Abstract
In Caenorhabditis elegans, histone acetyltransferase CBP-1 counteracts the repressive activity of the histone deacetylase HDA-1 to allow endoderm differentiation, which is specified by the E cell. In the sister MS cell, the endoderm fate is prevented by the action of an HMG box-containing protein, POP-1, through an unknown mechanism. In this study, we show that CBP-1, HDA-1 and POP-1 converge on end-1, an initial endoderm-determining gene. In the E lineage, an essential function of CBP-1 appears to be the activation of end-1 transcription. We further identify a molecular mechanism for the endoderm-suppressive effect of POP-1 in the MS lineage by demonstrating that POP-1 functions as a transcriptional repressor that inhibits inappropriate end-1 transcription. We provide evidence that POP-1 represses transcription via the recruitment of HDA-1 and UNC-37, the C.elegans homolog of the co-repressor Groucho. These findings demonstrate the importance of the interplay between acetyltransferases and deacetylases in the regulation of a critical cell fate-determining gene during development. Furthermore, they identify a strategy by which concerted actions of histone deacetylases and other co-repressors ensure maximal repression of inappropriate cell type-specific gene transcription.
Keywords: C.elegans embryonic development/CBP-1/histone acetylase/histone deacetylase/transcriptional repression
Introduction
During Caenorhabditis elegans early embryogenesis, the birth of two sister cells, the anterior MS and the posterior E cell, sets the stage for future differentiation of the mesoderm and endoderm tissues, respectively (reviewed in Priess, 1994; Schnabel and Priess, 1997; Newman-Smith and Rothman, 1998). Both positive and negative regulation are necessary to ensure the establishment of the MS and E fates. The positive factors that are involved in E fate determination include the maternally provided, converging Wnt and MAP kinase signals (Rocheleau et al., 1997, 1999; Thorpe et al., 1997; Meneghini et al., 1999) as well as transcriptional activators such as SKN-1 (Bowerman et al., 1992) and CBP-1, which is the C.elegans ortholog of CBP and p300, the two related mammalian coactivators and histone acetyltransferases (Bannister and Kouzarides, 1996; Ogryzko et al., 1996; Shi and Mello, 1998). In C.elegans, CBP-1 is essential for the execution of most of the differentiation programs during embryogenesis, including endoderm differentiation, which is abrogated in embryos lacking CBP-1 (Shi and Mello, 1998). Interestingly, inhibition of HDA-1, the C.elegans homolog of the mammalian histone deacetylase HDAC1 (Taunton et al., 1996; Shi and Mello, 1998), in embryos lacking CBP-1 can restore endoderm differentiation, suggesting that CBP-1 promotes differentiation by counteracting the repressive activity of HDA-1 (Shi and Mello, 1998). However, the molecular mechanisms underlying the functional interaction between CBP-1 and HDA-1 during endoderm differentiation are poorly understood. For instance, it is unclear at which steps during endoderm differentiation CBP-1 and HDA-1 act to exert their antagonistic activity. An attractive model is that CBP-1 may be important for the activation of the initial endoderm-determining genes in the E cell, while HDA-1 represses these genes.
The MS and E cell fate determination process also requires mechanisms that ensure the suppression of the E fate in the MS cell. A mutation in the pop-1 gene results in the conversion of an MS to an E cell (Lin et al., 1995), indicating an essential function for POP-1 in the MS and E fate decision process. To date, POP-1 is the only protein that has been genetically identified as a suppressor of the E fate in the MS descendants (Lin et al., 1995). POP-1 is an HMG box-containing protein that belongs to the TCF/LEF family of transcription factors whose founding members were first described in Drosophila and mammals (reviewed in Roose and Clevers, 1999). Members of the TCF/LEF family have been shown to play roles in multiple biological processes including cell fate determination, pattern formation and differentiation. The homology between POP-1 and other TCF/LEF members is restricted mainly to the HMG box, which mediates sequence-specific DNA binding as shown for TCF/LEF1 (Roose and Clevers, 1999). Therefore, POP-1 may bind similar sequence motifs and function as a transcriptional regulator. Recently, a truncated POP-1 protein containing the HMG box has been shown to bind a TCF target sequence (Korswagen et al., 2000). Despite the critical role POP-1 plays in the determination of the MS and E cell fates, virtually nothing is known about how POP-1 exerts its biological function at the molecular level.
Studies in Drosophila and mammals have shown that the ability of TCF/LEF proteins to activate or repress transcription is dictated by the Wnt signaling pathway, which controls the recruitment of coactivators and corepressors (reviewed in Roose and Clevers, 1999; Barker et al., 2000). In the absence of Wnt activity, TCF/LEF proteins mediate transcriptional repression through the recruitment of a corepressor Groucho (reviewed in Fisher and Caudy, 1998; Roose et al., 1998; Barker et al., 2000; Chen and Courey, 2000). Interestingly, genetic and biochemical interactions between Groucho and histone deacetylase during Drosophila embryogenesis have been identified recently (Chen et al., 1999). Neither Groucho nor histone deacetylases bind DNA directly, and each is brought to promoters by interactions with sequence-specific DNA-binding transcription factors, and functions as a transcriptional corepressor. In C.elegans, the Groucho homolog UNC-37 has been shown to play a role in post-embryonic neuronal cell fate determination (Pflugrad et al., 1997). However, the roles for UNC-37 and the histone deacetylase HDA-1 in embryogenesis and in the regulation of genes important for cell fate specification are largely unknown.
In this study, we show that the actions of CBP-1 and HDA-1 converge on end-1, a critical initial endoderm-determining gene whose product is sufficient to induce endoderm differentiation (Zhu et al., 1997, 1998). In the E lineage, a primary function of CBP-1 is to overcome inhibition by HDA-1 in order to activate end-1. This demonstrates the importance of the interplay between a histone acetylase and a histone deacetylase in regulating end-1 and provides a molecular basis for the requirement for CBP-1 in endoderm differentiation. We have also investigated factors that are involved in restricting end-1 transcription to E descendants. We find that POP-1 functions as a transcriptional repressor. Inhibition of either POP-1 alone, or HDA-1 and UNC-37 together results in ectopic activation of end-1::GFP in MS descendants, suggesting that these proteins play important roles in suppressing cell type-specific gene transcription during C.elegans embryogenesis. We provide biochemical and functional evidence that HDA-1 and UNC-37 act as corepressors for POP-1. Our findings support a model whereby POP-1 represses inappropriate transcription of the end-1 promoter via the recruitment of HDA-1 and UNC-37. These findings identify a strategy by which concerted actions of histone deacetylases and other corepressors may be necessary to ensure maximal repression of inappropriate cell type-specific gene transcription. This combinatorial scheme appears to be conserved in multiple organisms.
Results
CBP-1 and HDA-1 converge on the endoderm-specific end-1 promoter
Our previous study showed that CBP-1 is required for endoderm differentiation and that inhibition of hda-1 expression in an embryo lacking CBP-1 restored endoderm differentiation (Shi and Mello, 1998). We wished to identify molecular mechanisms underlying this functional interaction between CBP-1 and HDA-1 by determining the step(s) at which they exert their endoderm differentiation-promoting and inhibiting activities, respectively. One model is that CBP-1 is required early in this process to activate initial endoderm-determining genes. We tested this model by examining the effect of inhibiting cbp-1 expression on end-1, which is one such early endoderm-determining gene. Transcription of the end-1 gene is activated specifically in the E but not MS or other cell lineages. The end-1 gene encodes a GATA-like transcription factor which has been shown to be sufficient to initiate endoderm differentiation (Zhu et al., 1997, 1998). A transgenic C.elegans line was generated by integration of a reporter plasmid with green fluorescent protein (GFP) under the control of the end-1 promoter (see Materials and methods). This end-1 promoter fragment faithfully directs GFP expression in a temporal and spatial pattern identical to that of the endogenous end-1 gene (Kaltenbach et al., 2000). We used this transgenic C.elegans line to determine whether end-1 promoter activity is regulated by CBP-1 and HDA-1 as well as by factors that restrict end-1 transcription to the E descendants.
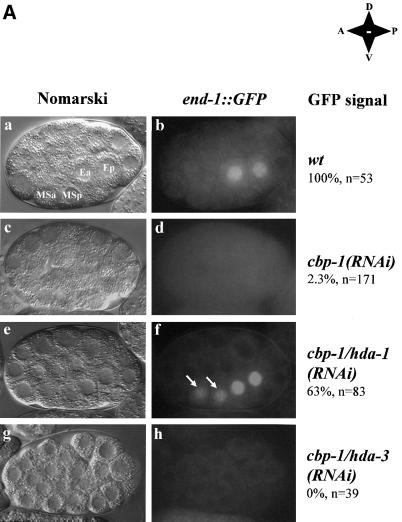
We used the RNA-mediated interference (RNAi) approach (Fire et al., 1998) to inhibit cbp-1 and hda-1 expression. As expected, in a wild-type embryo, the end-1::GFP signal is detected only in E descendants (Figure 1A, panel b). However, in a cbp-1(RNAi) embryo, the GFP signal is completely absent (Figure 1A, panel d). This suggests that CBP-1 is necessary for the activation of the end-1 promoter in E descendants, and the result is consistent with the previous finding that CBP-1 is essential for endoderm differentiation (Shi and Mello, 1998). Significantly, inhibition of hda-1 in the cbp-1(RNAi) embryo restores the end-1 promoter activity in E descendants (Figure 1A, panel f). In addition, end-1::GFP is ectopically activated in MS derived cells in ∼60% of the early embryos (Figure 1A, panel f, MS descendants indicated by arrows). In fact, ectopic activation of end-1::GFP is also observed in a similar percentage of the early embryos when hda-1 alone is inhibited (Figure 2F, detailed below), suggesting a role for hda-1 in suppressing end-1::GFP in MS derived cells.
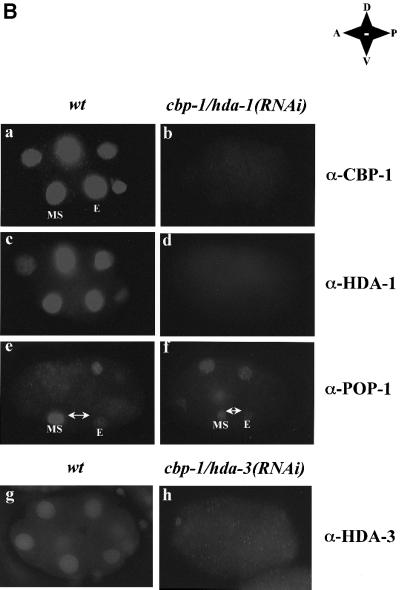
Fig. 1. Inhibition of hda-1 restores the promoter activity of an endoderm-specific reporter in cbp-1(RNAi) embryos. (A) CBP-1 and HDA-1 are involved in the regulation of the end-1 promoter activity in C.elegans embryos. Transgenic C.elegans carrying an integrated end-1::GFP reporter plasmid were injected with dsRNA for either cbp-1 alone or cbp-1 and hda-1 together. The stage of embryos derived from the injected animals was scored using Nomarski optics (a, c, e and g). Embryos were analyzed for the presence of the reporter plasmid-derived GFP signal at the 2E-cell stage (b, d and f). The MS descendents are indicated by arrows. Wild-type, cbp-1(RNAi), cbp-1/hda-1(RNAi), cbp-1/hda-3(RNAi) embryos are labeled as such on the right. The two E and two MS descendents are labeled as Ea, Ep, MSa and MSp, respectively. In addition, the anterior–posterior, dorsal–ventral orientations of the embryos are shown. Wt: wildtype; A: anterior; P: posterior; D: dorsal; V: ventral. (B) Protein expression in cbp-1/hda-1, and cbp-1/hda-3 RNAi embryos. Early wild-type (wt) and cbp-1/hda-1(RNAi) embryos were stained with CBP-1, HDA-1 and POP-1 polyclonal antibodies. While CBP-1 and HDA-1 were present in the nucleus of all cells in the wild-type embryos (a and c), both proteins were absent in cbp-1/hda-1 double RNAi embryos (b, n = 114 and d, n = 53). The embryos in a and b or c and d were equally exposed in order to visualize the outline of the RNAi embryos (b and d). As a result, the wild-type embryos were overexposed (a and c). cbp-1/hda-1(RNAi) embryos were also stained with POP-1 antibodies. Both wild-type (e) and the RNAi embryos (f, n = 5) displayed an asymmetric distribution of POP-1. Although the signal intensity in the MS cell varies, the POP-1 asymmetric expression pattern is unaffected in the cbp-1/hda-1(RNAi) embryos. MS and E cells are indicated in a, e and f. Wild-type and cbp-1/hda-3(RNAi) embryos were stained with HDA-3 antibodies in g and h. HDA-3 is expressed in all cells in the wild-type embryos (g) but was absent in the RNAi embryos (h, n = 62).
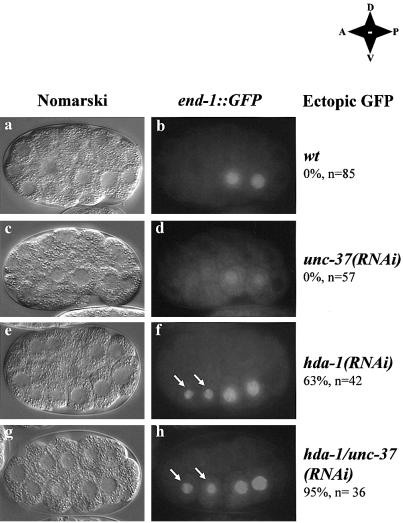
Fig. 2. Inhibition of hda-1 and unc-37 results in ectopic expression of end-1::GFP. Double-stranded hda-1 and unc-37 RNA was injected either singly or together into the end-1::GFP reporter strain. Embryos at the 2E cell stage were identified using Nomarski microscopy (a, c, e and g) and analyzed for the presence of the end-1::GFP signal (b, d, f and h). MS descendents in f and h are indicated by arrows. Note that GFP signals in the MS descendents of either the hda-1(RNAi) or hda-1/unc-37(RNAi) embryos are generally weaker than those in the E descendents. The reason for this difference is unclear at the present time. Embryos that ectopically express end-1::GFP are scored and indicated by percentage. Wild-type as well as (RNAi) embryos are labeled as such on the right. In addition, the anterior-posterior, dorsal-ventral orientations of the embryos are shown. Wt: wild-type; A: anterior; P: posterior; D: dorsal; V: ventral. n: number of embryos counted.
To ensure that restoration of the end-1 promoter activity in the cbp-1/hda-1(RNAi) embryos was not a result of non-specific weakening of the RNAi machinery due to the inclusion of a second double-stranded (ds) RNA (hda-1), we examined CBP-1 and HDA-1 expression and found both proteins to be efficiently inhibited in these double RNAi embryos (Figure 1B, panels a–d). The HMG box protein POP-1 plays an essential role in E and MS cell fate determination (Lin et al., 1995), and in suppression of end-1 promoter activity in the MS descendants (Figure 3A). Alteration of its expression may therefore affect E and MS cell differentiation. Thus we also examined POP-1 expression in the cbp-1/hda-1(RNAi) embryos. Consistent with the previous report (Lin et al., 1995), we found POP-1 expressed at a higher level in the MS cell than the E cell (Figure 1B, panel e). Importantly, this asymmetric expression pattern was maintained in the cbp-1/hda-1(RNAi) embryos (Figure 1B, panel f). Therefore, restoration of the end-1 promoter activity in the cbp-1/hda-1(RNAi) embryos was also not due to an alteration of the POP-1 expression pattern.
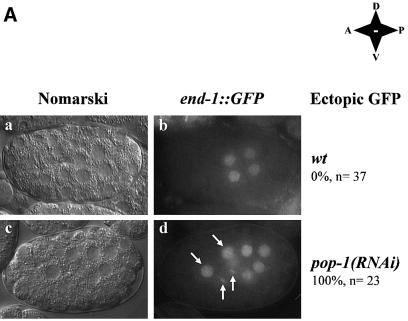
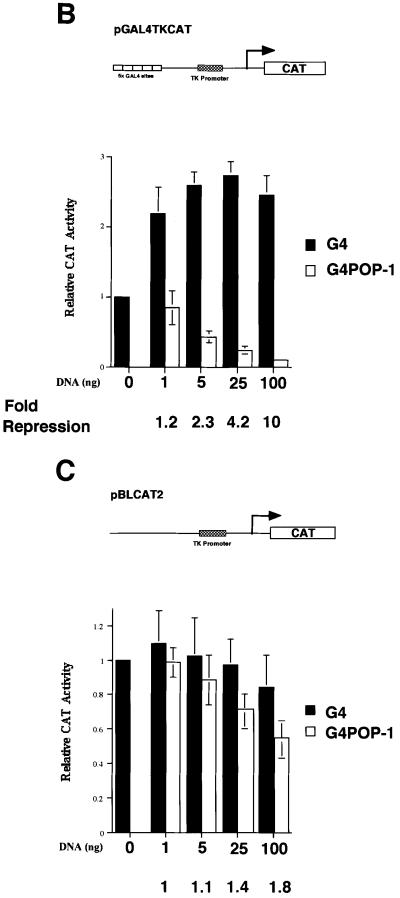
Fig. 3. POP-1 is a transcriptional repressor. (A) Inhibition of pop-1 restores the end-1 promoter activity in cbp-1(RNAi) embryos. Embryos at the 4E cell stage from hermaphrodite end-1::GFP mothers injected with pop-1 dsRNA were identified by light microscopy (a and c) and scored for the presence of the reporter plasmid-derived GFP signal (b and d). MS descendents are indicated by white arrows. Wild-type as well as RNAi embryos are labeled as such on the right. In addition, the anterior–posterior, dorsal–ventral orientations of the embryos are shown. Wt: wildtype; A: anterior; P: posterior; D: dorsal; V: ventral. (B) POP-1 represses transcription when targeted to a heterologous promoter. Various amounts of either pGAL4 (G4) or pGAL4–POP-1 (G4POP-1) containing the entire coding region of POP-1 were transfected into C33A cells, together with the reporter plasmid pGAL4-TKCAT. The schematic diagram of the reporter gene is shown above. The CAT activity in the absence of G4 or G4POP-1 is designated as 1. The black and open bars represent relative CAT activities in the presence of G4 or G4POP-1, respectively. The relative CAT activities are presented with the mean ± SD from three independent transfections. The fold repression by G4POP-1 is shown at the bottom. (C) G4POP-1 cannot repress transcription efficiently when not targeted to the promoter. The same set of CAT assays was carried out with G4POP-1 and the reporter plasmid TKCAT without the G4 binding sites.
As a further control, we examined the effect of inhibition of an hda-1 related gene, hda-3 (Shi and Mello, 1998), on the end-1 promoter activity in cbp-1(RNAi) embryos. We coinjected cbp-1 and hda-3 dsRNA and found that, although HDA-3 expression was inhibited efficiently (Figure 1B, compare panels h and g), neither the end-1 promoter activity (Figure 1A, panel h) nor endoderm differentiation (data not shown) was restored in cbp-1/hda-3(RNAi) embryos. This is consistent with the previous finding that inhibition of hda-3 does not affect endoderm differentiation or C.elegans development as a whole (Shi and Mello, 1998). Thus, these findings suggest that the actions of CBP-1 and HDA-1 in endoderm differentiation converge on end-1, an initial endoderm-determining gene. Furthermore, HDA-1 seems to repress end-1::GFP in MS descendants and may therefore play a role in restricting end-1::GFP expression to the E descendants.
Heat shock-induced END-1 expression can induce endodermal fate in embryos lacking CBP-1
The above demonstration that CBP-1 is required for the end-1 promoter activity suggests that an essential function of CBP-1 in endoderm differentiation may be the activation of end-1 transcription. To determine whether END-1 can induce endodermal fate in the absence of the coactivator CBP-1, we performed RNAi experiments in transgenic C.elegans JR663, in which END-1 can be inducibly expressed from end-1 cDNA under the control of a heat shock promoter (Zhu et al., 1998). As shown in Table I, before heat shock, endoderm differentiation in cbp-1(RNAi) embryos, scored by the presence of gut granules from three independent experiments, varied from 0 to 17%. However, after heat shock induction of END-1, the percentage of embryos with gut granules increased significantly (66–89%). To ensure that the process of heat shock did not restore CBP-1 expression, we stained some of these embryos with CBP-1 antibodies. We found no evidence for CBP-1 expression in cbp-1(RNAi) embryos that had been subjected to heat shock (n = 95), suggesting that CBP-1 expression was not restored by heat shock. These findings indicate that END-1 is sufficient to induce gut differentiation in the absence of CBP-1. Based on the previous report that END-1 is sufficient to induce endodermal fate in all cell lineages (Zhu et al., 1998), it is likely that all cell lineages including the E blastomere in the hs::end-1;cbp-1(RNAi) embryos contributed to gut formation. Taken together, the above results are consistent with the hypothesis that an essential function of CBP-1 in endoderm differentiation is to activate transcription of the end-1 gene.
Table I. Heat shock-induced END-1 induces gut differentiation in cbp-1(RNAi) embryos.
| Experiment | Percentage of dead embryos with gut granules (n) in hs-end-1 (JR663); cbp-1(RNAi) animals |
|
|---|---|---|
| Non-HS | HS | |
| 1 | 0% (32) | 66% (35) |
| 2 | 17% (48) | 89% (19) |
| 3 | 9% (212) | 84% (55) |
| Average | 8.7% (292) | 79.7% (109) |
JR663 animals were injected with double-stranded cbp-1 RNA. Twelve hours later, they were transferred to a fresh plate and allowed to lay eggs for 4 h. The embryos were then subjected to a 30 min heat shock at 33°C. The eggs were allowed to develop overnight and then scored for gut differentiation. Some of the embryos were then retrieved for staining with CBP-1 antibodies. HS, heat shock; n, number of embryos.
had-1 and unc-37 are involved in repression of the end-1 promoter in vivo
We next determined whether hda-1 is involved in repressing end-1::GFP in non-E descendants. In addition, we investigated unc-37 as another potential negative regulator since its Drosophila homolog Groucho has been shown to physically interact with histone deacetylase Rpd3p (Chen et al., 1999). Genetically, Drosophila animals carrying a strong loss-of-function allele of groucho appear to synergize with a rpd3 mutation to increase embryonic lethality (Chen et al., 1999). However, the molecular targets of this concerted action of Groucho and Rpd3p are unclear. We wished to determine whether hda-1 and unc-37 collaborate to repress the end-1 promoter, and thereby contribute to the E and MS cell fate determination.
We used RNAi to inhibit hda-1 and unc-37 expression, either alone or together. In the unc-37(RNAi) embryos, although the end-1::GFP signal is somewhat reduced compared with that in the wild-type embryos, the expression pattern is essentially wild-type throughout embryogenesis (Figure 2d). In the hda-1(RNAi) embryos, 63% (n = 42) of the RNAi embryos displayed ectopic end-1::GFP expression in MS descendants (Figure 2f, MS descendants are indicated by arrows). The incomplete ectopic activation of end-1::GFP in the hda-1(RNAi) embryo could be due to either incomplete RNAi or functional redundancy of hda-1 with hda-2 and hda-3, two highly related histone deacetylases (Shi and Mello, 1998). However, we failed to detect HDA-1 expression in either hda-1 or cbp/hda-1(RNAi) embryos, suggesting efficient RNAi (Figure 1B, panel d and data not shown). To address the redundancy issue, we found that inhibition of hda-2 and hda-3, either alone or together, did not affect embryonic development or end-1::GFP expression. When all three hda genes were inhibited by RNAi, the embryos displayed phenotypes indistinguishable from those of hda-1(RNAi) embryos (Shi and Mello, 1998). Together, these experiments suggest that incomplete induction of ectopic end-1::GFP in the hda-1(RNAi) embryos was unlikely to be due to inefficient inhibition of HDA-1 expression, or functional redundancy of hda-1 with hda-2 and hda-3.
Significantly, inhibition of both hda-1 and unc-37 together resulted in ectopic activation of end-1::GFP in nearly all the double RNAi embryos (95%, n = 36). This collaboration was only seen between unc-37 and hda-1, but not unc-37 with hda-2 or hda-3 (data not shown). An example of an hda-1/unc-37(RNAi) embryo is shown in Figure 2h. In this embryo, the expression of both HDA-1 and UNC-37 is inhibited, which resulted in ectopic expression of end-1::GFP in the two MS descendants (Figure 2h, MS descendants indicated by arrows). Furthermore, we observed that in later stage embryos in which either hda-1 alone, or hda-1 and unc-37 together are inhibited, end-1::GFP is found ectopically expressed in multiple cell types in addition to cells of the MS lineage (data not shown). Taken together, these results suggest that the collaboration of HDA-1 and UNC-37 plays an important role in the repression of end-1::GFP in MS descendants and perhaps in other non-E descendants as well as later in embryonic development.
POP-1 is a transcriptional repressor and can repress transcription of a reporter gene through the end-1 promoter
Histone deacetylases and Groucho are transcriptional corepressors that do not possess DNA-binding activities. How then are HDA-1 and UNC-37 recruited to the end-1 promoter to repress transcription? Genetically, the HMG box-containing POP-1 acts as a repressor of the E fate in the MS descendants (Lin et al., 1995). Therefore, one attractive possibility is that POP-1 may function as a transcriptional repressor that represses E-specific genes such as end-1 through recruitment of HDA-1 and UNC-37. To test this hypothesis, we first asked whether inhibition of pop-1, similar to inhibition of hda-1 and unc-37, results in ectopic end-1 transcription. As shown in Figure 3A, the end-1::GFP signal is ectopically induced in the pop-1(RNAi) embryos, suggesting de-repression of end-1 transcription in the absence of POP-1. This is consistent with previous data demonstrating that a lack of POP-1 results in an MS to E transformation (Lin et al., 1995). Ectopic activation of end-1::GFP is restricted to cells of the MS lineage in pop-1(RNAi) embryos. This is in contrast to the results in which hda-1 and unc-37 are inhibited, in which case ectopic activation is also observed in non-MS descendants later in development (data not shown). This finding suggests that POP-1 may function as a transcriptional repressor that represses end-1 transcription in MS descendants.
To determine whether POP-1 functions as a transcriptional repressor, we analyzed the ability of POP-1 to repress transcription in mammalian cells using the GAL4 fusion protein-based assay. We fused the GAL4 DNA-binding domain (G4, amino acids 1–149) in-frame to full-length POP-1 at its N-terminus to generate a GAL4–POP-1 fusion protein (G4POP-1). As shown in Figure 3B, G4POP-1 exhibits a dose-dependent repression of the GAL4-TKCAT reporter gene. At the highest concentration tested, G4POP-1 repressed GAL4-TKCAT 10-fold (Figure 3B). This repression is largely dependent on targeting G4POP-1 to the promoter of the reporter gene since G4POP-1 had marginal effects on a TKCAT reporter that lacks the G4 binding sites (Figure 3C, 1.8-fold repression at the highest dose of G4POP-1). Thus, POP-1 can efficiently repress transcription when targeted to the promoter, consistent with previous findings that the POP-1 homologs in other species can function as transcriptional repressors (reviewed in Roose and Clevers, 1999).
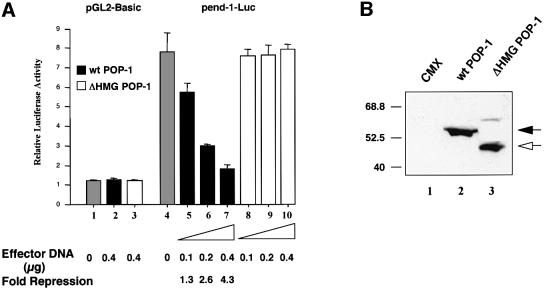
The fact that the end-1 promoter activity was de-repressed in the MS descendants in the absence of POP-1 (Figure 3A) provided significant evidence linking POP-1 to end-1 repression in vivo. The observation that POP-1 could repress transcription when targeted to a heterologous promoter (Figure 3B) further suggested that repression of end-1 by POP-1 is likely to be occurring at the transcriptional level. However, it remained unclear whether this was a direct or indirect effect of POP-1. We thus determined whether POP-1 functions to directly repress end-1 transcription. A 1.7 kb end-1 promoter fragment used in the C.elegans embryo assays (Figure 1 and Materials and methods) was fused to the luciferase (Luc) gene and used as a reporter (pend-1-Luc). As shown in Figure 4A, while POP-1 had no effect on the parental Luc vector (lane 2), it repressed pend-1-Luc in a dose-dependent manner (compare lanes 5–7 with lane 4). Significantly, a POP-1 mutant lacking the DNA-binding HMG box (ΔHMG POP-1) failed to repress pend-1-Luc, suggesting that binding of POP-1 to the end-1 promoter is necessary for POP-1 repression (Figure 4A, lanes 8–10). As a control, we found ΔHMG POP-1 expressed at a level comparable to that of the wild-type POP-1 (Figure 4B, compare lanes 2 and 3). Equal transfection efficiencies were ensured by monitoring the GFP signals expressed from the cotransfected GFP plasmid (data not shown). In a separate experiment, a GFP-tagged POP-1 was found to bind an extrachromosomal array containing the end-1 promoter (M.Maduro and J.Rothman, unpublished observation), providing evidence that POP-1 binds the end-1 promoter in C.elegans embryos. Taken together, these findings strongly suggest that POP-1 functions as a transcriptional repressor that can directly repress end-1 transcription in C.elegans embryos. Since end-1 transcription is positively regulated by CBP-1, likely to be recruited by the activator SKN-1 (Shi and Mello, 1998; Walker et al., 2000), our results provide molecular evidence that the opposing activities of POP-1 and CBP-1 converge on the end-1 promoter.
Fig. 4. POP-1 represses transcription of a reporter gene through the end-1 promoter. (A) Wild-type POP-1 but not a mutant lacking the HMG box represses the end-1 promoter. Increasing amounts (indicated at the bottom) of either wild-type or mutant ΔHMG POP-1 (subcloned into CMX; effector plasmids) were transfected into HeLa cells, together with the reporter plasmids pGL2-Basic (0.8 µg, lanes 1–3) or pend-1-Luc (0.8 µg, lanes 4–10). The total amount of CMV promoter-based vector was maintained constant in all transfection reactions through the addition of the parental CMX plasmid. The total amount of DNA was normalized to 1.5 µg per transfection by cotransfection of the Bluescript plasmid. The luciferase activity of the parent vector pGL2-Basic is designated as 1. The gray bars represent relative luciferase activities of pGL2-Basic (lane 1) and pend-1-Luc (lane 4), respectively. The black bars represent pGL2-Basic (lane 2) and pend-1-Luc activities (lanes 5–7) in the presence of wild-type POP-1, while the open bars indicate their respective activities in the presence of the mutant ΔHMG POP-1 (lanes 3 and 8–10). The relative luciferase activities are presented as the mean ± SD (n = 3). The fold repression by POP-1 is shown at the bottom. (B) Wild-type and ΔHMG POP-1 are expressed at comparable levels. Western blotting was performed to compare the expression levels of both wild-type and ΔHMG POP-1. HeLa cells were transfected with equivalent amounts of either the parental CMX plasmid or the corresponding wild-type and ΔHMG POP-1 expression vectors, as indicated at the top of the gel. Transfection efficiency was monitored by the GFP signals from the cotransfected GFP plasmid. Forty micrograms of whole-cell extracts were then resolved in a 10% SDS gel and transferred to a nitrocellulose membrane. The membrane was probed with the α-POP-1 polyclonal antibodies. The positions of the wild-type and ΔHMG POP-1 proteins are indicated by a black and an open arrow, respectively. The molecular weight markers (in kDa) are shown on the left.
Physical interactions among POP-1, HDA-1 and UNC-37
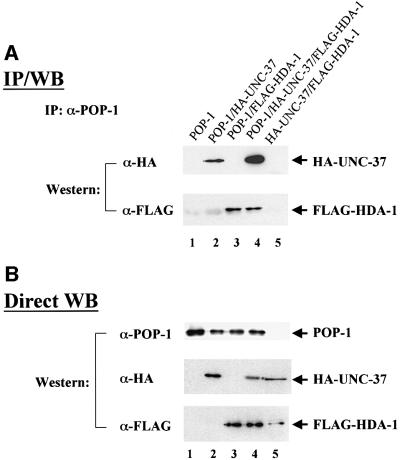
As described above, inhibition of either pop-1 or both hda-1 and unc-37 together causes ectopic expression of end-1::GFP in MS descendants in nearly all RNAi embryos during early embryogenesis. These findings suggest that HDA-1 and UNC-37 may function as corepressors for POP-1. This hypothesis predicts physical interactions among POP-1, HDA-1 and UNC-37. To address this issue, we performed co-immunoprecipitation (Co-IP) experiments. The three C.elegans genes were expressed in HeLa cells either alone or in combination, as indicated in Figure 5. HDA-1 was tagged with the FLAG and UNC-37 with the HA epitope, respectively. Cell lysates were prepared from the transfected cells and the POP-1 protein was immunoprecipitated with α-POP-1 polyclonal antibodies (gift of Rueyling Lin). The immunoprecipitates were then probed in western blots with either HA or FLAG monoclonal antibodies to detect the presence of UNC-37 and/or HDA-1 coprecipitated with POP-1. As shown in Figure 5A, UNC-37 can be co-immunoprecipitated with POP-1 (compare lane 2 with lanes 1, 3 and 5), as can HDA-1 (lanes 3 and 4). We have consistently observed greater amounts of UNC-37 co-immunoprecipitated with POP-1 in the presence of HDA-1 (Figure 5A, compare lanes 2 and 4). This is not due to increased protein levels, as western blotting shows that all three proteins are expressed at similar levels with different combinations of input plasmids (Figure 5B).
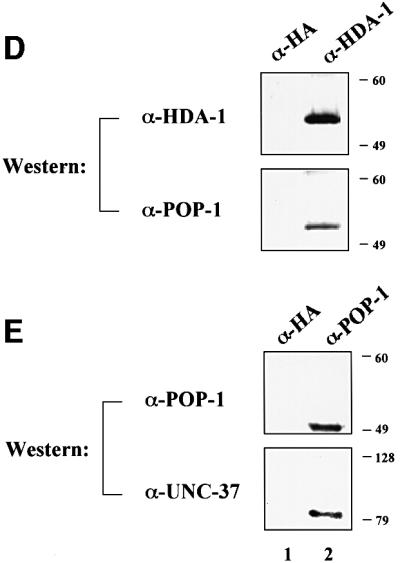
Fig. 5. Physical interactions among POP-1, HDA-1 and UNC-37. (A) POP-1 interacts with HDA-1 and UNC-37. To identify physical interactions among POP-1, HDA-1 and UNC-37, immunoprecipitation coupled with western blotting experiments were carried out. HeLa cells were transfected with various combinations of the input plasmids as indicated at the top. HDA-1 and UNC-37 were tagged with FLAG and HA epitope, respectively. The POP-1 immunoprecipitates were separated on SDS gels and probed with either FLAG or HA antibodies to detect FLAG-HDA-1 and HA-UNC-37, which are labeled and indicated as such on the right. IP: immunoprecipitation; WB: western blotting. (B) POP-1, HDA-1 and UNC-37 are expressed at similar levels in cells with different combinations of transfected plasmids. The lysates from the above transfected cells were probed directly with α-POP-1 polyclonal antibodies, α-FLAG and α-HA monoclonal antibodies. The POP-1, HDA-1 and UNC-37 bands are indicated by arrows and are labeled as such on the right. (C) HDA-1 interacts with UNC-37. Flag-tagged HDA-1 plasmid was transfected with MYC-tagged UNC-37 either in the presence or absence of the pop-1 plasmid into HeLa cells. Lysates were immunoprecipitated with α-HDA-1 antibodies or α-HA polyclonal antibodies as indicated. They were subsequently probed by western blotting with α-MYC antibodies to visualize co-precipitated MYC-UNC-37, which is indicated by an arrow and labeled as such. (D and E) POP-1 interacts with HDA-1 and UNC-37 in C.elegans embryos. Immunoprecipitation coupled with western blotting experiments were performed to investigate the ability of POP-1 to interact with HDA-1 and UNC-37 in C.elegans embryos. Total protein extracts from worm embryos were immunoprecipitated with the antibodies indicated at the top. The immunoprecipitates were resolved on an SDS gel and analyzed by western blotting with the antibodies indicated on the left. (D) Upper panel, α-HDA-1; lower panel, α-POP-1. (E) Upper panel, α-POP-1; lower panel, α-UNC-37. The molecular weight markers (in kDa) are shown on the right.
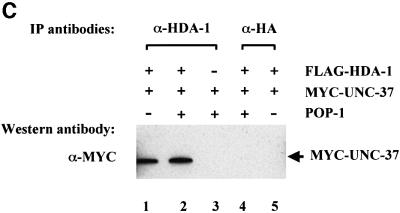
In addition to the individual interactions of HDA-1 and UNC-37 with POP-1, we also identified an interaction between HDA-1 and UNC-37. As shown in Figure 5C, the MYC-tagged UNC-37 can be co-immunoprecipitated by α-HDA1 polyclonal antibodies (lane 1). This interaction is dependent on the presence of transfected hda-1 plasmid since in its absence α-HDA1 polyclonal antibodies failed to bring down MYC-UNC37 (Figure 5C, lane 3). The interaction between HDA-1 and UNC-37 is not affected by POP-1, as similar amounts of MYC-UNC-37 are co-immunoprecipitated by α-HDA1 polyclonal antibodies in the presence or absence of the cotransfected pop-1 plasmid (Figure 5C, compare lanes 1 and 2). Lastly, unrelated polyclonal antibodies (α-HA polyclonal) failed to co-immunoprecipitate MYC-UNC-37 (Figure 5C, lanes 4 and 5). Taken together, we have detected not only interactions of POP-1 with HDA-1 and UNC-37, but also an interaction between HDA-1 and UNC-37. The current data cannot distinguish between the possibilities of a ternary complex involving all three proteins or two binary complexes, i.e. POP-1–HDA–1 and POP-1–UNC-37. However, we favor the ternary complex model, based on the protein interaction results and the fact that inhibition of both HDA-1 and UNC-37 is required to induce full ectopic expression of end-1::GFP (Figure 2).
To determine whether POP-1 interacts with HDA-1 and UNC-37 under physiological conditions in C.elegans, we carried out co-immunoprecipitation experiments using protein extracts isolated from mixed stage embryos. As shown in Figure 5D, POP-1 can be co-immunoprecipitated with HDA-1 polyclonal antibodies but not the unrelated antibodies that recognize the HA epitope. In addition, UNC-37 can be co-immunoprecipitated with POP-1 polyclonal antibodies (Figure 5E). These findings are consistent with the results obtained in mammalian cell culture (Figure 5A and B). Significantly, they further demonstrate that physical interactions between POP-1, HDA-1 and UNC-37 occur under physiological conditions.
UNC-37 and HDA-1 enhance POP-1-mediated transcriptional repression
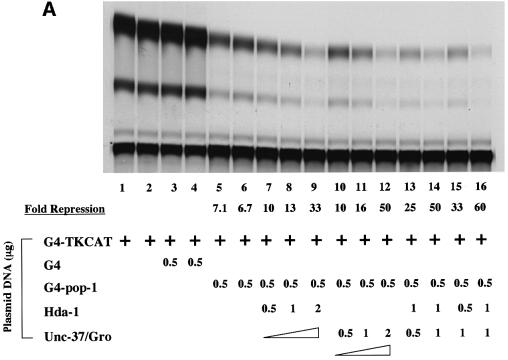
A predicted outcome of the interaction between POP-1 and these corepressors is an enhancement of repression mediated by POP-1. We addressed this issue using the transient transfection-based reporter gene assay described above (Figure 3). As shown in Figure 6A, G4POP-1 repressed the G4-TKCAT target gene by ∼7-fold (lanes 5 and 6). Interestingly, cotransfection of CMV-FLAG-HDA1 or CMV-HA-UNC-37 further increased G4POP-1-mediated repression in a dose-dependent manner (lanes 7–9 for HDA-1, 10-, 13- and 33-fold repression, and lanes 10–12 for UNC-37, 10-, 16- and 50-fold repression). This repression is dependent on the presence of POP-1, since HDA-1 and UNC-37, when expressed either alone or together, had very little effect on the activity of the G4TK promoter (Figure 6B). As shown in Figure 6A, the enhancement of POP-1-mediated repression by HDA-1 and UNC-37 is largely additive (compare lanes 13 and 14 with lane 8; lanes 15 and 16 with lane 11). These data are consistent with the findings obtained from the analysis in the C.elegans embryos (Figure 2), and lend further support to the hypothesis that HDA-1 and UNC-37 function as corepressors for POP-1 to repress end-1 promoter in the MS descendants.
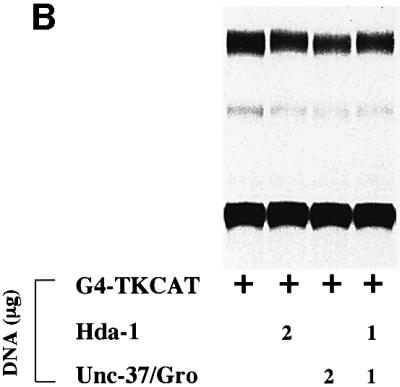
Fig. 6. HDA-1 and UNC-37 enhance transcriptional repression mediated by POP-1. (A) HDA-1 and UNC-37 enhance POP-1 repression. pG4 or pG4-POP1 was cotransfected with different combinations and amounts of pFLAG-HDA-1 or pHA-UNC-37 into HeLa cells together with the reporter plasmid pG4-TKCAT. The amount of transfected plasmid DNAs and the combination of plasmids used in each transfection are indicated. The extent of acetylation in various reactions was determined relative to that for pG4-TKCAT alone. The fold repression (average of three independent experiments) by G4POP-1 alone as well as in the presence of HDA-1 and UNC-37 singly or together is shown. The amount of G4pop-1 DNA transfected was such that POP-1-mediated repression was in the linear range under the assay conditions. (B) HDA-1 and UNC-37 do not affect G4TKCAT on their own. A representative CAT assay result is shown. The plasmids and the amount used in each transfection are indicated at the bottom.
Discussion
During C.elegans early embryogenesis, the birth of the two sister cells, E and MS, from the EMS cell sets the stage for the development of future endoderm and mesoderm tissues. Determination of the fates of the E and MS descendants involves positive regulatory signals that specify the E fate and negative regulators that inhibit the E fate in the MS cell. In this study we have provided molecular insights into both aspects of this decision making process. We find that in the E cell, the endoderm-promoting activity of CBP-1 lies in its ability to overcome the repressive activity of HDA-1 on activation of end-1, a critical initial endoderm-determining gene. Furthermore, we find that POP-1, HDA-1 and UNC-37 play essential roles in restricting end-1 promoter activity to E descendants. We provide multiple lines of evidence that POP-1 functions as a transcriptional repressor while HDA-1 and UNC-37 act as corepressors. Taken together, our findings suggest a model whereby a critical function of CBP-1 in endoderm differentiation is the activation of end-1 transcription, while a POP-1 repressor complex involving POP-1, HDA-1 and UNC-37 plays a pivotal role in repressing the end-1 promoter. Significantly, the collaboration of the two corepressors, HDA-1 and UNC-37, is essential to ensure maximal repression of inappropriate end-1 transcription.
CBP-1 in endoderm differentiation
Several models can be postulated to explain the requirement of CBP-1 in endoderm differentiation. CBP-1 may be necessary for the activation of early endoderm-determining genes or it may be required for activation of genes at later steps of the differentiation process. Our findings support the model in which CBP-1 is essential for transcriptional activation of the first zygotic endoderm-determining gene end-1 (Figure 1A) and its related gene end-3 (Maduro et al., 2001; M.Victor and Y.Shi, unpublished result). Interestingly and perhaps somewhat surprisingly, ectopically expressed END-1 is sufficient to induce gut differentiation in embryos lacking CBP-1 (Table I). Although we cannot rule out the possibility that undetectable levels of CBP-1 in the hs::end-1; cbp-1(RNAi) embryos may be sufficient to support transcription important for endoderm differentiation, our findings are highly suggestive that the critical function of CBP-1 in this process is the initiation of the zygotic endoderm transcription program, but CBP-1 may not be essential for subsequent transcriptional events.
Genetic and biochemical interactions among POP-1, HDA-1 and UNC-37
The model that maximal repression of the end-1 promoter requires recruitment of both HDA-1 and UNC-37 by POP-1 is supported by both genetic and biochemical studies. As we have shown in this study, inhibition of pop-1 or both hda-1 and unc-37 results in a fully penetrant ectopic activation of end-1::GFP, suggesting that these proteins are likely to function together to mediate repression. While ectopic activation of end-1::GFP is restricted to the MS descendants in pop-1(RNAi) embryos, it is more widespread in the hda-1/unc-37(RNAi) embryos, although ectopic expression appears to be similarly restricted to MS descendants in the early embryos. The fact that end-1::GFP is ectopically activated in non-MS descendants later in development upon inhibition of hda-1 and unc-37 suggests that HDA-1 and UNC-37 are likely to be recruited by other unknown repressors. These results suggest that HDA-1 and UNC-37 may play a general role in mediating repression of cell type-specific genes. Finally, we found no evidence that hda-1 or hda-1/cbp-1 double RNAi embryos make gut from the MS blastomeres. As transcriptional corepressors, it is likely that HDA-1 and UNC-37 are recruited by other unknown transcription factors, in addition to POP-1, and are thus involved in the regulation of genes that do not involve POP-1. We postulate that in the absence of HDA-1 and UNC-37, disregulation of these genes, which normally do not participate in suppression of the endodermal fate, may interfere with gut differentiation in the MS cells. Alternatively, although hs-end-1 can induce ectopic endodermal differentiation in multiple cell lineages, the unlikely possibility that the MS cell is not competent to give rise to endoderm unless POP-1 is absent cannot be ruled out at the present time.
HDA-1 and UNC-37, as corepressors for POP-1, are predicted to interact physically with POP-1 and to enhance repression mediated by POP-1. Indeed, both HDA-1 and UNC-37 interact with POP-1. In addition, over-expression of HDA-1 or UNC-37 enhances repression mediated by POP-1 in a transient transfection-based reporter gene assay, reinforcing the idea that these proteins are corepressors for POP-1. This is consistent with recent studies documenting biochemical interactions of TCF/LEF family members with histone deacetylase and Groucho (Chen et al., 1999; Billin et al., 2000). Significantly, our findings identified a molecular target, i.e. the end-1 promoter, whose repression requires the collaboration of these two corepressors. These results highlight the critical importance of concerted actions of multiple corepressors in restricting inappropriate cell type-specific gene transcription.
Mechanisms of repression by POP-1, HDA-1 and UNC-37
The mechanistic basis for recruitment of both HDA-1 and UNC-37 in order to achieve full repression of end-1::GFP is currently unclear. Studies in other organisms have shown that histone deacetylases contribute to the condensation of the local chromatin structure. Groucho, on the other hand, has also been shown to bind histones (Palaparti et al., 1997; Flores-Saaib and Courey, 2000) and perhaps functions analogously as the yeast corepressor TUP1 (Flores-Saaib and Courey, 2000). Interestingly, Groucho binds hypoacetylated histone H3 with the highest affinity (Flores-Saaib and Courey, 2000). Thus, it is conceivable that HDA-1 may be recruited by POP-1 to deacetylate histones, and the hypoacetylated histones can then be recognized by UNC-37. Together, these proteins may facilitate the formation of a higher order repressive chromatin structure, resulting in transcriptional repression of the target genes.
The idea that multiple protein complexes with distinct biochemical activities are recruited to promoters to ensure the formation of a repressive chromatin complex was recently suggested by studies of Ume6p-mediated repression in yeast. Ume6p, a sequence-specific DNA-binding protein, has been shown to recruit both yeast histone deacetylase Rpd3p and Isw2, a component of the Swi/Snf complex, to achieve optimal repression of meiotic genes (Goldmark et al., 2000). It is believed that Isw2 promotes formation of nuclease-inaccessible chromatin structure and changes the positioning of the nucleosome at the target promoter. At the same time, Rpd3p removes the acetyl group from histone tails, thereby further enhancing the repressed state of chromatin at the promoter region. Similar to the findings described in this report, mutation of either gene alone in yeast does not result in full repression of the target genes. This is because in the rpd3 single mutant, repression of the target genes still occurs due to properly positioned nucleosomes. On the other hand, although the nucleosomes are mis-positioned in the isw2 single mutant, deacetylation by Rpd3p can still keep the target genes repressed, albeit to a lesser extent. It will be interesting to determine whether UNC-37 provides a function similar to that of Isw2 in collaborating with histone deacetylase for maximal repression. One possibility is that UNC-37 may recruit an Isw2-like protein in C.elegans.
In summary, we have identified a molecular basis for the opposing biological activities of CBP-1 and HDA-1 by demonstrating that their actions converge on end-1, an initial endoderm-determining gene. Significantly, a critical function for CBP-1 in endoderm differentiation appears to be the activation of the first endoderm-determining gene end-1, which initiates the transcriptional cascade important for endoderm differentiation. In addition, we have shown that POP-1 functions as a transcriptional repressor that restricts the end-1 promoter activity to the E descendants. Our findings support a model whereby POP-1 represses transcription by recruiting the corepressors HDA-1 and UNC-37, and the recruitment of both corepressors is essential for maximal repression. These findings can be integrated into the context of E and MS cell fate determination by the following model. We propose that in the E cell, CBP-1 is required for activation of end-1 and is capable of overcoming the repressive effect of POP-1 and HDA-1 on the end-1 promoter, leading to its activation and subsequent endoderm differentiation. In the MS cell, a POP-1 repressor complex including POP-1, HDA-1 and UNC-37 identified in this study plays a dominant role in repressing end-1 and perhaps other E-specific promoter activities. One possible explanation is that CBP-1 simply cannot overcome the dominant activity of the POP-1 repressor complex, due to the abundance of POP-1 in the MS cell, as opposed to the lower level of POP-1 in the E cell as a result of Wnt activity (Lin et al., 1995, 1998; Meneghini et al., 1999; Rocheleau et al., 1999). A more interesting possibility is that CBP-1 is differentially regulated in E versus MS descendants. While CBP-1 is fully active in the E cell, the absence of the Wnt signaling pathway and/or the presence of an inhibitory activity may prevent CBP-1 from activating end-1 in the MS lineage.
Materials and methods
Strains and plasmids
Strain JR 553 (wIs28 {pJZ21 + [rol-6D (su1006)]}) in the C.elegans Bristol strain N2 genetic background was used to analyze end-1::GFP expression. PJZ21 is an end-1::NLS::GFP::lacZ fusion containing 1.7 kb of end-1 promoter sequence and the end-1 cDNA (lacking the last 30 amino acids of the END-1 ORF) cloned into pPD96.04 (kind gift from Andy Fire). JR663 (N2, wIs48) carry the end-1 cDNA under the control of a heat-inducible promoter (Zhu et al., 1998). Caenorhabditis elegans culturing, handling and manipulation were as described (Brenner, 1974).
The full length pop-1 was isolated by PCR using plasmid pRL159 (gift from Rueyling Lin) as a template with BamHI and XbaI cloning sites added at the 5′ and 3′ ends, respectively. It was subcloned in-frame into a pcDNA3 vector that carries the GAL4 DNA-binding domain, or a CMV promoter-based mammalian expression plasmid CMX. The ΔHMG POP-1 lacks the HMG box DNA-binding domain spanning amino acid residues 188 to 281, and the corresponding DNA fragment was also subcloned into CMX for expression in mammalian cells. The pend-1-Luc reporter was generated by subcloning 1.7 kb of the end-1 promoter sequence into the pGL2-Basic vector (Promega) which contains the firefly luciferase cDNA. hda-1 and unc-37 were also isolated by PCR using plasmids yk109d9 (hda-1) and D5 (unc-37) as templates (gifts from Yuji Kohara and David Miller, respectively) and subcloned into an HA tag-, FLAG tag- or MYC tag-containing CMV promoter-based expression vector, respectively. Ce-hda-3 contains the full-length hda-3 cDNA isolated from a mixed stage C.elegans cDNA library (kind gift from Bob Barstead). All plasmids were verified by sequencing.
Antibodies
The regions corresponding to amino acids 374–460 of HDA-1 and amino acids 370–460 of HDA-3 were amplified by PCR from the cDNA clones yk109d9 and Ce-hda-3, respectively. They were then subcloned into pGEX-4T2 (Pharmacia). Purified GST fusion proteins were used for the production and subsequent affinity purification of rabbit polyclonal antibodies by Santa Cruz Biotechnology. The generation of α-UNC-37 (gift from David Miller) and α-POP-1 antibodies (gift from Reuyling Lin) is described elsewhere (Lin et al., 1995; Pflugrad et al., 1997). Monoclonal antibodies used for the immunoprecipitation and western blotting experiments shown in Figure 5A–C were purchased from the following companies: α-MYC (Santa Cruz Biotechnology), α-HA (Covance/BabCO), α-FLAG (Sigma) and α-GAL4 (mono- and polyclonal, Santa Cruz Biotechnology).
RNA-mediated interference (RNAi)
dsRNA was prepared by simultaneous T3/T7 in vitro transcription using the RiboMAXTM system (Promega) according to the manufacturer’s instructions. PCR templates for in vitro transcription were derived from yk6f6 (cbp-1), yk109d9 (hda-1), Ce-had-3 (hda-3), D5 (unc-37) and GAL4-POP-1 (pop-1), respectively. RNA was injected into the body cavity of young adult hermaphrodites as described before (Fire et al., 1998; Shi and Mello, 1998) at concentrations ranging from 0.8 to 1.0 µg/µl. The same final concentrations were used in double injections as for the respective single injections. Animals were allowed to recover overnight, transferred to a new plate and allowed to lay eggs for 3 h. They were assayed for embryonic lethality the next day, and only experiments resulting in >95% embryonic lethality were scored. Mother hermaphrodites were cut open and the embryos were mounted on agar pads for examination by light and fluorescent microscopy (24 h after injection). For the end-1::GFP analyses shown in Figures 1 and 2, we observed a similar increase in the number of embryos with ectopic GFP signals when end-1::GFP expression was analyzed 48 h (instead of 24 h) after RNA injection. Unc-37(RNAi) embryo gave no ectopic expression (n = 11). Of the hda-1(RNAi) (n = 13) and hda-1/unc-37(RNAi) (n = 18) embryos, 38 and 72% were found to have ectopic GFP signals, respectively. Therefore, it is unlikely that incomplete activation of the end-1 promoter activity in MS descendants analyzed 24 h after injection was due to the persistence of undetectable levels of HDA-1 protein.
Microscopy
Methods for mounting and viewing C.elegans embryos using Nomarski microscopy have been described previously (Sulston et al., 1983). The identity of differentiated cell types in experimental embryos was assigned based on morphological criteria using the light microscope. GFP signals were scored using fluorescence microscopy. Light and fluorescent light micrographs were taken electronically using a CCD black/white video camera. Photographs were processed using Adobe Photoshop™.
Heat shock experiments
Animals from strain JR 663 were injected with cbp-1 dsRNA, allowed to recover for 12 h and then transferred to a fresh plate. They were allowed to lay eggs for 4 h and these embryos were subjected to a 33°C heat shock for 30 min. Eggs were allowed to develop overnight at 15°C and scored for the presence of birefringent gut granules using polarized light as described previously (Laufer et al., 1980). Among all the controls described in the text, we also found that heat shock did not lead to an increase in gut granules in N2 wt worms treated with cbp-1 dsRNA (data not shown).
Preparation of worm embryonic extracts andco-immunoprecipitation
Total protein extracts from worm embryos were prepared by sonication of the embryos in co-IP buffer (50 mM Tris pH 7.9; 100 mM KCl; 20% glycerol; 3 mM MgCl2; 0.1% NP-40; 1 mM DTT and protease inhibitors). Four hundred micrograms of protein extracts were incubated either with α-POP-1, α-HDA-1 or α-HA antibodies (Covance/BabCO). In order to avoid cross reaction with the IgG heavy chains during the western blotting procedure, all antibodies used for immunoprecipitation were first coupled to protein G beads. After three washes in co-IP buffer, the immunoprecipitates were resolved in a 10% SDS–polyacrylamide gel, transferred to a nitrocellulose membrane and probed with antibodies. Immuno-detection of the proteins was performed as previously described (Johnstone et al., 1996).
Transfection, CAT and luciferase assays
C33A and HeLa cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and antibiotics. Cells were transfected by the calcium phosphate precipitation method as previously described (Shi et al., 1991). The total amount of DNA was normalized with plasmid PSP72 for each transfection. Cells were harvested 48 h after addition of the precipitates. Transfections were carried out with at least two independent DNA preparations and repeated at least three times. For chloramphenicol acetyltransferase (CAT) assays, whole-cell extracts were prepared as described (Shi et al., 1991). TLC plates were analyzed on a Molecular Dynamics ‘Storm860TM’ PhosphorImager using ImageQuantTM 1.1 software. The amount of cell extract used was such that the CAT activity was within the linear range. For luciferase assays, HeLa cells were harvested 36 h after and luciferase activity was determined as described (See et al., 2001). The luciferase activities were normalized on the basis of β-galactosidase activity of cotransfected CMV β-gal vector.
Acknowledgments
Acknowledgements
We thank Keith Blackwell, Azad Bonni and Grace Gill for critical reading of the manuscript and members of the Shi laboratory for helpful discussion. We gratefully acknowledge Rueyling Lin and David Miller for POP-1 and UNC-37 antibodies and plasmids, Yuji Kohara for the many EST clones used in the studies and Bob Barstead for a cDNA library. Dominica Calvo was supported by a fellowship from the Spanish government and from the Taplin Funds for Discovery. M.V. is supported by a Taplin postdoctoral fellowship. This work was supported by a grant from the NIH (GM58012) to Y.S.
References
- Bannister A.J. and Kouzarides,T. (1996) The CBP co-activator is a histone acetyltransferase. Nature, 384, 641–643. [DOI] [PubMed] [Google Scholar]
- Barker N., Morin,P.J. and Clevers,H. (2000) The Yin-Yang of TCF/β-catenin signaling. Adv. Cancer Res., 77, 1–24. [DOI] [PubMed] [Google Scholar]
- Billin A.N., Thirlwell,H. and Ayer,D.E. (2000) β-catenin-histone deacetylase interactions regulate the transition of LEF1 from a transcriptional repressor to an activator. Mol. Cell. Biol., 20, 6882–6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman B., Eaton,B.A. and Priess,J.R. (1992) skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C.elegans embryo. Cell, 68, 1061–1075. [DOI] [PubMed] [Google Scholar]
- Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics, 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G. and Courey,A.J. (2000) Groucho/TLE family proteins and transcriptional repression. Gene, 249, 1–16. [DOI] [PubMed] [Google Scholar]
- Chen G., Fernandez,J., Mische,S. and Courey,A.J. (1999) A functional interaction between the histone deacetylase Rpd3 and the corepressor Groucho in Drosophila development. Genes. Dev., 13, 2218–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu,S., Montgomery,M.K., Kostas,S., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Fisher A.L. and Caudy,M. (1998) Groucho proteins: transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes Dev., 12, 1931–1940. [DOI] [PubMed] [Google Scholar]
- Flores-Saaib R.D. and Courey,A.J. (2000) Analysis of Groucho-histone interactions suggests mechanistic similarities between Groucho- and Tup-mediated repression. Nucleic Acids Res., 28, 4189–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmark J.P., Fazzio,T.G., Estep,P.W., Church,G.M. and Tsukiyama,T. (2000) The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell, 103, 423–433. [DOI] [PubMed] [Google Scholar]
- Johnstone R.W. (1996) A novel repressor, par-4, modulates transcription and growth suppression functions of the Wilms’ tumor suppressor WT. Mol. Cell. Biol., 16, 6945–6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbach L., Horner,M., Rothman,J.H. and Mango,S.E. (2000) The TBP-like factor CeTLF is required to activate RNA polymerase II transcription during C.elegans embryogenesis. Mol. Cell, 6, 705–713. [DOI] [PubMed] [Google Scholar]
- Korswagen H.C., Herman,M.A. and Clevers,H.C. (2000) Distinct β-catenins mediate adhesion and signalling functions in C.elegans. Nature, 406, 527–532. [DOI] [PubMed] [Google Scholar]
- Laufer J.S., Bazzicalupo,P. and Wood,W.B. (1980) Segregation of developmental potential in early embryos of Caenorhabditis elegans. Cell, 19, 569–577. [DOI] [PubMed] [Google Scholar]
- Lin R., Hill,R.J. and Priess,J.R. (1998) POP-1 and anterior-posterior fate decisions in C.elegans embryos. Cell, 92, 229–239. [DOI] [PubMed] [Google Scholar]
- Lin R., Thompson,S. and Priess,J.R. (1995) pop-1 encodes an HMG box protein required for the specification of a mesoderm precursor in early C.elegans embryos. Cell, 83, 599–609. [DOI] [PubMed] [Google Scholar]
- Maduro M.F., Meneghini,M.D., Bowerman,B., Broitman-Maduro,G. and Rothman,J.H. (2001) Restriction of mesoendoderm to a single blastomere by the combined action of SKN-1 and a GSK-3β homolog is mediated by MED-1 and -2 in C.elegans. Mol. Cell, 7, 475–485. [DOI] [PubMed] [Google Scholar]
- Meneghini M.D., Ishitani,T., Carter,J.C., Hisamoto,N., Ninomiya-Tsuji,J., Thorpe,C.J., Hamill,D.R., Matsumoto,K. and Bowerman,B. (1999) MAP kinase and Wnt pathways converge to downregulate an HMG-domain repressor in Caenorhabditis elegans. Nature, 399, 793–797. [DOI] [PubMed] [Google Scholar]
- Newman-Smith E.D. and Rothman,J.H. (1998) The maternal-to-zygotic transition in embryonic patterning of Caenorhabditis elegans. Curr. Opin. Genet. Dev., 8, 472–480. [DOI] [PubMed] [Google Scholar]
- Ogryzko V., Schiltz,R.L., Russanova,V., Howard,B.H. and Nakatani,Y. (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell, 87, 953–959. [DOI] [PubMed] [Google Scholar]
- Palaparti A., Baratz,A. and Stifani,S. (1997) The Groucho/transducin-like enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J. Biol. Chem., 272, 26604–26610. [DOI] [PubMed] [Google Scholar]
- Pflugrad A., Meir,J.Y., Barnes,T.M. and Miller,D.M.,III (1997) The Groucho-like transcription factor UNC-37 functions with the neural specificity gene unc-4 to govern motor neuron identity in C.elegans. Development, 124, 1699–1709. [DOI] [PubMed] [Google Scholar]
- Priess J.R. (1994) Establishment of initial asymmetry in early Caenorhabditis elegans embryos. Curr. Opin. Genet. Dev., 4, 563–568. [DOI] [PubMed] [Google Scholar]
- Rocheleau C., Downs,W.D., Lin,R.L., Wittman,C., Bei,Y.X., Ali,M., Priess,J.R. and Mello,C.C. (1997) Wnt signaling and an APC related gene specify endoderm in early C.elegans. Cell, 90, 707–716. [DOI] [PubMed] [Google Scholar]
- Rocheleau C.E., Yasuda,J., Shin,T.H., Lin,R., Sawa,H., Okano,H., Priess,J.R., Davis,R.J. and Mello,C.C. (1999) WRM-1 activates the LIT-1 protein kinase to transduce anterior/posterior polarity signals in C.elegans. Cell, 97, 717–726. [DOI] [PubMed] [Google Scholar]
- Roose J. and Clevers,H. (1999) TCF transcription factors: molecular switches in carcinogenesis. Biochim. Biophys. Acta, 1424, M23–37. [DOI] [PubMed] [Google Scholar]
- Roose J., Molenaar,M., Peterson,J., Hurenkamp,J., Brantjes,H., Moerer,P., van de Wetering,M., Destree,O. and Clevers,H. (1998) The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature, 395, 608–612. [DOI] [PubMed] [Google Scholar]
- Schnabel R. and Priess,J.R. (1997) Specification of cell fates in the early embryo. In Riddle,D.L., Blumenthal,T., Meyer,B.J. and Priess,J.R. (eds), C.elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- See R.H., Calvo,D., Shi,Y.J., Kawa,H., Luke,M.P.-S., Yuan,Z.M. and Shi,Y. (2001) Stimulation of p300-mediated transcription by the kinase MEKK1. J. Biol. Chem., 276, 16310–16317. [DOI] [PubMed] [Google Scholar]
- Shi Y. and Mello,C. (1998) A CBP/p300 homolog specifies multiple differentiation pathways in Caenorhabditis elegans. Genes Dev., 12, 943–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Seto,E., Chang,L.S. and Shenk,T. (1991) Transcriptional repression by YY1, a human GLI-Kruppel related protein, and relief of repression by adenovirus E1A protein. Cell, 67, 377–388. [DOI] [PubMed] [Google Scholar]
- Sulston J.E., Schierenberg,E., White,J.G. and Thomson,J.N. (1983) The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol., 100, 64–119. [DOI] [PubMed] [Google Scholar]
- Taunton J., Hassig,C.A. and Schreiber,S.L. (1996) A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science, 272, 408–411. [DOI] [PubMed] [Google Scholar]
- Thorpe C.J., Schlesinger,A., Carter,J.C. and Bowerman,B. (1997) Wnt signaling polarizes an early C.elegans blastomere to distinguish endoderm and mesoderm. Cell, 90, 695–705. [DOI] [PubMed] [Google Scholar]
- Walker A.K., See,R.H., Batchelder,C., Kophengnavong,T., Gronniger,J.T., Shi,Y. and Blackwell,T.K. (2000) A conserved transcription motif suggesting functional parallels between C.elegans SKN-1 and Cap’n’Collar-related proteins. J. Biol. Chem., 275, 22166–22171. [DOI] [PubMed] [Google Scholar]
- Zhu J., Fukushige,T., McGhee,J.D. and Rothman,J.H. (1998) Reprogramming of early embryonic blastomeres into endodermal progenitors by a Caenorhabditis elegans GATA factor. Genes Dev., 12, 3809–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.W., Hill,R.J., Heid,P.J., Fukuyama,M., Sugimoto,S., Priess,J.R. and Rothman,J.H. (1997) end-1 encodes an apparent GATA factor that specifies the endoderm precursor in Caenorhabditis elegans embryos. Genes Dev., 11, 2883–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]