleotide of the guide siRNA (see also Figure 5A).
Sequence effects and 2′-deoxy substitutions in the 3′ overhang
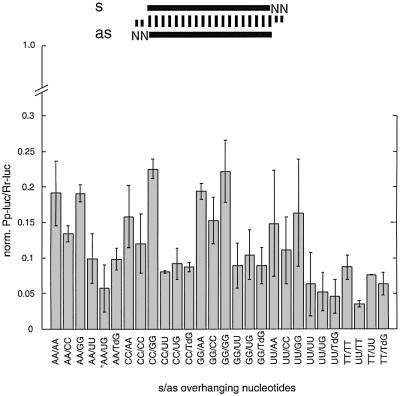
The 2 nt 3′ overhang is critical for siRNA function. We wanted to know whether the sequence of the overhanging nucleotides contributes to target recognition or is only a feature required for reconstitution of the endonuclease complex (RISC or siRNP). We synthesized sense and antisense siRNAs with AA, CC, GG, UU and UG 3′ overhangs and included the 2′-deoxy modifications TdG and TT (T, 2′-deoxythymidine; dG, 2′-deoxyguanosine). The wild-type siRNAs contained AA in the sense 3′ overhang and UG in the antisense 3′ overhang (AA/UG). All siRNA duplexes were functional in the interference assay and reduced target expression at least 5-fold (Figure 7). The most efficient siRNA duplexes, which reduced target expression >10-fold, were of the sequence type NN/UG, NN/UU, NN/TdG and NN/TT (N, any nucleotide). siRNA duplexes with an antisense siRNA 3′ overhang of AA, CC or GG were less active by a factor of 2–4 when compared with the wild-type sequence UG or the mutant UU. This reduction in RNAi efficiency is likely to be due to the contribution of the penultimate 3′ nucleotide to sequence-specific target recognition, as the 3′-terminal nucleotide was changed from G to U without effect.
Fig. 7. Sequence variation of the 3′ overhang of siRNA duplexes. The 2 nt 3′ overhang (NN, in grey) was changed in sequence and composition as indicated (T, 2′-deoxythymidine; dG, 2′-deoxyguanosine; asterisk, wild-type siRNA duplex). Normalized interference ratios were determined as described in Figure 1. The wild-type sequence is the same as depicted in Figure 4.
Changes in the sequence of the 3′ overhang of the sense siRNA did not reveal any sequence-dependent effects, which was not surprising because the sense siRNA is not expected to contribute to the sequence-specific recognition of the sense target mRNA.
Sequence specificity of target recognition
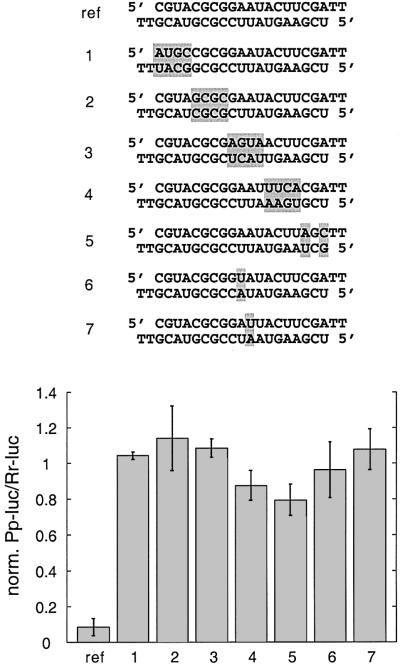
In order to examine the sequence specificity of target recognition, we introduced sequence changes into the paired segments of siRNA duplexes and determined the efficiency of silencing. Sequence changes were introduced by inverting short segments of 3 or 4 nt or inducing point mutations (Figure 8). The sequence changes in one siRNA strand were compensated for in the complementary siRNA strand to avoid perturbing the base-paired siRNA duplex structure. The sequence of all 2 nt 3′ overhangs was TT to reduce costs of synthesis. The TT/TT reference siRNA duplex was comparable in RNAi to the wild-type siRNA duplex AA/UG (Figure 7). The ability to mediate reporter mRNA destruction was quantified using the translation-based luminescence assay. Duplexes of siRNAs with inverted sequence segments showed dramatically reduced ability for targeting the firefly luciferase reporter (Figure 8). The sequence changes located between the 3′ end and the middle of the antisense siRNA completely abolished target RNA recognition, but mutations near the 5′ end of the antisense siRNA exhibited a small degree of silencing. Transversion of the AU base pair located directly opposite the predicted target RNA cleavage site or 1 nt further away from the predicted site prevented target RNA cleavage, therefore indicating that a single mutation within the centre of a siRNA duplex discriminates between mismatched targets.
Fig. 8. Sequence specificity of target recognition. The sequences of the mismatched siRNA duplexes are shown, modified sequence segments or single nucleotides are shaded in grey. The reference duplex (ref) and the siRNA duplexes 1–7 contain 2′-deoxythymidine 2 nt overhangs. The silencing efficiency of the thymidine-modified reference duplex was comparable to the wild-type sequence (Figure 7). Normalized interference ratios were determined as described in Figure 1.
Discussion
siRNAs are valuable reagents for inactivation of gene expression, not only in insect cells but also in mammalian cells, with a great potential for therapeutic application (Elbashir et al., 2001a). We have systematically analysed the structural determinants of siRNA duplexes required to promote efficient target RNA degradation in D.melanogaster embryo lysate, thus providing rules for the design of most potent siRNA duplexes. A perfect siRNA duplex is able to silence gene expression with an efficiency comparable to a 500 bp dsRNA, given that comparable quantities of total RNA are used.
The siRNA user guide
Efficiently silencing siRNA duplexes are composed of 21 nt sense and 21 nt antisense siRNAs and must be selected to form a 19 bp double helix with 2 nt 3′-overhanging ends. 2′-deoxy substitutions of the 2 nt 3′-overhanging ribonucleotides do not affect RNAi, but help to reduce the costs of RNA synthesis and may enhance RNase resistance of siRNA duplexes. More extensive 2′-deoxy or 2′-O-methyl modifications reduce the ability of siRNAs to mediate RNAi, probably by interfering with protein association for siRNP assembly.
Target recognition is a highly sequence-specific process, mediated by the siRNA complementary to the target. The 3′-most nucleotide of the guide siRNA does not contribute to the specificity of target recognition, while the penultimate nucleotide of the 3′ overhang affects target RNA cleavage and a mismatch reduces RNAi 2- to 4-fold. The 5′ end of the guide siRNA also appears more permissive for mismatched target RNA recognition when compared with the 3′ end. Nucleotides in the centre of the siRNA, located opposite to the target RNA cleavage site, are important specificity determinants and even single nucleotide changes reduce RNAi to undetectable levels. This suggests that siRNA duplexes may be able to discriminate mutant or polymorphic alleles in gene targeting experiments, which may become an important feature for future therapeutic developments.
Sense and antisense siRNAs, when associated with the protein components of the endonuclease complex or its commitment complex, were suggested to play distinct roles; the relative orientation of the siRNA duplex in this complex defines which strand can be used for target recognition (Elbashir et al., 2001b). Synthetic siRNA duplexes with an equal number of overhanging nucleotides have dyad symmetry with respect to the double-helical structure, but not with respect to sequence. The association of siRNA duplexes with the RNAi proteins in the D.melanogaster lysate leads to the formation of two asymmetric complexes. In such hypothetical complexes, the chiral environment is distinct for sense and antisense siRNA, hence their function. The prediction obviously does not apply to palindromic siRNA sequences or to RNAi proteins that could associate as homodimers. To minimize sequence effects that may affect the ratio of sense- and antisense-targeting siRNPs, we suggest using siRNA sequences with identical 3′-overhanging sequences. We recommend adjusting the sequence of the overhang of the sense siRNA to that of the antisense 3′ overhang because the sense siRNA does not have a target in typical knock-down experiments. Asymmetry in the reconstitution of sense- and antisense-cleaving siRNPs could be, partially, responsible for the variation in RNAi efficiency observed for various 21 nt siRNA duplexes with 2 nt 3′ overhangs used in this study (Figure 1). Alternatively, the nucleotide sequence at the target site and/or the accessibility of the target RNA structure may be responsible for the variation in efficiency observed for these siRNA duplexes. It should be noted that all siRNAs used in this study are derived from a short region of one gene. Thus, it is more likely that differences in siRNA efficiency are a consequence of the primary sequences of the siRNAs and the respective target sites, rather than the secondary or tertiary structure of the targeted RNA.
Natural siRNAs versus synthetic siRNAs
In D.melanogaster, siRNA duplexes are produced in vitro and in vivo from long dsRNAs (Hammond et al., 2000; Yang et al., 2000; Zamore et al., 2000). About 45% of these short RNAs are precisely 21 nt long, 28% are 22 nt long and a few percent are shorter or longer RNAs (Elbashir et al., 2001b). This length distribution correlates with our finding that 21 nt siRNA duplexes are the most efficient mediators of mRNA degradation. Beside the length, the paired structure and overhang are also important. This structural feature may explain why siRNA duplexes isolated from the dsRNA processing reaction under denaturing conditions were less potent for RNAi than longer dsRNAs that were processed to siRNAs during the targeting reaction (Zamore et al., 2000). Presumably, denaturation followed by renaturation favoured the formation of the thermodynamically more stable, blunt-ended, but less active, siRNA duplexes. Isolation of siRNAs under native conditions does not reduce siRNA activity (Nykänen et al., 2001).
Production of siRNAs from long dsRNA requires the RNase III enzyme dicer (Bernstein et al., 2001). Dicer is a bidentate RNase III, which also contains an ATP-dependent RNA helicase domain and a PAZ domain, presumably important for dsRNA unwinding and mediation of protein–protein interactions, respectively (Cerutti et al., 2000; Bernstein et al., 2001). Dicer is evolutionarily conserved in worms, flies, plants, fungi and mammals (Matsuda et al., 2000), and has a second cellular function important for the development of these organisms (Ray et al., 1996; Jacobsen et al., 1999; Grishok et al., 2001; Hutvágner et al., 2001; Knight and Bass, 2001). At present, it is uncertain whether dicer activity in species other than D.melanogaster produces siRNAs of predominantly 21 nt in length. The estimates of siRNA size vary in the literature between 21 and 25 nt (Hamilton and Baulcombe, 1999; Hammond et al., 2000; Hutvágner et al., 2000; Parrish et al., 2000; Yang et al., 2000; Zamore et al., 2000; Elbashir et al., 2001b).
In a recent study of the effect of siRNA length in mammalian cells (primary mouse embryonic fibroblasts, 293 and HeLa cells), duplexes of 21–27 nt siRNAs with 2 nt 3′ overhangs were directed against different co-transfected reporter genes (Caplen et al., 2001). Duplexes of 22 and 23 nt siRNAs were found to be slightly more efficient in triggering sequence-specific gene silencing than 21 nt siRNA duplexes. In our hands, using the dual luciferase assay system in HeLa cells, 21 nt siRNA duplexes with 2 nt 3′ overhang are 2- to 3-fold more efficient than 20 or 22–25 nt siRNA duplexes (data not shown), therefore recapitulating the results obtained from the D.melanogaster biochemical system. In contrast to the D.melanogaster system, siRNA duplexes >23 nt in length are still triggering some RNAi in HeLa cells and also in C.elegans (Caplen et al., 2001). However, it remains to be determined whether the RNA strands finally incorporated into the active endonuclease complex are of the initially provided length. It is possible that exonucleases present in C.elegans and mammalian cells trim longer siRNAs to their optimum length and that these exonucleases are absent from D.melanogaster lysate.
The functional anatomy of long dsRNAs as a trigger for RNAi was analysed previously in C.elegans (Parrish et al., 2000). Activation of RNAi by injection of long dsRNA requires at least two steps: dsRNA processing by dicer RNase III and siRNP or RISC formation. Substitution of one of the strands of the long dsRNA by DNA abolished RNAi and even the substitution of C by dC or U by dT in only one of the strands caused a substantial decrease in RNAi. Because introduction of 2′-fluoro modifications into long RNA had no effect, it was suggested that an A-form double helical structure was important for triggering RNAi (Parrish et al., 2000). We have been able to substitute eight ribose residues of a siRNA duplex by 2′-deoxyribose residues without substantial reduction of RNAi, although it should be noted the 2′-deoxy modifications were clustered at the 3′ end of the siRNAs, including the 2 nt 3′ overhangs. It is possible that the four 2′-deoxy modifications, which are located in the paired region at the end of the helix, do not affect the overall A-form helical structure and do not strongly compromise RISC formation. Complete modification of one or both siRNA strands by 2′-deoxyribose, however, abolished RNAi. Interestingly, substitution by 2′-O-methylribose, which adopts the ribose sugar pucker, also abolished RNAi, probably because methylation of the 2′-hydroxyls blocked hydrogen bond formation or introduced steric hindrance.
It was recently demonstrated that a 5′-phosphate on the target-complementary strand of a siRNA duplex is required for siRNA function and that ATP is used to maintain the 5′-phosphates of the siRNAs (Nykänen et al., 2001). However, 5′-phosphorylation of fully 2′-deoxy- or 2′-O-methyl-modified siRNA strands was not able to restore RNAi (data not shown). Unmodified siRNA duplexes with free 5′-hydroxyls and 2 nt 3′ overhangs are readily phosphorylated in D.melanogaster embryo lysate (Nykänen et al., 2001). In this respect, it should be noted that our reported RNAi efficiencies were determined by pre-incubating the siRNA duplexes for 15 min in D.melanogaster lysate before adding target and control mRNAs, thus providing sufficient time for 5′-phosphorylation of siRNA duplexes to occur. Comparison of the RNAi efficiencies of 5′-phosphorylated and 5′-non-phosphorylated siRNAs (for duplexes shown in Figures 1E, F and 2C) did not reveal any sizeable differences (data not shown).
Conclusions
We have performed an extensive analysis of the length, sequence and structure of siRNA duplexes in D.melanogaster embryo lysate. Duplexes of 21 nt siRNAs with 2 nt 3′ overhangs were shown to be the most efficient triggers of RNAi-based mRNA degradation. The target recognition is a highly sequence-specific process, although not all positions of a guide siRNA contribute equally to specificity. These results are important for the design of efficient siRNAs in order to silence genes in D.melanogaster and provide a basis for similar studies in other organisms.
Materials and methods
RNA preparation and RNAi assay
Chemical RNA synthesis, annealing and luciferase-based RNAi assays were performed as described previously (Tuschl et al., 1999; Zamore et al., 2000; Elbashir et al., 2001b). Synthetic RNAs were gel purified after deprotection. The formation of siRNA duplexes was verified by agarose gel electrophoresis using 4% NuSieve GTG agarose (BMA, Rockland, ME) in 0.5× TBE buffer. All siRNA duplexes were directed against firefly luciferase and the luciferase mRNA sequence was derived from pGEM-luc (DDBJ/EMBL/GenBank accession No. X65316) as described (Tuschl et al., 1999). The siRNA duplexes were incubated in a D.melanogaster RNAi/translation reaction for 15 min prior to addition of mRNAs. Translation-based RNAi assays were performed at least in triplicate.
For mapping of sense target RNA cleavage, a 177 nt transcript was generated, corresponding to the firefly luciferase sequence between positions 113 and 273 relative to the start codon, followed by the 17 nt complement of the SP6 promoter sequence (Elbashir et al., 2001b). For mapping of antisense target RNA cleavage, a 166 nt transcript was produced from a template, which was amplified from plasmid sequence by PCR using the 5′ primer TAATACGACTCACTATAGAGCCCATA TCGTTTCATA (T7 promoter underlined) and 3′ primer AGAG GATGGAACCGCTGG. The target sequence corresponds to the complement of the firefly luciferase sequence between positions 50 and 215 relative to the start codon. Guanylyl transferase labelling was performed as described previously (Zamore et al., 2000). For mapping of target RNA cleavage, 100 nM siRNA duplex was incubated with 5–10 nM target RNA in D.melanogaster embryo lysate under standard conditions (Zamore et al., 2000) for 2 h at 25°C. The reaction was stopped by the addition of 8 vols of proteinase K buffer [200 mM Tris–HCl pH 7.5, 25 mM EDTA, 300 mM NaCl, 2% (w/v) SDS]. Proteinase K (dissolved in water; Merck) was added to a final concentration of 0.6 mg/ml. The reactions were then incubated for 15 min at 65°C, extracted with phenol/chloroform/isoamyl alcohol (25:24:1) and precipitated with 3 vols of ethanol. Samples were loaded on 6% sequencing gels. Length standards were generated by partial RNase T1 digestion and partial base hydrolysis of the cap-labelled sense or antisense target RNAs.
Acknowledgments
Acknowledgements
We acknowledge Heike Taubner and Jutta Meyer for technical assistance, Phil Zamore for critical comments on the manuscript and H.Jäckle and R.Lührmann for support. This work was funded by a BMBF Biofuture grant, No. 0311856.
References
- Ambros V. (2000) Control of developmental timing in Caenorhabditis elegans. Curr. Opin. Genet. Dev., 10, 428–433. [DOI] [PubMed] [Google Scholar]
- Bass B.L. (2000) Double-stranded RNA as a template for gene silencing. Cell, 101, 235–238. [DOI] [PubMed] [Google Scholar]
- Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. [DOI] [PubMed] [Google Scholar]
- Caplen N.J., Parrish,S., Imani,F., Fire,A. and Morgan,R.A. (2001) Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl Acad. Sci. USA, 98, 9742–9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew R.W. (2001) Gene silencing by double-stranded RNA. Curr. Opin. Cell Biol., 13, 244–248. [DOI] [PubMed] [Google Scholar]
- Cerutti L., Mian,N. and Bateman,A. (2000) Domains in gene silencing and cell differentiation proteins: the novel PAZ domain and redefinition of the piwi domain. Trends Biochem. Sci., 25, 481–482. [DOI] [PubMed] [Google Scholar]
- Clemens J.C., Worby,C.A., Simonson-Leff,N., Muda,M., Maehama,T., Hemmings,B.A. and Dixon,J.E. (2000) Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl Acad. Sci. USA, 97, 6499–6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C. and Macino,G. (2000) Post-transcriptional gene silencing across kingdoms. Curr. Opin. Genet. Dev., 10, 638–643. [DOI] [PubMed] [Google Scholar]
- Dalmay T., Hamilton,A., Rudd,S., Angell,S. and Baulcombe,D.C. (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell, 101, 543–553. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001a) Duplexes of 21-nucleotide RNAs mediate RNA interference in mammalian cell culture. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Lendeckel,W. and Tuschl,T. (2001b) RNA interference is mediated by 21 and 22 nt RNAs. Genes Dev., 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A. (1999) RNA-triggered gene silencing. Trends Genet., 15, 358–363. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Fraser A.G., Kamath,R.S., Zipperlen,P., Martinez-Campos,M., Sohrmann,M. and Ahringer,J. (2000) Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature, 408, 325–330. [DOI] [PubMed] [Google Scholar]
- Gönczy P. et al. (2000) Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature, 408, 331–336. [DOI] [PubMed] [Google Scholar]
- Grishok A. et al. (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell, 106, 23–34. [DOI] [PubMed] [Google Scholar]
- Hamilton A.J. and Baulcombe,D.C. (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science, 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Bernstein,E., Beach,D. and Hannon,G.J. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature, 404, 293–296. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Boettcher,S., Caudy,A.A., Kobayashi,R. and Hannon,G.J. (2001a) Argonaute2, a link between genetic and biochemical analyses of RNAi. Science, 293, 1146–1150. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Caudy,A.A. and Hannon,G.J. (2001b) Post-transcriptional gene silencing by double-stranded RNA. Nature Rev. Genet., 2, 110–1119. [DOI] [PubMed] [Google Scholar]
- Hutvágner G., Mlynarova,L. and Nap,J.P. (2000) Detailed characterization of the posttranscriptional gene-silencing-related small RNA in a GUS gene-silenced tobacco. RNA, 6, 1445–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G., McLachlan,J., Bálint,É., Tuschl,T. and Zamore,P.D. (2001) A cellular function for the RNA interference enzyme dicer in small temporal RNA maturation. Science, 293, 834–838. [DOI] [PubMed] [Google Scholar]
- Jacobsen S.E., Running,M.P. and Meyerowitz,M.E. (1999) Disruption of an RNA helicase/RNase III gene in Arabidopsis causes unregulated cell division in floral meristems. Development, 126, 5231–5243. [DOI] [PubMed] [Google Scholar]
- Knight S.W. and Bass,B.L. (2001) A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in C. elegans. Science, 293, 2269–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam G. and Thummel,C.S. (2000) Inducible expression of double-stranded RNA directs specific genetic interference in Drosophila. Curr. Biol., 10, 957–963. [DOI] [PubMed] [Google Scholar]
- Maeda I., Kohara,Y., Yamamoto,M. and Sugimoto,A. (2001) Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr. Biol., 11, 171–176. [DOI] [PubMed] [Google Scholar]
- Matsuda S., Ichigotani,Y., Okuda,T., Irimura,T., Nakatsugawa,S. and Hamaguchi,M. (2000) Molecular cloning and characterization of a novel human gene (HERNA) which encodes a putative RNA-helicase. Biochim. Biophys. Acta, 1490, 163–169. [DOI] [PubMed] [Google Scholar]
- Moss E.G. (2000) Non-coding RNAs: lightning strikes twice. Curr. Biol., 10, R436–R439. [DOI] [PubMed] [Google Scholar]
- Mourrain P. et al. (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell, 101, 533–542. [DOI] [PubMed] [Google Scholar]
- Nykänen A., Haley,B. and Zamore,P.D. (2001) Nucleotide cofactor requirements and small interfering RNA conformation in the RNA interference pathway. Cell, 107, 309–321. [DOI] [PubMed] [Google Scholar]
- Olsen P.H. and Ambros,V. (1999) The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol., 216, 671–680. [DOI] [PubMed] [Google Scholar]
- Palauqui J.C., Elmayan,T., Pollien,J.M. and Vaucheret,H. (1997) Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J., 16, 4738–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish S., Fleenor,J., Xu,S., Mello,C. and Fire,A. (2000) Functional anatomy of a dsRNA trigger: differential requirement for the two trigger strands in RNA interference. Mol. Cell, 6, 1077–1087. [DOI] [PubMed] [Google Scholar]
- Pasquinelli A.E. et al. (2000) Conservation of the sequence of let-7 heterochronic regulatory RNA. Nature, 408, 86–89. [DOI] [PubMed] [Google Scholar]
- Piano F., Schetterdagger,A.J., Mangone,M., Stein,L. and Kemphues,K.J. (2000) RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr. Biol., 10, 1619–1622. [DOI] [PubMed] [Google Scholar]
- Ray A., Lang,J.D., Golden,T. and Ray,S. (1996) Short integument (SIN1), a gene required for ovule development in Arabidopsis, also controls flowering time. Development, 122, 2631–2638. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Slack,F.J., Basson,M., Pasquinelli,A.E., Bettinger,J.C., Rougvie,A.E., Horvitz,H.R. and Ruvkun,G. (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature, 403, 901–906. [DOI] [PubMed] [Google Scholar]
- Sharp P.A. (2001) RNA interference 2001. Genes Dev., 15, 485–490. [DOI] [PubMed] [Google Scholar]
- Smardon A., Spoerke,J., Stacey,S., Klein,M., Mackin,N. and Maine,E. (2000) EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol., 10, 169–178. [DOI] [PubMed] [Google Scholar]
- Stark G.R., Kerr,I.M., Williams,B.R., Silverman,R.H. and Schreiber,R.D. (1998) How cells respond to interferons. Annu. Rev. Biochem., 67, 227–264. [DOI] [PubMed] [Google Scholar]
- Tuschl T. (2001) RNA interference and small interfering RNAs. ChemBioChem, 2, 239–245. [DOI] [PubMed] [Google Scholar]
- Tuschl T., Zamore,P.D., Lehmann,R., Bartel,D.P. and Sharp,P.A. (1999) Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev., 13, 3191–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui-Tei K., Zenno,S., Miyata,Y. and Saigo,K. (2000) Sensitive assay of RNA interference in Drosophila and Chinese hamster cultured cells using firefly luciferase gene as target. FEBS Lett., 479, 79–82. [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2001) RNA silencing as a plant immune system against viruses. Trends Genet., 17, 449–459. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Vain,P., Angell,S. and Baulcombe,D.C. (1998) Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell, 95, 177–187. [DOI] [PubMed] [Google Scholar]
- Waterhouse P.M., Wang,M.B. and Lough,T. (2001) Gene silencing as an adaptive defence against viruses. Nature, 411, 834–842. [DOI] [PubMed] [Google Scholar]
- Wightman B., Ha,I. and Ruvkun,G. (1993) Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell, 75, 855–862. [DOI] [PubMed] [Google Scholar]
- Yang D., Lu,H. and Erickson,J.W. (2000) Evidence that processed small dsRNAs may mediate sequence-specific mRNA degradation during RNAi in Drosophila embryos. Curr. Biol., 10, 1191–1200. [DOI] [PubMed] [Google Scholar]
- Zamore P.D., Tuschl,T., Sharp,P.A. and Bartel,D.P. (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell, 101, 25–33. [DOI] [PubMed] [Google Scholar]