Abstract
Mobile group II introns encode reverse transcriptases that also function as intron-specific splicing factors (maturases). We showed previously that the reverse transcriptase/maturase encoded by the Lactococcus lactis Ll.LtrB intron has a high affinity binding site at the beginning of its own coding region in an idiosyncratic structure, DIVa. Here, we identify potential secondary binding sites in conserved regions of the catalytic core and show via chemical modification experiments that binding of the maturase induces the formation of key tertiary interactions required for RNA splicing. The interaction with conserved as well as idiosyncratic regions explains how maturases in some organisms could evolve into general group II intron splicing factors, potentially mirroring a key step in the evolution of spliceosomal introns.
Keywords: reverse transcriptase/ribozyme/RNA catalysis/RNA folding/RNA–protein interaction
Introduction
Group II introns found in organelle and bacterial genomes splice via a lariat intermediate and are thought to be ancestors of nuclear pre-mRNA introns (Michel and Ferat, 1995). The group II intron splicing reactions are RNA catalyzed and require a conserved RNA structure formed by six interacting double-helical domains (DI–VI; Michel and Ferat, 1995). An evolutionary relationship between group II and spliceosomal introns is supported by similarities in reaction mechanisms and splice site consensus sequences, by analogies between the structure and function of group II intron domains and snRNAs, by the ability of the ‘snRNA-like’ group II intron domains to associate in trans and by the discovery that some group II introns are mobile genetic elements, suggesting how introns could have spread in nuclear genomes (Michel and Ferat, 1995; Lambowitz et al., 1999). Together, these findings suggest a scenario in which mobile group II introns invaded nuclear genomes and then degenerated, with group II intron domains evolving into snRNAs. If so, further studies of mobile group II introns may provide insight into how present-day splicing mechanisms evolved.
The best studied mobile group II introns are the yeast mtDNA introns aI1 and aI2 and the Lactococcus lactis Ll.LtrB intron (reviewed in Lambowitz et al., 1999). These introns encode reverse transcriptases that function both in intron mobility and as maturases in RNA splicing. The group II intron-encoded proteins (IEPs) are typically encoded in DIV, outside the intron’s catalytic core, and have three major conserved domains: an N-terminal reverse transcriptase domain, domain X associated with splicing or maturase activity and a C-terminal DNA endonuclease domain, which functions in intron mobility. After translation, the IEP binds specifically to the intron RNA to promote splicing and then remains associated with the excised intron lariat RNA to form a ribonucleoprotein (RNP) complex that catalyzes intron mobility. In the major mobility pathway, retrohoming, the intron RNA in this RNP reverse splices directly into a specific target site in double-stranded DNA and is reverse transcribed by the IEP. In addition to homing, group II introns can also retrotranspose to ectopic sites that resemble the normal homing site at low frequency, leading to the dispersal of the introns to different genomic locations (Dickson et al., 2001; and references therein).
The splicing activity of the group II IEPs was first demonstrated genetically for the two yeast mtDNA introns and subsequently both genetically and biochemically for the L.lactis Ll.LtrB intron (see Lambowitz et al., 1999). All three IEPs function specifically in splicing the intron in which they are encoded, although the yeast aI1 and aI2 maturases appear to have some cross-reactivity. Other group II introns encode related proteins that have, in some cases, been inferred to function in splicing. Many of these proteins contain all three conserved domains, but some lack the C-terminal endonuclease domain, while others, such as the MatK proteins encoded by chloroplast tRNALys introns, retain only a well conserved domain X and remnants of the reverse transcriptase domain (Mohr et al., 1993). The latter proteins are presumed to have lost dispensable mobility functions, but retain the essential splicing function.
Although all three characterized maturases are intron-specific splicing factors, several investigators have suggested that putative maturases in some organisms function in splicing multiple group II introns. Suggestive evidence for such a generalized role of group II intron maturases comes from experiments showing that inhibition of chloroplast protein synthesis in higher plants blocks the splicing of multiple group II introns, including some that do not themselves encode maturases (Vogel et al., 1999; and references therein), and that maturase-like proteins, no longer encoded within introns, are conserved in the chloroplast DNAs of non-photosynthetic plants that still retain introns in essential genes (Wolfe et al., 1992; Copertino et al., 1994). Highly degenerate Euglena group II introns, referred to as group III introns, are also hypothesized to use a common splicing apparatus that includes a maturase encoded by one of the introns (Copertino et al., 1994). The evolution of an intron-specific maturase to function in splicing multiple group II introns may parallel a step in the evolution of a common cellular splicing machinery for spliceosomal introns.
To study the maturase-promoted splicing reaction biochemically, we have developed an experimental system based on the L.lactis Ll.LtrB intron, where it is possible to obtain large amounts of the purified IEP (denoted LtrA protein) via expression in Escherichia coli (Matsuura et al., 1997). We showed that this purified maturase is sufficient to promote the splicing of the Ll.LtrB intron in vitro at low Mg2+ concentrations, where the intron RNA cannot by itself fold into the catalytically active RNA structure (Saldanha et al., 1999). The protein binds specifically to the Ll.LtrB intron, but not to other group II introns, as expected for an intron-specific splicing factor. Deletion analysis showed that the LtrA protein has a primary high affinity binding site in intron subdomain DIVa, an idiosyncratic structure near the beginning of its own coding region, with other regions of DIV also contributing to binding (Wank et al., 1999). Although LtrA does not bind tightly to the intron when DIV is deleted, its binding to DIV is enhanced by both upstream and downstream regions of the intron, and the interaction of LtrA with DIV positions the reverse transcriptase active site to initiate cDNA synthesis just downstream of the intron in the 3′ exon, as required for intron mobility. These findings suggested that after binding to DIV, the maturase might make weaker secondary contacts with 5′ and 3′ regions of the intron to promote the folding of the catalytic core. Supporting this hypothesis, intron RNAs deleted for the primary high affinity binding site in DIVa show residual maturase-dependent splicing in vitro and in vivo, indicating that the weaker, secondary contacts are sufficient to promote splicing even in the absence of high affinity binding to domain IV (Wank et al., 1999; X.Cui and A.M.Lambowitz, in preparation).
Here, we analyzed the interaction of the maturase with the Ll.LtrB RNA in detail by quantitative binding assays and RNA footprinting. Our results identify potential secondary binding sites in conserved regions of the catalytic core and demonstrate directly that the binding of the maturase induces an RNA conformational change that results in the formation of key tertiary interactions required for catalytic activity. These findings show how the maturase functions and have implications for the evolution of splicing mechanisms.
Results
Specific binding of the maturase to 5′ and 3′ segments of the group II intron
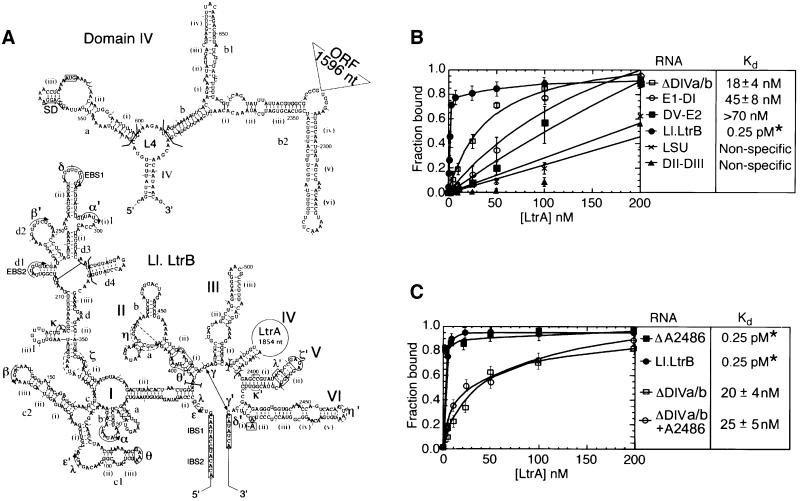
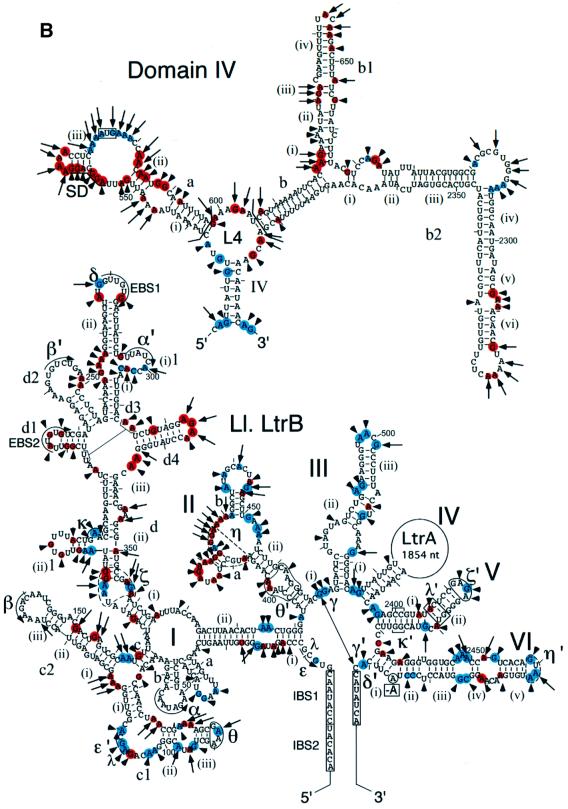
Figure 1A shows a secondary structure model for the 902 nucleotide ΔORF derivative of the Ll.LtrB intron used in this study. The DIV subsegment of this intron, shown in detail in Figure 1A, contains the high affinity maturase-binding site in DIVa. An in vitro transcript containing the ΔORF Ll.LtrB intron and flanking exons is referred to as Ll.LtrB RNA (see Materials and methods). To detect weaker, secondary binding sites, we first carried out equilibrium binding assays with a derivative of the Ll.LtrB RNA that has a minimal DIV deleted for both DIVa and DIVb (ΔDIVa/b; Figure 1B). In these experiments, small amounts of 32P-labeled RNAs were incubated with increasing concentrations of LtrA protein, and binding was assayed by retention of the complex on a nitrocellulose filter. The assays were done under previously established protein-dependent splicing conditions in reaction medium containing 450 mM NaCl and 5 mM Mg2+ at 30°C, the relatively high salt concentration being necessary to minimize non-specific binding of the LtrA protein (Saldanha et al., 1999).
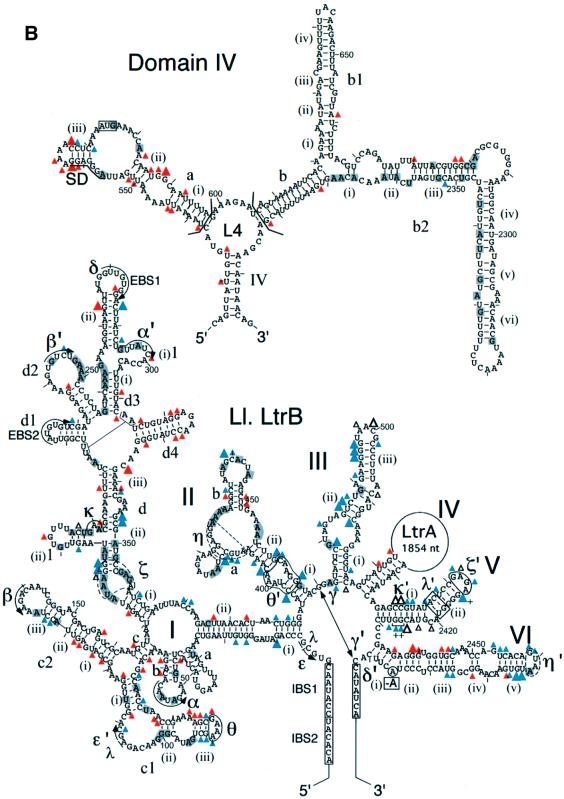
Fig. 1. Secondary structure model of the Ll.LtrB intron and equilibrium binding assays. (A) Predicted secondary structure of the 902 nucleotide ΔORF derivative of the Ll.LtrB intron. The structure is based on the generalized group IIA intron structure in Toor et al. (2001). Nucleotide residues involved in the EBS–IBS pairings and tertiary interactions (Greek letters) are marked. Tertiary interactions are modeled based on sequence and structure comparisons with other group II introns and have not been verified experimentally for the Ll.LtrB intron. The η–η′ interaction does not fit the consensus for other group IIA introns and was best modeled as a ‘group IIB-type’ interaction (Chanfreau and Jacquier, 1996; Costa et al., 1997). The ζ receptor for the unusual GAGA tetraloop at the top of DV (ζ′) is shown as an 11 nucleotide sequence based on the 11 nucleotide GAAA receptor (Costa and Michel, 1995). The η, η′ and ζ elements are indicated in dashed lines to denote these uncertainties. The boundaries of deletions or subsegments used in different experiments are demarcated by brackets, and the branch point adenosine deleted in the ΔA2486 mutant is boxed. The predicted secondary structure of DIV is shown above, with the location of the deleted 1596 nucleotide LtrA ORF segment indicated to the upper right. Subdomain DIVa, the high affinity binding site for the LtrA protein, contains the putative Shine–Dalgarno (SD) sequence (underlined) and initiation codon (boxed) of the LtrA ORF. (B and C) Equilibrium binding assays. 32P-labeled RNAs were incubated with increasing concentrations of LtrA protein for 1 h at 30°C in reaction medium containing (B) 450 mM NaCl/5 mM MgCl2 or (C) 500 mM NH4Cl/5 mM MgCl2. Binding was assayed by the retention of the 32P-labeled RNA on a nitrocellulose filter. The Kd values are the mean ± SD for three experiments for each RNA. The specific binding of LtrA to the ΔDIVa/b intron was not detected previously in reaction media containing bovine serum albumin (Wank et al., 1999). *The Kd value for the wild-type Ll.LtrB RNA is too low to be determined by equilibrium binding; the value 0.25 pM indicated in the figure is the apparent Kd measured as koff/kon (Wank et al., 1999).
Figure 1B shows that LtrA does in fact bind specifically to the ΔDIVa/b RNA with an apparent Kd of 18 ± 4 nM. This binding was much weaker than that of the wild-type Ll.LtrB RNA, containing the high affinity binding site (apparent Kd measured as koff/kon = 0.25 pM; Wank et al., 1999), but significantly better than that of a non-specific RNA containing the Neurospora crassa mt LSU group I intron. A 5′ segment of Ll.LtrB RNA consisting of E1-DI also bound specifically with an apparent Kd of 45 ± 8 nM, while a 3′ segment consisting of DV-E2 bound less tightly (apparent Kd >70 nM), but again more strongly than the non-specific RNA. In contrast, a segment containing only DII and DIII did not bind better than the non-specific RNA. Together, these findings show that LtrA can bind specifically to the Ll.LtrB RNA in the absence of its high affinity DIVa binding site and suggest at least two secondary binding sites in the 5′ and 3′ segments of the Ll.LtrB RNA.
RNA footprinting strategy
To investigate the interaction of the LtrA protein with the intron RNA in detail, we carried out RNA footprinting, using iodine cleavage of phosphorothioate-containing RNAs to assess phosphodiester backbone interactions, and chemical probing with dimethyl sulfate (DMS) and kethoxal to assess base interactions. In general, the binding of a protein can result in RNA protection directly, or indirectly by inducing the formation of higher order RNA structure. To help distinguish these possibilities, we compared modification protection patterns of the Ll.LtrB RNA under protein-dependent and self-splicing conditions in the presence or absence of the LtrA protein. Protections observed in the absence of LtrA at low or high Mg2+ concentration are assigned to RNA structure. Potential protein protection sites were identified as positions that remained accessible in the folded RNA under self-splicing conditions and were protected only in the presence of the LtrA protein, with the best candidates assigned based on additional structural information (see Discussion). The reaction conditions were modified slightly, with NH4+ instead of Na+ as the monovalent cation to optimize self-splicing.
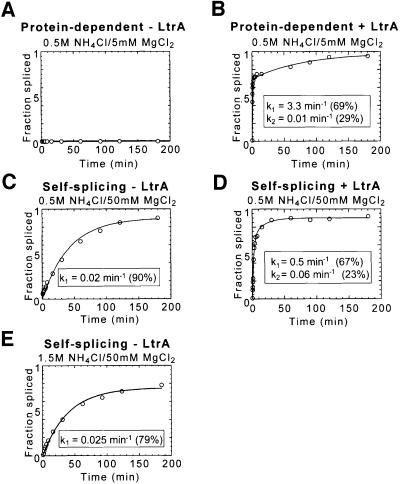
Figure 2 shows splicing time courses for the wild-type Ll.LtrB intron under the different conditions used in RNA footprinting. Under protein-dependent conditions (0.5 M NH4Cl/5 mM MgCl2), the wild-type Ll.LtrB RNA showed no detectable splicing in the absence of LtrA, but was spliced efficiently in its presence (Figure 2A and B). The protein-dependent splicing reaction was biphasic, with the fast phase (69%) occurring at 3.3/min and the slow phase (29%) at 0.01/min. The biphasic kinetics under protein excess conditions were attributed to a more slowly folding RNA conformer (Saldanha et al., 1999). When the Mg2+ concentration was increased (0.5 M NH4Cl/50 mM MgCl2), the intron RNA folds into the active structure and self-splices in the absence of LtrA (Figure 2C). This self-splicing occurred at a rate of 0.02/min, considerably slower than protein-dependent splicing at 5 mM Mg2+. LtrA could still bind under these conditions (koff = 0.001/min compared with 0.002/min at 5 mM Mg2+) and increased the reaction rate ∼25-fold [fast phase, 0.5/min (67%); slow phase, 0.06/min (23%); Figure 2D]. Finally, under higher salt conditions (1.5 M NH4Cl/50 mM MgCl2), the rate of self-splicing was maximal (0.025/min; Figure 2E), but the LtrA protein had no effect, presumably reflecting that its binding is impeded under these conditions (not shown). We note that even under the higher salt conditions, the maximal rate of self-splicing is still ∼130-fold slower than the rate of protein-dependent splicing at 5 mM Mg2+.
Fig. 2. RNA splicing of the Ll.LtrB intron under protein-dependent and self-splicing conditions. Splicing time courses were carried out with 32P-labeled Ll.LtrB RNA (20 nM) at 30°C in different reaction media used for RNA structure mapping and footprinting. (A and B) 0.5 M NH4Cl/5 mM Mg2+ (protein-dependent conditions) in the absence or presence of 200 nM LtrA, respectively. (C and D) 0.5 M NH4Cl/50 mM Mg2+ (self-splicing conditions) in the absence or presence of 200 nM LtrA, respectively. (E) 1.5 M NH4Cl/50 mM Mg2+ (maximal self-splicing conditions).
To prevent splicing during footprinting, we used a mutant Ll.LtrB RNA deleted for the branch point adenosine in DVI (ΔA2486). This mutation inhibits splicing completely, with the mutant RNA showing only slow residual hydrolysis at the splice sites (<5% after 3 h; not shown). As desired, the mutation did not significantly affect the binding of the LtrA to Ll.LtrB RNA in koff experiments (not shown), nor the weaker binding of LtrA to the ΔDIVa/b RNA in equilibrium binding experiments under the footprinting conditions (Kds = 20–25 nM; Figure 1C). Other branch point mutants (A2486 changed to U, C or G) or DV ‘active site’ mutants (G2399A, G2399C) showed residual and for the G2399 mutants surprisingly high splicing activity in the presence of the LtrA protein and would therefore produce unsuitably heterogeneous RNA populations during the footprinting (not shown). This problem is particularly cogent for group II introns, which are thought to undergo active site rearrangements to position the different nucleophiles used in the two steps of splicing (Chanfreau and Jacquier, 1996). The branch point deletion used here presumably traps the RNA structure prior to the first step of splicing. Although it is formally possible that this modification could affect the local structure at the active site, the binding assays and results below indicate that the RNA construct can form most if not all of the expected wild-type tertiary structure at 50 mM Mg2+ or at 5 mM Mg2+ in the presence of the LtrA protein. The ability to form most of the correct tertiary structure at 50 mM Mg2+ is pertinent to the interpretation of the footprinting experiments.
Iodine footprinting
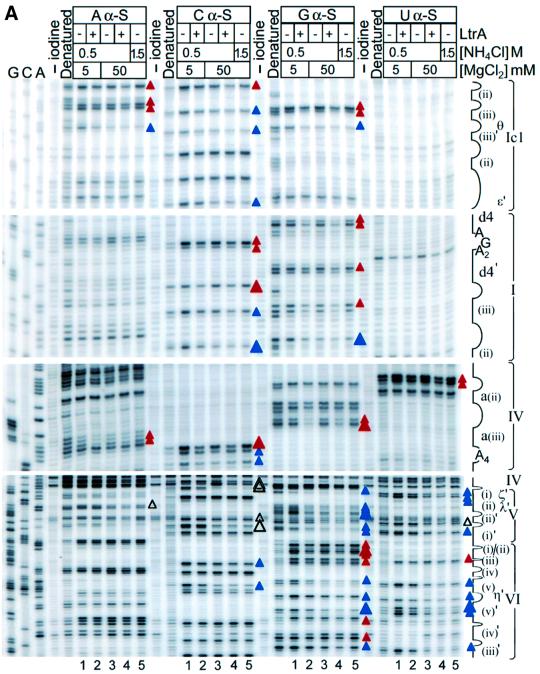
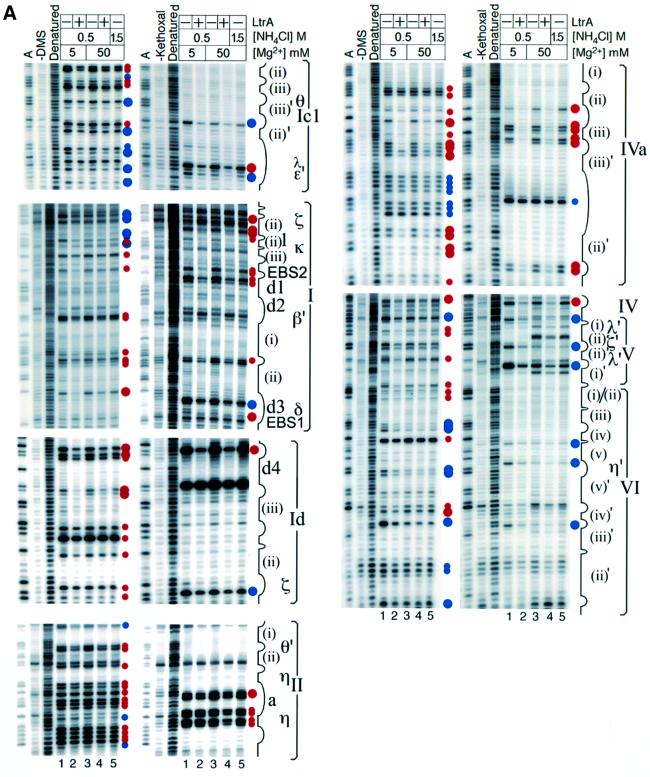
Figure 3 shows iodine cleavage of the phosphodiester backbone of Ll.LtrB RNA in the reaction media described above in the presence or absence of the LtrA protein. For these experiments, four separate transcripts, each substituted with 5% of a different phosphorothioate, were incubated with or without LtrA in the different reaction media, and then cleaved with iodine under conditions that give on average less than one cleavage per molecule. Cleavage sites were mapped by primer extension, using 5′-labeled primers complementary to different positions in the Ll.LtrB RNA. Controls showed that each of the 5% phosphorothioate substitutions had little if any effect on the rate of protein-dependent or self-splicing of the wild-type Ll.LtrB RNA (not shown). Because the experiments are carried out at RNA and protein concentrations well above thr Kd, individual phosphorothioate substitutions are not expected to have a large effect on the footprinting pattern, although perturbations due to some substitutions cannot be totally excluded. Representative iodine footprinting data are shown in Figure 3A, and the results are summarized in Figure 3B.
Fig. 3. Iodine cleavage of the Ll.LtrB RNA in the presence or absence of LtrA protein. Four Ll.LtrB-ΔA2486 RNAs (20 nM), each substituted with 5% of a different phosphorothioate (αS A, C, G, U), were incubated in different reaction media at 30°C in the presence or absence of 50 nM LtrA protein and then cleaved with iodine. Cleavage sites were mapped by primer extension, using 5′-labeled primers complementary to different positions in the Ll.LtrB RNA. (A) Representative gels. Lanes: G, C, A, dideoxy sequencing ladders obtained from plasmid pGMΔORF-ΔA2486 using the same 5′-labeled primers; ‘–iodine’, RNA incubated in reaction medium containing 0.5 M NH4Cl/50 mM Mg2+ without iodine; ‘Denatured’, RNA cleaved with iodine under denaturing conditions (see Materials and methods). Lanes 1–5, iodine cleavage reactions; lanes 1 and 2, 0.5 M NH4Cl/5 mM Mg2+ (protein-dependent conditions) in the absence and presence of LtrA, respectively; lanes 3 and 4, 0.5 M NH4Cl/50 mM Mg2+ (self-splicing conditions) in the absence and presence of LtrA, respectively; lane 5, 1.5 M NH4Cl/50 mM Mg2+ (maximal self-splicing conditions). Landmarks in the intron RNA are indicated to the right of the gel. (B) Summary of iodine cleavage protection patterns. Gray shading indicates the positions protected by RNA structure in reaction medium containing 5 mM Mg2+ (lane 1) compared with denatured RNA (>25% decrease in band intensity). Large and small blue or open triangles indicate positions strongly (>50%) or moderately (49–25%) protected by RNA structure at 50 mM Mg2+ (lanes 4 and 5) compared with 5 mM Mg2+ (lane 1). Blue triangles indicate positions protected at 50 mM Mg2+ (lanes 3–5) and also protected at 5 mM Mg2+ in the presence of LtrA (lane 2), presumably reflecting the formation of all or part the active RNA structure in the presence of the protein. Open triangles indicate a subset of positions protected at 50 mM Mg2+ (lanes 3–5), but not at 5 mM Mg2+ in the presence or absence of LtrA (lanes 1 and 2), possibly reflecting the formation of aberrant RNA structures or interactions at 50 mM Mg2+. Large and small red triangles (protein-dependent protections) indicate positions that remained accessible at 50 mM Mg2+ (lanes 3 and 5) and were strongly (>50%) or moderately (49–25%) protected only in the presence of LtrA protein (lanes 2 and 4). + indicates positions that were protected (>25%) in the free RNA at 50 mM Mg2+ (lane 3) and showed increased protection after addition of LtrA (lane 4), reflecting either strengthened RNA structure or additional protection by the protein. Assignments are based on quantiation of two complete data sets for all positions and three data sets for the DV–VI region. All positions refer to the 5′ phosphate of the marked nucleotide residue. Positions that could not be monitored because of consistent strong reverse transcription stops were C-6, C-1, G1, A393, U2322, U2328, U2340, U2365 and U2375.
First, the results show that some regions of phosphodiester backbone are protected by RNA structure, even under protein-dependent conditions (0.5 M NH4Cl/5 mM Mg2+). These regions, highlighted in gray shading in Figure 3B, are found primarily in DI and DII, with additional protections scattered in DIII, DIV and DVI. Under self-splicing conditions (0.5 M NH4Cl/50 mM Mg2+), many of these protections were intensified and a number of additional positions in DI, DII, DIII, DV and DVI were protected (Figure 3, blue or open triangles). The protection pattern was essentially the same under the high salt conditions (1.5 M NH4Cl/50 mM Mg2+). In general, the regions that were protected at 50 mM Mg2+ (blue or open triangles plus gray shading) neighbor sites of known tertiary interactions and include DIb (α), DIc1 (θ and ε′), DIc2 (β), DId(i)–(iii) (ζ and κ), DId2 (β′), DId3 (α′), DII (η and θ′), one side of DIII, parts of DV (κ′, λ′ and ζ′) and the top of DVI (η′). Together, these results suggest that some tertiary structure forms at low Mg2+ concentrations, but that higher Mg2+ concentrations are required to support additional tertiary structure, including many of the long-range interactions between different domains.
Most positions that were protected in the free RNA at 50 mM Mg2+ were protected to a similar extent at 5 mM Mg2+ in the presence of LtrA (Figure 3, blue triangles), presumably reflecting the formation of all or part of the active RNA structure in the presence of the protein. However, a few positions (open triangles), mostly in domain V, were protected at 50 mM Mg2+ (Figure 3A, lanes 3–5), but not at 5 mM Mg2+, in either the presence or absence of LtrA (lanes 1 and 2). These protections may reflect the formation of aberrant phosphate backbone structures or interactions at 50 mM Mg2+ that could contribute to the slow rate of self-splicing compared with protein-dependent splicing. We also note that DV is only partially protected at 5 mM Mg2+ in the presence of LtrA, even though most other tertiary structure has formed. In constrast, in a ribozyme derived from the yeast aI5γ intron, DV was completely protected from hydroxyl radical cleavage in reaction medium containing 500 mM KCl/100 mM Mg2+ (Swisher et al., 2001). This discrepancy could reflect differences between group IIA and IIB introns or the nature of the RNA constructs, a different stage of the reaction trapped by the branch point deletion in our construct and/or the formation of aberrant or inappropriately rigid RNA structures at high Mg2+.
Superimposed over the RNA protections, candidate protein protection sites were identified as positions that remained accessible at 50 mM Mg2+ and were protected only in the presence of LtrA protein (red triangles). These sites were found mainly in DI, DIV and DVI, with a few additional sites in DII. In DIV, most of the putative protein protection sites were clustered in the previously identified high affinity binding site DIVa, with some additional sites scattered in the DIV stem, L4, DIVb, DIVb1 and DIVb2, in agreement with the finding that these regions also contribute to LtrA binding (Wank et al., 1999).
In DI, sites protected in the presence of LtrA protein were found in subdomains b, c1, c2, d(ii)–(iii) and d4, with a few additional sites scattered elsewhere. DIb, c1, c2 and d(ii)–(iii) are conserved group II intron structures involved in long-range tertiary interactions (α–α′, β–β′, ε–ε′, θ–θ′ and κ–κ′), while DId4 is an idiosyncratic structure found only in subgroup IIA introns (Toor et al., 2001). Outside of DI, other clusters of protein-induced protections were found in DIIb and in DVI on the side opposite the branch point adenosine. Somewhat surprisingly, no phosphate backbone protections were observed in proximal regions of the 5′ or 3′ exons. As indicated previously, binding of the protein positions the reverse transcriptase active site to initiate reverse transcription in the 3′ exon (Wank et al., 1999), but this interaction may require the presence of an appropriate DNA primer.
Chemical modification with DMS and kethoxal
Figure 4 shows chemical modification experiments with DMS and kethoxal under the same conditions, with modification sites mapped as reverse transcription stops one base prior to the modified nucleotide. Reverse transcription detects DMS modification at the N1 position of unpaired adenines and the N3 position of unpaired cytidines, and kethoxal modifications at the N1 and N2 positions of unpaired guanines. Almost all of the DMS modifications were at adenines, due to inefficient modification of cytidines under the conditions used.
Fig. 4. DMS and kethoxal modification in the presence or absence of LtrA protein. Ll.LtrB-ΔA2486 RNA (20 nM) was incubated in different reaction media at 30°C in the presence or absence of 50 nM LtrA protein, and then modified with DMS or kethoxal. Modification sites were mapped by primer extension, using 5′-labeled primers complementary to different positions in Ll.LtrB RNA. (A) Representative gels. Lanes: A, dideoxy A sequencing ladder obtained using the same 5′-labeled primers; ‘–DMS’ or ‘–kethoxal’, RNA incubated without DMS or kethoxal; Denatured, RNA modified under denaturing conditions (see Materials and methods). Lanes 1–5, modification reactions; lanes 1 and 2, 0.5 M NH4Cl/5 mM Mg2+ (protein-dependent conditions) in the absence and presence of LtrA, respectively; lanes 3 and 4, 0.5 M NH4Cl/50 mM Mg2+ (self-splicing condition) in the absence and presence of LtrA, respectively; lane 5, 1.5 M NH4Cl/50 mM Mg2+ (maximal self-splicing conditions). Landmarks in the intron are indicated to the right of the gel. (B) Summary of DMS and kethoxal modification protection patterns. Long and short arrows indicate strong and moderate modifications at 5 mM Mg2+ (>10-fold and 2- to 10-fold increase in band intensity compared with the unmodified lane, respectively). Large and small blue circles indicate positions strongly (>50%) or moderately (49–25%) protected by RNA structure at 50 mM Mg2+ (lanes 3 and 5) compared with 5 mM Mg2+ (lane 1). Large and small red circles indicate positions that remained accessible at 50 mM Mg2+ (lanes 3 and 5) and were strongly (>50%) or moderately (25–49%) protected only in the presence of the LtrA protein (lanes 2 and 4). Positions that could not be monitored because of consistent strong reverse transcription stops were C-6, C-1, G1 and A393.
The chemical modification data show first that most of the conserved group II intron secondary structure is formed under protein-dependent conditions at 5 mM Mg2+. Strong and moderate modification sites, indicated by long and short arrows, respectively, were confined largely to ‘single-stranded’ loop or junction regions. Notable exceptions were the short stems DIVa(ii) and a(iii) and some GC base pairs in the elements tentatively identified as ζ and η, both of which function as tetraloop receptors. These same positions were also modified at 50 mM Mg2+, but showed increased protection in the presence of the LtrA protein, reflecting either direct protection by the protein or stabilization of the base pairs. With respect to tertiary interactions, key bases involved in the IBS1–EBS1, α–α′, β–β′ and θ–θ′ interactions were protected at 5 mM Mg2+, suggesting that DI and DII are at least partially folded under these conditions.
At 50 mM Mg2+, additional bases, highlighted in blue, were protected. As discussed below, a number of these bases are in loop or junction regions involved in conserved long-range tertiary interactions required for folding of the catalytic core. All of the bases that were protected by RNA structure at 50 mM Mg2+ were similarly protected at 5 mM Mg2+ in the presence of the LtrA protein, presumably reflecting induction of the active RNA structure by the protein. Curiously, IBS2 was largely protected in the free RNA, while EBS2 was modified at both 5 and 50 mM Mg2+ and became protected only in the presence of the LtrA protein (see bases A223, G225 and G227). A possible explanation is that IBS2 is occluded in an alternate structure in the free RNA at both 5 and 50 mM Mg2+, and the correct pairing is formed only in the presence of the protein.
Bases that remained accessible under self-splicing conditions and were protected only in the presence of the LtrA protein are highlighted in red. As in the iodine footprinting, strong protein protections were found in the high affinity binding site DIVa and include all four bases of the terminal A4 tetraloop, as well as bases in stem DIVa(iii), the adjoining JIVa(ii)/(iii) loop, the short stem (ii) and JIVa(i)/(ii). A number of these protected bases were confirmed by in vitro selection and mutagenesis to be critical for LtrA binding, and these experiments also showed that LtrA stabilizes the short stem DIVa(iii) (R.N.Singh, R.J.Saldanha, L.M.D’Souza and A.M.Lambowitz, in preparation).
Outside DIV, clusters of protein-dependent base protections were found in DI, II, V and VI. In DI, these protein-dependent base protections were interspersed with protein-dependent phosphate backbone protections in subdomains c1/c2, d(i)–(iii) and d4 (see Figure 5). In DII, a cluster of moderate protein-dependent protections was found neighboring the element tentatively identified as η. In DV, the few protein-dependent base protections were clustered in the elbow region near λ′ and may reflect stabilization of the λ–λ′ interaction in the presence of the protein. Finally, in DVI, protein-dependent base protections were interspersed with phosphate backbone protections on the side opposite the branch point adenosine. This surface presumably is exposed to the solvent and seems a good candidate for a bona fide protein interaction site.
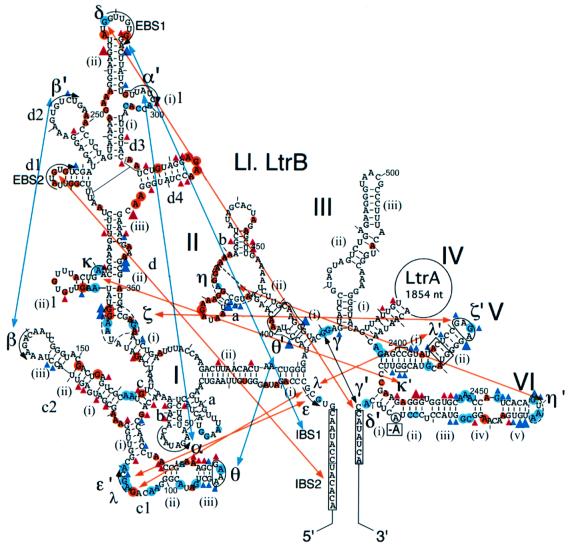
Fig. 5. Protein-induced RNA conformational changes detected by chemical modification. Phosphate backbone protections are indicated by red and blue triangles, and base protections by red and blue circles, as in Figures 3 and 4. Blue indicates positions that were protected by RNA structure at 50 mM Mg2+ and at 5 mM Mg2+ in the presence of LtrA protein. Only those blue positions in or near conserved tertiary interaction sites are shown; the complete sets are shown in Figures 3 and 4. Red (protein-dependent protections) indicates positions that were protected at 5 or 50 mM Mg2+ only in the presence of LtrA protein. Large and small symbols indicate strong and moderate protections, respectively. Blue lines indicate tertiary interactions that form stably at 5 mM Mg2+ in the absence of LtrA, and red lines indicate tertiary interactions that do not form stably at 5 mM Mg2+, but are induced by LtrA binding or by 50 mM Mg2+.
Protein-induced RNA conformational changes
Previous studies of self-splicing group II introns identified a number of conserved tertiary interactions required for folding of the catalytic core (Michel and Ferat, 1995; Swisher et al., 2001; and references therein). The chemical modification experiments provide information about the status of the nucleotide residues involved in these interactions and thus can be used to assess whether or not they form at low or high Mg2+ in the presence or absence of LtrA. The critical data are summarized in Figure 5. As judged by the protection of key bases, some tertiary interactions, including IBS1–EBS1, α–α′, β–β′ and θ–θ′, form even at 5 mM Mg2+ in the absence of the LtrA protein. In contrast, a second set of long-range interactions (δ–δ′, ε–ε′, γ–γ′, κ–κ′, λ–λ′, ζ–ζ′), do not form stably at 5 mM Mg2+, but are induced by binding of the LtrA protein or by 50 mM Mg2+. A similar protection pattern was also seen for a number of bases in DIII, possibly reflecting tertiary interactions between DIII and DI and/or DV (Figures 3 and 4; see Konforti et al., 1998; and references therein). As noted above, the LtrA protein may also play a role in stabilizing base pairing in the elements tentatively identified as ζ, the receptor for the terminal tetraloop of DV, and η, the receptor for the terminal tetraloop of DVI during the second step of splicing (Chanfreau and Jacquier, 1996). The suboptimal formation of these structures at 50 mM Mg2+ may also contribute to the slow rate of self-splicing compared with protein-dependent splicing. We conclude that the binding of the maturase promotes the formation of conserved long-range RNA tertiary interactions that are required for splicing.
Discussion
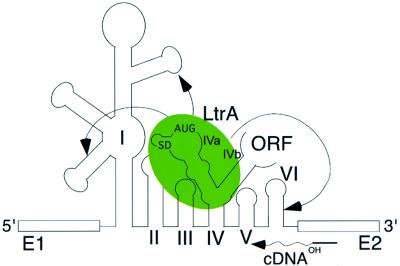
Together, our results suggest a model for maturase function shown in Figure 6. At low (5 mM) Mg2+ concentrations, where splicing is dependent on the LtrA protein, the RNA structure mapping shows that the Ll.LtrB RNA by itself forms most of the conserved group II intron secondary structure and some tertiary structure, particularly in DI and DII. According to the model, the maturase binds to this partially folded intron via its high affinity binding site in DIVa and then makes secondary contacts with conserved regions to promote the final folding of the catalytic core. When the high affinity binding site is deleted, the maturase can still promote splicing by binding directly to the catalytic core, but this splicing is inefficient under physiological conditions. The chemical modification experiments show that the maturase-induced conformational change includes the formation of a number of conserved long-range tertiary interactions, which do not form stably in the free RNA at low Mg2+ concentrations (Figure 5), and additional UV-cross-linking experiments support these findings by showing a strong protein-enhanced cross-link between U191 near κ in domain I and C2421 near κ′ in domain V (J.W.Noah and A.M.Lambowitz, unpublished). The relatively weak binding of LtrA to regions outside of DIV (apparent Kd for the ΔDIVa/b intron = ∼20 nM) may reflect either that the secondary contacts to these regions are linked to an energetically unfavorable conformational change or that they make only a small energetic contribution to the final folding of the intron RNA. Finally, although efficient splicing of the wild-type Ll.LtrB RNA requires the binding of LtrA to DIV, it remains possible that a minor component of the reaction occurs by direct binding of the maturase to the catalytic core, with a slower conformational change to the active RNA structure.
Fig. 6. Model for maturase-promoted RNA splicing. The LtrA protein binds first to its primary binding site DIVa and then makes secondary contacts (arrows) with upstream and downstream regions of the intron to promote folding of the catalytic core. The binding of LtrA to DIVa also positions the protein to initiate cDNA synthesis from a DNA primer in the 3′ exon, as required for intron mobility (Wank et al., 1999).
The high affinity binding of the maturase to DIVa contributes to protein-dependent splicing in two ways: it decreases the Kd for binding of LtrA to the Ll.LtrB RNA by ∼105-fold, and it increases the rate of splicing at saturating protein concentrations, presumably by facilitating the rate-limiting, protein-induced conformational to the active RNA structure (Saldanha et al., 1999; Wank et al., 1999). In vivo splicing of the ΔDIVa/b intron was only 8–10% that of the wild-type intron, demonstrating that the high affinity binding of LtrA to DIVa is necessary for efficient splicing at intracellular RNA and protein concentrations (X.Cui and A.M.Lambowitz, in preparation) Similarly, recent findings show that the yeast aI2 maturase binds to the cognate aI2 DIVa structure and that deletion of this structure inhibits in vivo splicing by 60% (M.Chao, A.M.Lambowitz and P.S.Perlman, in preparation) Thus, the high affinity binding to DIVa and its requirement for efficient splicing in vivo are likely to be general characteristics of these group II intron maturases. In addition to splicing, we showed previously that the binding of LtrA to DIVa positions the protein to initiate reverse transcription in the 3′ exon, as required for intron mobility (Wank et al., 1999), and experiments with the yeast aI2 intron show that the deletion of DIVa is essential for stable binding of the maturase to the excised intron lariat RNA and for intron mobility (M.Chao, A.M.Lambowitz and P.S.Perlman, in preparation).
Our results provide considerable additional detail about the high affinity LtrA binding site in DIVa. We find that the putative protein protection sites in this region are clustered toward the distal end and include all four bases of the terminal A4 loop of DIVa(iii), the distal AGG residues in stem IVa(iii) and the AGG in the adjoining JIVa(ii)/(iii). In agreement with the footprinting, a number of these bases, including the first two As of the terminal tetraloop, the two Gs at the distal end of the stem and the AG in JIVa(ii)/(iii), were invariant in in vitro selection for LtrA binding to the DIVa substructure and were confirmed by mutagenesis to be critical for recognition by the LtrA protein (R.N.Singh, R.J.Saldanha, L.M.D’Souza and A.M.Lambowitz, in preparation). Notably, a number of bases in the Shine–Dalgarno sequence, including the A and G in the short stem (iii), were modified at 5 mM Mg2+, but protected in the presence of LtrA. These findings suggest that the binding of LtrA may sequester the Shine–Dalgarno sequence to autoregulate its own translation, and experiments with reporter gene constructs show that this is in fact the case (R.N.Singh, R.J.Saldanha, L.M.D’Souza and A.M.Lambowitz, in preparation).
Although the high affinity binding of LtrA to DIVa is essential for efficient splicing, its weaker interactions with other regions of the catalytic core are still sufficient to promote splicing when the high affinity binding site is deleted (Wank et al., 1999). The equilibrium binding experiments here show that the maturase binds specifically to intron segments E1–DI and DV–E2, and the RNA footprinting experiments identify potential protein protection sites in these regions, as well as in DII, which did not bind specifically as part of the DII–DIII segment in the equilibrium binding experiments. The latter discrepancy could reflect either that DII does not fold properly in the absence of other regions of the RNA or that the DII sites do not make a strong contribution to protein binding. The interaction of the maturase with conserved as well as idiosyncratic regions of the intron RNA accounts for the cross-specificity of some maturases and suggests how maturases in some organisms could have evolved into general group II intron splicing factors (see Introduction).
The RNA footprinting localized the potential protein protection sites to different regions of domains I, II, V and VI. By definition, all of these sites are accessible in the folded RNA under self-splicing conditions, but we cannot exclude that some of the protein-dependent protections reflect formation or stabilization of additional RNA structures in the presence of the protein. In DI, the most likely protein-binding sites are those with clusters of phosphate backbone and base protections in regions with few RNA protections, particularly in DIc2 and DId4. DIc2 is a conserved structure in group II introns, whereas DId4 is found only in subgroup IIA introns. The deletion of DId4 in the context of the ΔDIVa/b intron results in only a 1.5- to 2-fold increase in Kd for LtrA (Kd = 43 ± 9 nM), implying that this structure individually makes only a small energetic contribution to binding (not shown). In DII, the candidate protein protection sites surround the sequence tentatively identified as η, which is a potential docking site for the DVI terminal loop in the second step of splicing (Chanfreau and Jacquier, 1996). In DV, the few base protections in the elbow region near element λ may reflect stabilization of the λ–λ′ interaction. In contrast, there are a number of protein-dependent phosphate backbone and base protections throughout DVI, and most lie on the solvent-accessible side opposite the branch point adenosine, as expected for bona fide protein protection sites. In the three-dimensional fold, DI, II and VI associate via the θ–θ′ and η–η′ interactions, and the protein-binding sites in these domains may form one binding surface, with the protein stabilizing the correct relative orientation of the domains. Our results indicate that the maturase must contain at least one binding site for DIVa and one or more additional binding sites that interact with conserved regions of the catalytic core. We are also struck by the number of base protections in purine-rich loops [the terminal A4 loop and J(ii)/(iii) of DIVa, the terminal AGAA loop of DId, and internal loops in DIIa], raising the possibility that this is a preferred recognition motif for the maturase.
The mechanism found here in which the maturase binds to a high affinity binding site outside the catalytic core and makes weaker contacts with the catalytic core differs from that for the well-studied group I intron splicing factors CYT-18 and CBP2, which bind strongly to the catalytic core to stabilize its active structure (reviewed in Lambowitz et al., 1999). This difference may reflect that group I introns use a single active site conformation for both steps of splicing, whereas group II introns may require a more flexible arrangement to accommodate structural rearrangements between the two steps of splicing (Chanfreau and Jacquier, 1996). After group II intron splicing, additional structural rearrangements in the RNP complex are required for different steps in intron mobility in which the bound IEP first contacts the DNA target site via specific base and phosphate backbone interactions, then stabilizes the active RNA structure for both steps of reverse splicing, cleaves the bottom strand of the DNA target site and finally repositions the reverse transcriptase domain to use the 3′ end of the cleaved bottom strand as a primer for reverse transcription (Singh and Lambowitz, 2001). These conformational changes may necessitate a mechanism in which one region of the IEP remains anchored to the RNA via its high affinity binding to DIVa, while the remainder is relatively free to interact with different RNA or DNA sites. With respect to splicing, group II introns must position first the 2′ OH of the branch point adenosine and then the 3′ OH of the 5′ exon at the active site, and the intron lariat produced during the first step may generate a somewhat different protein-binding site for the second step (Chanfreau and Jacquier, 1996). Spliceosomal introns also undergo conformational changes between the different steps of splicing and may likewise employ flexible, anchored interactions to facilitate protein rearrangements in some cases. In addition, this more complex system makes extensive use of ‘DExD/H box’ ATPases to mediate structural rearrangements (Staley and Guthrie, 1998).
Finally, with respect to evolution, phylogenetic analysis of a large number of recently sequenced group II introns indicates that association with the IEP is longstanding and preceded the subdivision of different group II intron structural subclasses, with little if any exchange of IEPs between different introns (Toor et al., 2001). We suggested previously that the reverse transcriptase first became associated with the intron to promote intron mobility and then adapted secondarily to function in RNA splicing (Wank et al., 1999), but it remains possible that group II introns evolved from a pre-existing retrotransposable element that acquired self-splicing capability (Curcio and Belfort, 1996). In either case, the prolonged co-evolution of the maturase with its intron RNA would lead to a situation in which maturases evolved to recognize idiosyncratic features of different group II introns (e.g. specific features of DIVa or DId4), while at the same time retaining contacts with conserved regions that are required to promote splicing. If the intron was dependent on the IEP for splicing, then the IEP could not be lost unless a suitable substitute protein was available. The most likely initial substitutes, perhaps exemplified by the situation in chloroplasts, would be other group II intron maturases, which still recognize conserved features of the catalytic core. In addition, some chloroplast introns appear to use both a maturase and a cellular protein, Crs1P, to facilitate splicing (Jenkins et al., 1997), and it is possible that such cellular partners or other proteins could eventually replace the maturase to support splicing. If mobile group II introns were the evolutionary ancestors of spliceosomal introns, similar transitions to dependence on trans-acting proteins would have had to occur as part of the emergence of a general splicing apparatus, a key step in the evolution of spliceosomal introns.
Materials and methods
Recombinant plasmids
pGMΔORF contains a 902 nucleotide ΔORF version of the L.lactis Ll.LtrB intron cloned behind the phage T3 promoter in pBSKS+ (Saldanha et al., 1999). pGMΔORF-ΔDIVa/b, pGMΔORF-ΔA2486 and pGMΔORF-ΔDIVa/b+A2486 are derivatives deleted for subdomains DIVa and b (Wank et al., 1999) and/or the branch point adenosine (A2486).
In vitro transcription
Plasmids were linearized with BamHI and transcribed with T3 RNA polymerase (Wank et al., 1999). Wild-type Ll.LtrB RNA is made up of 1214 nucleotides and consists of a 214 nucleotide 5′ exon, the 902 nucleotide ΔORF intron and a 98 nucleotide 3′ exon. Subsegments of Ll.LtrB RNA were synthesized with T7 RNA polymerase from DNA templates generated from pGMΔORF by PCR using a 5′ primer containing the T7 promoter sequence in conjunction with an appropriate downstream primer. Phosphorothioate-containing RNAs were synthesized with transcription mixes containing 5% of the specified NTPαS (Glen Research). RNAs were renatured by heating to 55°C for 1–5 min and then slowly cooling to 30°C, immediately prior to use.
Preparation of LtrA protein
The LtrA protein was expressed in E.coli BL21(DE3) using an intein-based affinity purification system (Impact; New England Biolabs) (Saldanha et al., 1999). For iodine footprinting, the protein was purified further using a 1 ml HiTrap SP ion exchange column (Amersham-Pharmacia) to remove residual dithiothreitol, which interferes with the analysis. For this additional purification, 1 ml of sample (∼2 mg protein) was loaded after dilution with 5 ml of column buffer (100 mM NaCl, 50 mM HEPES–KOH pH 7.8). The column was then washed with 10 ml of buffer and eluted with 10 ml of 1 M NaCl, 50 mM HEPES–KOH pH 7.8. Fractions (250 µl) were collected in chilled tubes and stored at –70°C.
RNA binding and splicing assays
For equilibrium binding assays, 32P-labeled RNA (5–50 pM) was incubated with increasing concentrations of LtrA protein in 200 µl of reaction medium containing 40 mM Tris–HCl pH 7.5 and specified concentrations of MgCl2 and NaCl or NH4Cl for 1 h at 30°C, then filtered through a nitrocellulose membrane to bind RNP complexes. The filters were washed three times with 1 ml of reaction medium, dried and counted for Cerenkov radioactivity. koff values were measured by nitrocellulose filter binding, as described (Saldanha et al., 1999). RNA splicing reactions were carried out with 32P-labeled Ll.LtrB RNA or its derivatives at 30°C in reaction media containing 40 mM Tris–HCl pH 7.5 and specified concentrations of NH4Cl and MgCl2. Splicing products were analyzed in a denaturing 6% polyacrylamide gel, which was dried and quantitated with a PhosphorImager. The fraction spliced was calculated as the ratio of splicing products to total RNA, and the data were best fit to equations with either one or two exponentials to obtain kobs (Saldanha et al., 1999).
RNA structure mapping and footprinting
RNA structure mapping and footprinting were carried out with 20 nM RNA and 50 nM LtrA protein in reaction media containing 40 mM HEPES-KOH pH 7.8 and specified concentrations of NH4Cl and MgCl2 at 30°C. LtrA was pre-incubated with the RNA for 30 min to allow complex formation. Iodine cleavage reactions with phosphorothioate-containing RNAs were initiated by adding 5 µl of a fresh iodine stock solution (400 µM in 100% ethanol; Sigma), incubated for 5 min at 30°C and terminated by adding 20 µg of yeast tRNA carrier (Sigma), followed by ethanol precipitation in the presence of 3 M sodium acetate (pH 5.3). Chemical modification reactions were initiated by adding 25 µl of diluted DMS (1:20) or kethoxal (1:100) and incubated for 5–10 min at 30°C. Denatured RNAs were prepared by heating 20 nM RNA in 0.5 ml of distilled water or 0.5 M NH4Cl to 100°C for 5 min, and then cooling rapidly to 50°C, prior to cleavage or modification for 30 s. In all cases, the conditions gave, on average, less than one cleavage or modification per molecule. Cleavage and modification sites were mapped by primer extension using SuperScript II or AMV reverse transcriptase (Invitrogen), essentially as described (Caprara et al., 1996), except that the reactions were carried out at 40°C for 30 min. Under these conditions, the primer extension products in the iodine cleavage experiments typically have one extra nucleotide residue added to their 3′ end by the reverse transcriptase. Primer extension products were analyzed in a denaturing 6% polyacrylamide gel, which was dried and quantitated with a PhosphorImager. Lanes were normalized to correct for unequal loading, using either full-length RNA or reverse transcriptase strong stops common to all lanes. Criteria for modifications and protections are described in the legends of Figures 3 and 4.
Acknowledgments
Acknowledgements
We thank François Michel, Anna Pyle and colleagues, Navtev Toor, Steven Zimmerly, Georg Mohr and Roland Saldanha for comments on the manuscript, and Ken Johnson for help with kinetics. This work was supported by NIH grant GM37949. J.W.N. is supported by an NIH post-doctoral fellowship.
References
- Caprara M.G., Mohr,G. and Lambowitz,A.M. (1996) A tyrosyl-tRNA synthetase protein induces tertiary folding of the group I intron catalytic core. J. Mol. Biol., 257, 512–531. [DOI] [PubMed] [Google Scholar]
- Chanfreau G. and Jacquier,A. (1996) An RNA conformational change between the two chemical steps of group II self-splicing. EMBO J., 15, 3466–3476. [PMC free article] [PubMed] [Google Scholar]
- Copertino D.W., Hall,E.T., Van Hook,F.W., Jenkins,K.P. and Hallick,R.B. (1994) A group III twintron encoding a maturase-like gene excises through lariat intermediates. Nucleic Acids Res., 22, 1029–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M. and Michel,F. (1995) Frequent use of the same tertiary motif by self-folding RNAs. EMBO J., 14, 1276–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M., DÉme,E., Jacquier,A. and Michel,F. (1997) Multiple tertiary interactions involving domain II of group II self-splicing introns. J. Mol. Biol., 267, 520–536. [DOI] [PubMed] [Google Scholar]
- Curcio M.J. and Belfort,M. (1996) Retrohoming: cDNA-mediated mobility of a bacterial group II intron requires a catalytic RNA. Cell, 84, 9–12. [DOI] [PubMed] [Google Scholar]
- Dickson L., Huang,H.-R., Liu,L., Matsuura,M., Lambowitz,A.M. and Perlman,P.S. (2001) Retrotransposition of a yeast group II intron occurs by reverse splicing directly into ectopic DNA sites. Proc. Natl Acad. Sci. USA, 98, 13207–13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins B.D., Kulhanek,D.J. and Barkan,A. (1997) Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell, 9, 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konforti B.B., Liu,Q. and Pyle,A.M. (1998) A map of the binding site for catalytic domain 5 in the core of a group II intron ribozyme. EMBO J., 17, 7105–7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz A.M., Caprara,M.G., Zimmerly,S. and Perlman,P.S. (1999) Group I and group II ribozymes as RNPs: clues to the past and guides to the future. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 451–485.
- Matsuura M. et al. (1997) A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev., 11, 2910–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F. and Ferat,J.L. (1995) Structure and activities of group II introns. Annu. Rev. Biochem., 64, 435–461. [DOI] [PubMed] [Google Scholar]
- Mohr G., Perlman,P.S. and Lambowitz,A.M. (1993) Evolutionary relationships among group II intron-encoded proteins and identification of a conserved domain that may be related to maturase function. Nucleic Acids Res., 21, 4991–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha R., Chen,B., Wank,H., Matsuura,M., Edwards,J. and Lambowitz,A.M. (1999) RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry, 38, 9069–9083. [DOI] [PubMed] [Google Scholar]
- Singh N.N. and Lambowitz,A.M. (2001) Interaction of a group II intron ribonucleoprotein endonuclease with its DNA target site investigated by DNA footprinting and modification interference. J. Mol. Biol., 309, 361–386. [DOI] [PubMed] [Google Scholar]
- Staley J.P. and Guthrie,C. (1998) Mechanical devices of the spliceosome: motors, clocks, springs and things. Cell, 92, 315–326. [DOI] [PubMed] [Google Scholar]
- Swisher J., Duarte,C.M., Su,L.J. and Pyle,A.M. (2001) Visualizing the solvent-inaccessible core of a group II intron ribozyme. EMBO J., 20, 2051–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N., Hausner,G. and Zimmerly,S. (2001) Co-evolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA, 7, 1142–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J., Böerner,T. and Hess,W.R. (1999) Comparative analysis of splicing of the complete set of chloroplast group II introns in three higher plant mutants. Nucleic Acids Res., 27, 3866–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wank H., SanFilippo,J., Singh,R.N., Matsuura,M. and Lambowitz,A.M. (1999) A reverse transcriptase/maturase promotes splicing by binding at its own coding segment in a group II intron RNA. Mol. Cell, 4, 239–250. [DOI] [PubMed] [Google Scholar]
- Wolfe K.H., Morden,C.W. and Palmer,J.D. (1992) Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc. Natl Acad. Sci. USA, 89, 10648–10652. [DOI] [PMC free article] [PubMed] [Google Scholar]