Abstract
The circadian timing system in mammals is composed of a master pacemaker in the suprachiasmatic nucleus (SCN) of the hypothalamus and slave clocks in most peripheral cell types. The phase of peripheral clocks can be completely uncoupled from the SCN pacemaker by restricted feeding. Thus, feeding time, while not affecting the phase of the SCN pacemaker, is a dominant Zeitgeber for peripheral circadian oscillators. Here we show that the phase resetting in peripheral clocks of nocturnal mice is slow when feeding time is changed from night to day and rapid when switched back from day to night. Unexpectedly, the inertia in daytime feeding-induced phase resetting of circadian gene expression in liver and kidney is not an intrinsic property of peripheral oscillators, but is caused by glucocorticoid signaling. Thus, glucocorticoid hormones inhibit the uncoupling of peripheral and central circadian oscillators by altered feeding time.
Keywords: adrenalectomy/circadian gene expression/clock/corticosterone/GR knockout
Introduction
In mammals, most physiology and behavior is subject to daily oscillations that are driven by an endogenous circadian timing system. Current evidence suggests that this system has a hierarchical structure: a master pacemaker located in the suprachiasmatic nucleus (SCN) of the hypothalamus synchronizes slave oscillators in most tissues (for reviews see Brown and Schibler, 1999; Reppert and Weaver, 2001; Ripperger and Schibler, 2001). As indicated by its name, the circadian clock can measure time only approximately and therefore has to be reset every day by external time cues (Zeitgebers). Light, the major Zeitgeber for the central pacemaker, entrains the phase of oscillations in SCN neurons via the retinohypothalamic tract, and glutamate appears to be the major neurotransmitter in this process (Ding et al., 1994; Shirakawa and Moore, 1994; Ebling, 1996). Light pulses delivered to animals kept in constant darkness during the subjective night not only elicit phase shifts, but also induce a surge of immediate early gene expression in SCN neurons. The products of these immediate early genes include the circadian clock components PER1 and PER2, and several transcription factors, such as c-Fos, Jun-B and EGR3 (Albrecht et al., 1997; Shearman et al., 1997; Shigeyoshi et al., 1997; Morris et al., 1998). It is likely that some of these regulatory proteins participate in the light-induced phase shifting.
Seven genes essential for mammalian circadian clock function have been identified. These include the two period isoforms mPer1 and mPer2 (Shearman et al., 1997; Sun et al., 1997; Tei et al., 1997; Zheng et al., 1999), the two cryptochrome isoforms mCry1 and mCry2 (Kume et al., 1999; van der Horst et al., 1999), two genes, Clock and Bmal1, encoding PAS helix–loop–helix transcription factors (King et al., 1997; Bunger et al., 2000) and Tau, encoding casein kinase 1ε (Lowrey et al., 2000). The products of these genes have been assembled into a model of a molecular oscillator, which is based on interlocked feedback loops in gene expression (Shearman et al., 2000). In this model, the two cryptochromes repress transcription of mCry and mPer genes via interference with the two PAS helix–loop–helix transcription factors Clock and Bmal1. The cyclic accumulation of mPER2 then acts to generate a rhythmic expression of Bmal1, whose oscillation may reinforce the negative feedback loop of mCry expression. Post-transcriptional regulation also appears to play an important role in rhythm generation (Keesler et al., 2000; Lowrey et al., 2000; Toh et al., 2001). Clock-controlled genes such as Arginine Vasopressin or Dbp, whose products display cyclic accumulation but are not required for clock function, can be regulated by the same interlocked feedback loops of gene expression that drive the rhythmic expression of bona fide pacemaker genes (Jin et al., 1999; Ripperger et al., 2000).
Circadian gene expression of clock and clock-controlled genes is not only observed in the SCN, but also in most peripheral tissues. Moreover, circadian gene expression persists in organ explants for several consecutive days and can be induced in immortalized rat and mouse tissue culture cells (Balsalobre et al., 1998; Yamazaki et al., 2000; Yagita et al., 2001). Hence, it appears that peripheral cells contain bona fide circadian timekeepers that can generate circadian oscillations during several consecutive days. The phase angle relationship between different mRNA accumulation cycles is very similar in SCN neurons and peripheral cell types, and mutations in essential clock genes affect these oscillators in a similar way. Therefore, central and peripheral oscillators are likely to function according to similar molecular principles (Balsalobre et al., 1998; Yagita et al., 2001).
In spite of their resemblance in molecular make-up, central and peripheral circadian clocks differ from each other in that only the former are self-sustained. Hence, it appears that the SCN must periodically re-entrain peripheral oscillators in order to prevent the dampening of circadian gene expression in organs such as liver, kidney, heart and pancreas (Yamazaki et al., 2000). The mechanisms by which the SCN accomplishes this task are not yet understood, but they are likely to be complex. In fact, ligands of the glucocorticoid receptor (Balsalobre et al., 2000a) and two other nuclear receptors, RARα and RXRα (McNamara et al., 2001), calcium ionophores and chemicals that activate either protein kinase A, protein kinase C and/or MAP kinases (Akashi and Nishida, 2000; Balsalobre et al., 2000b; Yagita and Okamura, 2000) can all trigger circadian gene expression in tissue culture cells. Moreover, some of these signals also provoke phase shifts in peripheral tissues of intact animals (e.g. Balsalobre et al., 2000a; McNamara et al., 2001). It is thus likely that multiple signaling pathways are used for phase resetting of circadian peripheral oscillators. Balsalobre et al. (2000a) have recently demonstrated that dexamethasone, a glucocorticoid receptor agonist, acts as a strong phase-shifting agent for peripheral oscillators in the mouse. However, since the phase of oscillators is the same in hepatocytes that do or do not express glucocorticoid receptors, glucocorticoid hormones cannot be the only Zeitgeber signal for peripheral clocks.
Feeding time has recently been shown to be a dominant Zeitgeber for rhythmic gene expression in tissues such as liver, pancreas, kidney and heart (Damiola et al., 2000; Stokkan et al., 2001). Thus, if nocturnal animals like mice or rats are fed exclusively during the day for extended periods, the phase of circadian gene expression in several examined organs becomes completely inverted. Interest ingly, however, restricted feeding has no influence on the phase of rhythmic gene expression in the SCN (Damiola et al., 2000; Stokkan et al., 2001). These observations raise the possibility that the SCN entrains peripheral clocks indirectly by imposing activity and resting phases, and thus feeding behavior. According to this model, food metabolites or signals elicited by feeding behavior or food processing would be the principal timing cues for peripheral oscillators.
Upon switching nocturnal animals to daytime feeding, it takes several days until the reversed phase is established (Damiola et al., 2000). This relatively slow phase adaptation is reminiscent of the time-consuming photic entrainment of the SCN master clock, which manifests itself in jet lag after long-distance east- or westbound trips. Here we demonstrate that the sluggish phase resetting of peripheral clocks by altered feeding time is not caused by an intrinsic inertia of circadian oscillators to adapt to the new Zeitgeber time, but by active signaling pathways that counteract rapid phase resetting. Thus, in the absence of glucocorticoid hormones or the glucocorticoid hormone receptor, feeding time can induce large phase shifts in peripheral tissues such as the liver, thereby inverting the phase of their oscillators within a few days.
Results
Daytime feeding phase-shifts peripheral oscillators faster in adrenalectomized compared with sham-operated mice
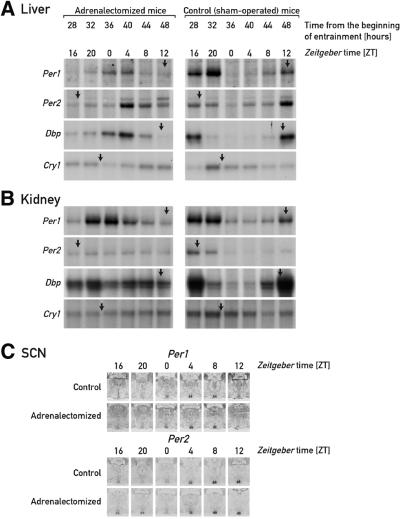
As shown previously, food is a dominant Zeitgeber for circadian oscillators in several mouse tissues, including liver, pancreas, kidney and heart. Mice, which are nocturnal animals, ingest ∼80% of their daily food intake during the night (N.Preitner, unpublished results). As a consequence, restricting feeding time to the dark period does not significantly alter the phase angle of cyclic gene expression (Damiola et al., 2000). However, when food is offered only during the light phase during several consecutive days, the phase of circadian gene expression of clock and clock-controlled genes becomes completely inverted after 7–10 days (Damiola et al., 2000). Among several tissues examined, this phase resetting occurred most rapidly in the liver. We wished to determine whether glucocorticoid hormones, which efficiently induce circadian gene expression in cultured cells and elicit potent phase shifts in peripheral tissues of intact animals, participate in the food-induced resetting of peripheral oscillators. To this end, the kinetics of phase changes in the rhythmic expression of the clock genes mPer1, mPer2 and mCry1, and the clock-controlled gene Dbp, were compared in livers and kidneys of normal (sham-operated) and adrenalectomized mice. As shown in Figure 1A and B, the accumulation profiles of the mRNA encoded by these genes are strikingly different during the second day after initiation of daytime feeding. In sham-operated animals (right panels), the peak and trough levels of some mRNAs are offset by only ∼4 h when compared with mice fed ad libitum or during the night (Figure 2A and C). For example, in control animals, Dbp mRNA, whose circadian transcription is regulated directly by components of the molecular oscillator (Ripperger et al., 2000), reaches peak and trough levels in liver and kidney at about Zeitgeber times ZT14 and ZT2, respectively, during the second day of daytime feeding (ZT0 = time when the lights are switched on). However, much larger phase shifts are observed in tissues of adrenalectomized mice. Thus, during the second day after restricted feeding, Dbp mRNA peak levels are observed in liver between ZT0 and ZT4, which is significantly closer to the time at which daytime feeding-induced phase resetting is completed (Figure 2B and Damiola et al., 2000).
Fig. 1. Accumulation of circadian transcripts in adrenalectomized and sham-operated mice at the onset of daytime feeding. Adrenalectomized mice and sham-operated mice, kept under a 12 h light–dark regimen (lights on ZT0), were switched from ad libitum to daytime feeding. On day D, food was removed at ZT12 and provided exclusively during the light phase of the following days. During the second day (D+1) of restricted feeding, animals were killed at 4 h intervals and the livers, kidneys and brains were collected for monitoring the accumulation of mRNAs specified by the three clock genes mPer1, mPer2 and mCry1, and the clock output gene Dbp. Relative transcript levels in liver (A) and kidney (B) were determined by RNase protection assays using radiolabeled antisense RNA probes. Tbp (Tatabox-binding protein) mRNAs served as a control for a transcript whose accumulation does not oscillate during the day (not shown). The approximate times at which peak levels of mRNA accumulation are observed in animals fed ad libitum or during the night are ZT12 (mPer1), ZT17 (mPer2), ZT11 (Dbp) and ZT22 (mCry1) (see Damiola et al., 2000 and Figure 2C). These times are indicated by arrows on top of the respective panels. The accumulation of mPer1 and mPer2 mRNA in the SCN was assessed by in situ hybridization to coronal brain sections, using 35S-labeled antisense RNA probes (C). Only the hypothalamus regions containing the SCN (small pairwise structures at the base of the hypothalamus) are depicted. At ZT20, the signal obtained for mPer2 mRNA is somewhat higher in the adrenalectomized animal compared with sham-operated animals, but this difference does not significantly change the phase of mPer2 mRNA accumulation.
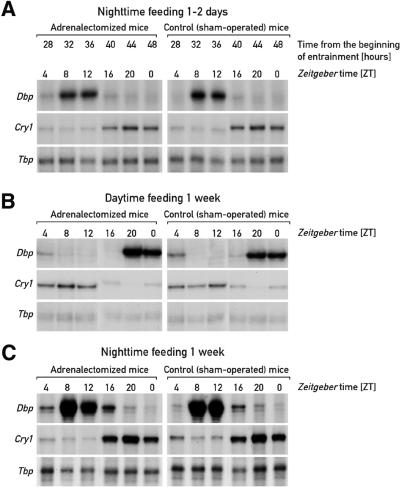
Fig. 2. Hepatic accumulation of mCry1 and Dbp transcripts in night-time- and daytime-fed adrenalectomized and sham-operated mice. Adrenalectomized mice and sham-operated mice fed ad libitum were kept under a 12 h light–dark regimen (lights on ZT0) before they were switched to night-time feeding [(A) and (C)] or daytime feeding (B). One day (A) or 1 week [(B) and (C)] after restricted feeding had been initiated, animals were killed at 4 h intervals and the livers were collected for monitoring the accumulation of Dbp and mCry1 transcripts. Tbp mRNA was included as a transcript whose accumulation is neither circadian- nor feeding time-dependent.
In kidney, phase reversal by daytime feeding takes somewhat longer than in liver (Damiola et al., 2000). Thus, as we have observed previously for the transition from ad libitum to daytime feeding, the expression of some genes (e.g. Dbp, mPer2 and mCry1) is not yet rhythmic in kidney during the second day of daytime feeding. Nevertheless, mPer1 mRNA, which in kidney is the sole examined transcript with a clearly cyclic accumulation during the second day of restricted feeding, already follows a profile close to that observed after complete phase reversal.
In contrast to the observations made in liver and kidney, daytime feeding does not affect circadian gene expression in the SCN (Damiola et al., 2000; Stokkan et al., 2001). As shown by the in situ hybridization experiments presented in Figure 1C, this holds true for rhythmic mPer1 and mPer2 mRNA accumulation in both sham-operated and adrenalectomized mice. Hence, as reported in previous studies (Balsalobre et al., 2000a; Damiola et al., 2000; Stokkan et al., 2001), neither feeding time nor glucocorticoid signaling appear to influence the phase of the central molecular pacemaker.
Similar experiments to those presented in Figure 1 were also performed with mice fed exclusively during the night, or for extended time periods during the day. The experiments presented in Figure 2 indicate that glucocorticoid signaling does not influence the phase of circadian gene expression in animals subjected to night-time feeding for either 1–2 days (Figure 2A) or 1 week (Figure 2C). This is in keeping with the previously published finding that the phases of circadian gene expression are indistinguishable in animals fed ad libitum or exclusively during the night (Damiola et al., 2000). We also noticed that after extended periods of daytime feeding, the steady-state phase of cyclic mRNA accumulation is the same in the presence or absence of glucocorticoid hormone signaling (Figure 2B).
We draw two major conclusions from the results presented thus far. First, glucocorticoid signaling strongly slows down—but does not prevent—the adaptation of circadian gene expression to a feeding schedule that is in conflict with the normal activity phase of the animal. Secondly, glucocorticoids are not required for the phase setting of circadian oscillators in liver and kidney under conditions in which food is available throughout the 24 h day (see also Damiola et al., 2000) or during the normal activity phase.
Daytime feeding changes the profile of daily glucocorticoid secretion
In animals fed ad libitum, glucocorticoid hormones are secreted by the adrenal gland in a circadian and episodic fashion (Carrillo et al., 1980; Tronche et al., 1998). The episodes, consisting of short bouts of hormone secretion (Shiraishi et al., 1984), are not synchronized between animals. The SCN pacemaker is believed to control the daily fluctuations of corticosterone levels via the hypothalamus–pituitary gland–adrenal axis and additional routes (Lejeune-Lenain et al., 1987). In both diurnal and nocturnal mammals, serum glucocorticoids reach peak levels around or just before the onset of the activity phase. Given the strong inhibition of food-induced phase resetting by glucocorticoid signaling, we wished to examine whether daytime feeding affects the temporal pattern of serum glucorticoids. To this end we monitored the serum concentrations of corticosterones at 4 h intervals around the clock in mice fed ad libitum (Figure 3A), in mice at the onset of daytime feeding (Figure 3C) and in mice exposed for 1 week to daytime feeding (Figure 3B). After 1 week, the inverted phase of circadian gene expression has been fully established in the peripheral organs examined. Because of the episodic nature of corticosterone secretion (Carrillo et al., 1980), the values obtained for the 5–7 individuals killed at a given time point vary greatly at times when these hormones reach high circadian levels, but are low in all animals at nadir times of glucocorticoid secretion. As a consequence, the circadian phase of glucocorticoid secretion is characterized best by the trough levels, which are highly reproducible. In accordance with published data (Cheifetz, 1971), corticosterone plasma levels are lowest at the dark–light transition (ZT0) and highest at the light–dark transition (ZT12) in animals kept under a 12 h light–dark regimen (Figure 3A). Interestingly the temporal glucocorticoid profile is bimodal in mice stably entrained to daytime feeding, with a peak of short duration between ZT20 and ZT4 (zenith at ZT0) and a broader peak between ZT4 and ZT20 (zenith between ZT8 and ZT12) (Figure 3B). The average levels of both peaks are in the range of 200 ng/ml, which resembles the average peak level measured for animals fed ad libitum.
Fig. 3. Diurnal corticosterone levels in animals fed ad libitum or during the day. Daytime-dependent corticosterone serum levels were determined in mice fed ad libitum (A), or in mice fed exclusively during the day for 7 days (B) or 1–2 days (C). For each time point, the blood was collected from five [(B) and (C)] to seven (A) individuals. As indicated in the text, due to the episodic nature of glucocorticoid secretion, the individual variability is small during nadir times but large during zenith times. In the steady-state profiles shown in (A) and (B), the values obtained for ZT16 and ZT20 are repeated to visualize the bimodal distributions.
Based on these results, we conclude that daytime feeding establishes a bimodal temporal pattern of cortico sterone secretion, of which only the first peak appears to depend upon feeding time. The second oscillation is akin to the glucocorticoid profile recorded in animals fed ad libitum in that it reaches maximal values just before the light–dark transition. This second wave of corticosterone secretion may be controlled primarily by the SCN pacemaker, which does not change its phase in daytime-fed animals. At the onset of daytime feeding, corticosterone levels can attain particularly high levels during the dark phase (Figure 3C). Conceivably, these elevated glucocorticoid concentrations could be responsible for slowing down daytime feeding-induced resetting of peripheral oscillators (see Discussion).
The inhibition of food-entrained phase resetting by glucocorticoids requires the glucocorticoid receptor and is tissue autonomous
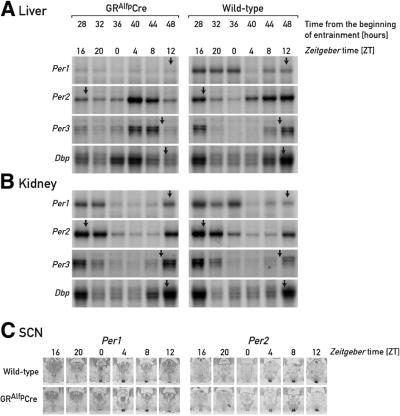
The observed inhibitory effect of glucocorticoid signaling on feeding time-induced phase resetting could be accomplished via direct cell-autonomous mechanisms or indirect mechanisms involving systemic signals whose production and/or release into the blood is controlled by glucocorticoid hormones. To discriminate between these two scenarios, we examined phase resetting of circadian gene expression in livers and kidneys of wild-type mice and GRAlfpCre mice, carrying glucocorticoid receptor null alleles exclusively in the hepatocytes (Kellendonk et al., 2000). As shown in Figure 4A (left panel), on the second day of daytime feeding the phases of circadian expression of Dbp, mPer1, mPer2 and mPer3 in the livers of GRAlfpCre mice are already shifted to values close to those observed 1 week after the animals had been subjected to the new feeding regimen. In contrast, the cyclic gene expression of these genes is only moderately advanced in the kidneys of GRAlfpCre mice (Figure 4B, left panel) and in kidneys or livers of wild-type mice (Figure 4A and B, right panels).
Fig. 4. Accumulation of circadian transcripts in GRAlfpCre and wild-type mice at the onset of daytime feeding. GRAlfpCre and wild-type mice were switched to daytime feeding as explained in the legend to Figure 1. The hepatocytes of GRAlfpCre mice harbor a glucocorticoid receptor null allele and thus cannot respond to corticosterone signaling. The relative mRNA levels were determined by RNase protection assays for liver (A) and kidney (B), and by in situ hybridization for the SCN (C). For comparison, the approximate peak times of mRNA accumulation in animals fed ad libitum or during the night are indicated by arrows (see Figure 1).
The expression of mPer1 behaves somewhat differently when compared with that of the other circadian genes examined. Under normal circumstances, it is high during the dark phase of the first day and low throughout the period of daytime feeding examined in the livers of GRAlfpCre mice. Relatively low levels of mPer1 mRNA have also been observed in the livers of adrenalectomized animals (Figure 1A). As outlined in the previous section, corticosterone levels are particularly high during the dark phase at the onset of daytime feeding, when mPer1 mRNA accumulates to high cellular concentrations. This correlation could indicate that mPer1 is directly regulated by glucocorticoid hormones (see Balsalobre et al., 2000a and Discussion). We noticed, however, that in kidneys of adrenalectomized animals, mPer1 expression is not significantly diminished when compared with sham-operated animals (Figure 1B). Thus, in contrast to the observations made with liver, in kidney glucocorticoid signaling does not appear to be required for high mPer1 mRNA expression.
We also compared the circadian mPer1 and mPer2 mRNA accumulation profiles in the SCN of GRAlfpCre and wild-type mice on the second day of daytime feeding (Figure 4C). As expected from the data presented in Figure 1, rhythmic gene expression is not significantly altered in the SCNs of mice subjected to restricted feeding, in spite of the high corticosterone levels measured during the dark phase at the onset of daytime feeding. In fact, neither glucocorticoid signaling nor restricted feeding causes phase shifts in circadian gene expression in SCN neurons (Balsalobre et al., 2000a; Damiola et al., 2000; Stokkan et al., 2001).
The switch from daytime to night-time feeding results in a rapid phase resetting of circadian gene expression in liver and kidney
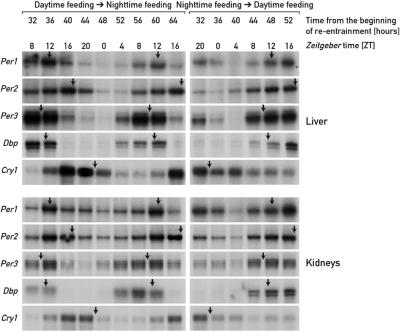
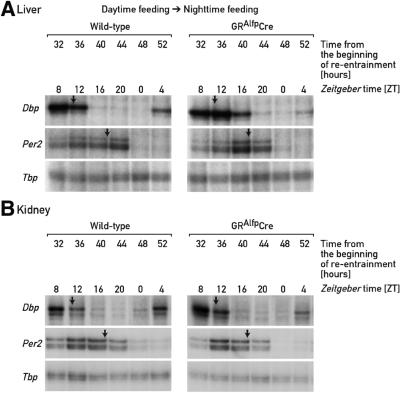
For nocturnal animals, daytime feeding is in conflict with their activity phase, and although feeding time is a dominant Zeitgeber for circadian clocks in several peripheral tissues, it takes several days to adapt the phase of these oscillators to the unusual feeding schedule. In the results above, we demonstrated that the inertia in this phase resetting is not an intrinsic property of peripheral oscillators, but is caused principally by glucocorticoid signaling. We wished to determine whether the phase resetting in peripheral organs proceeds with different kinetics when mice are switched from an abnormal to a normal feeding schedule than when they are switched in the reverse direction. To this end, we subjected mice to 8 days of daytime feeding and then switched them to night-time feeding. Circadian gene expression was recorded at 4 h intervals between 32 and 64 h after the switch from daytime to night-time feeding. The data depicted in Figure 5 show that, in liver, the night-time entrained phase of circadian gene expression is already approached 2–3 days after changing the feeding regimen. For example, on the second day after switching from daytime to night-time feeding, Dbp mRNA reaches zenith levels between ZT8 and ZT12, similar to what had been observed in animals fed ad libitum or during the night. The phase changes proceed with slightly slower kinetics in kidney, in which the phases of several mRNA accumulation profiles lag behind those monitored in liver by ∼2 h. We have previously noticed that circadian kidney gene expression also adapts somewhat more slowly to daytime feeding, at least for certain examined genes.
Fig. 5. Accumulation of circadian transcripts in mice switched from daytime to night-time feeding or vice versa. Mice kept under a 12 h LD regimen were entrained to daytime feeding or night-time feeding for 8 days, and then switched to night-time or daytime feeding, respectively. At the times after feeding time reversal (re-entrainment) indicated, liver and kidney RNAs were subjected to RNase protection assays with the probes indicated on the left-hand side of the panels. Tbp mRNA was included as a transcript whose accumulation is neither circadian- nor feeding time-dependent (not shown). For comparison, the approximate peak times of mRNA accumulation in animals fed ad libitum or during the night are indicated by arrows.
In control experiments, we have monitored circadian gene expression in mice switched from restricted night-time feeding to daytime feeding 2–3 days after reversal of the feeding regimen. The data presented in Figure 5 (right panels) are comparable to those reported for animals switched from unrestricted to daytime feeding (see Figure 1A and B and Damiola et al., 2000). They confirm that in animals with an intact hypothalamus–pituitary gland–adrenal axis, phase adaptations are reached slowly when the feeding schedule is set in conflict with the activity phase.
In order to test whether glucocorticoid signaling also affects the phase adaptation in circadian gene expression accompanying the switch back from daytime to night-time feeding, we compared the Dbp mRNA accumulation profile 2–3 days after changing the feeding regimen in livers and kidneys of wild-type and GRAlfpCre mice. Figure 6A demonstrates that the kinetics of phase resetting of Dbp and mPer2 mRNA accumulation are similar in wild-type and GRAlfpCre mice on the second day after swapping feeding time. In a control experiment we re-examined Dbp mRNA accumulation 2–3 days after swapping mice of the two genotypes from night-time to daytime feeding, and observed that the phase of circadian Dbp mRNA accumulation shifted considerably faster in the liver of GRAlfpCre mice (data not shown), as expected from the data presented in Figure 4. Moreover, no significant differences between the Dbp mRNA accumulation patterns could be noticed in the kidneys of wild-type and GRAlfpCre mice, which both express the glucocorticoid receptor (Figure 6B).
Fig. 6. Accumulation of Dbp and mPer2 mRNA in GRAlfpCre and wild-type mice switched from daytime to night-time feeding. Mice kept under a 12 h LD regimen were entrained to daytime feeding during 8 days, and then switched to night-time feeding. At the times after the feeding time reversal (re-entrainment) indicated, liver (A) and kidney (B) RNAs were subjected to RNase protection assays with Dbp, mPer2 and Tbp probes. Tbp mRNA was included as a transcript whose accumulation is neither circadian- nor feeding time-dependent. For comparison, the approximate peak times of mRNA accumulation in animals fed ad libitum or during the night are indicated by arrows. Note that glucocorticoid signaling [compare left and right autoradiographs in (A)] has little if any effect on the rapid phase adjustments in mRNA accumulation cycles observed in mice switched from daytime to night-time feeding.
Discussion
The circadian timing system in mammals is constructed in a hierarchical fashion: a master pacemaker localized in SCN neurons synchronizes slave clocks in most cell types of peripheral tissues (Yamazaki et al., 2000). While light is clearly the major Zeitgeber for the central clock, recent reports suggest that feeding time is the dominant timing cue for at least some peripheral clocks (Damiola et al., 2000; Stokkan et al., 2001). Thus, in nocturnal rodents, such as rats and mice, daytime feeding completely uncouples the phase of circadian gene expression in liver, kidney, heart and pancreas from that of the SCN pacemaker. These observations opened the possibility that the SCN may entrain peripheral clocks indirectly by setting the activity phase, and thus feeding time, if food availability is not in conflict with the normal activity phase. Similar to the phase resetting of the SCN pacemaker by large changes in the photoperiod, the phase adjustment of peripheral oscillators by feeding time is gradual rather than instantaneous. Even in the liver, in which food-induced phase shifting proceeds somewhat faster than in other tissues examined, several consecutive days of daytime feeding are required to reach the new and inverted steady-state phase (Damiola et al., 2000). The inertia in phase resetting could reflect the possibility that the limited daily phase-shifting capacity is an intrinsic property of peripheral clocks. However, the results presented in this study offer an alternative interpretation: namely, that a rapid phase adjustment in response to altered feeding time is inhibited by glucocorticoid hormone signaling. In fact, daytime feeding can induce large phase shifts in mice in which glucocorticoid signaling has been interrupted either by adrenalectomy or ablation of the glucocorticoid receptor gene.
In previous work, the glucocorticoid analog dexamethasone has been demonstrated to be a potent phase-shifting agent for circadian gene expression in peripheral mouse tissues (Balsalobre et al., 2000a). Moreover, dexamethasone efficiently stimulates circadian gene expression in rat-1 fibroblasts grown in tissue culture (Balsalobre et al., 2000b). For three reasons, glucocorticoids have been considered to be particularly well suited as Zeitgebers for peripheral oscillators used by the SCN to synchronize peripheral clocks. First, their cyclic secretion is under the control of the hypothalamus–pituitary gland–adrenal axis, which in turn is controlled by the SCN. Secondly, with the noteworthy exception of the SCN, the glucocorticoid receptor is expressed in all cell types examined (Rosenfeld et al., 1988; Balsalobre et al., 2000a), a pattern that is a prerequisite for an input component of the ubiquitous peripheral clocks. Thirdly, in the SCN of newborn animals, the glucocorticoid receptor gene is expressed, but it becomes silenced within the first weeks after birth (Rosenfeld et al., 1988), just before the animals display robust circadian gene expression in peripheral organs (S.A.Brown and U.Schibler, unpublished observations). In view of the different phase angles determined for circadian gene expression in the SCN and in peripheral tissues, at least some signaling pathways affecting the phase in the latter should not be operative in the former. However, although glucocorticoid hormones fulfill all of these prerequisites of Zeitgebers for peripheral clocks, the glucocorticoid receptor is dispensable for the synchronization of circadian liver gene expression under steady-state conditions. Thus, the steady-state phase of circadian liver gene expression is identical in GRAlfpCre mice, whose hepatocytes are homozygous for a glucocorticoid receptor null allele, and wild-type mice (Balsalobre et al., 2000a). Hence, glucocorticoid hormones cannot be the sole Zeitgeber for peripheral tissues.
Given the known connection between glucocorticoid signaling and fasting (Dallman et al., 1999), one might have anticipated that glucocorticoid signaling may play a role in the phase resetting of circadian gene expression by feeding time. However, Stokkan et al. (2001) have demonstrated that the rephasing of cyclic gene expression in rat liver cannot be mimicked by repeated daily injections of corticosterones at times when these hormones peak in animals subjected to restricted feeding. As shown here, glucocorticoids inhibit rather than promote the phase adjustments of peripheral oscillators to daytime feeding, and circadian gene expression can experience large phase shifts in response to restricted feeding, once the counteracting glucocorticoid signaling pathway is inactivated. That peripheral oscillators are capable of conducting very large phase shifts is also demonstrated by an experiment in which mice are first entrained to daytime feeding and then switched back to night-time feeding. This switch is accompanied by a very rapid phase reversal in circadian liver gene expression, both in the presence and absence of glucocorticoid signaling. Hence, it appears that glucocorticoid signaling slows down exclusively phase changes provoked by a feeding regimen that is in conflict with the daily activity phase, but has little effect when the phase is switched from an abnormal to a normal feeding schedule.
Glucocorticoid signaling could be used by the SCN master clock to prevent a rapid uncoupling of peripheral oscillators. However, our results demonstrate that the temporal pattern of serum glucocorticoid hormones changes significantly during daytime feeding, in that it follows a bimodal distribution in daytime-fed animals. In particular, at the onset of daytime feeding, corticosterone levels are unusually high, probably as a consequence of decreased glucose levels. It is, thus, difficult to decide whether the circadian glucocorticoid secretion controlled by the SCN or that provoked by restricted feeding is responsible for the observed phenotype. A further complication is introduced by the episodic nature of corticosterone secretion. As glucocorticoids are secreted in bouts, their levels vary dramatically between sera harvested from different individuals, and this makes the comparison of individuals difficult. Real-time monitoring of blood samples in individual animals would be a more reliable way to follow glucocorticoid secretion profiles, but unfortunately this method is difficult to apply to mice, given their small blood volume.
We noticed that in livers and kidneys of animals with intact glucocorticoid signaling, daytime feeding affects the expression of mPer1 more dramatically than that of other circadian genes. Thus, while the steady-state phases of mPer1 and Dbp mRNA accumulation are nearly identical (see Damiola et al., 2000), they are transiently different at the onset of daytime feeding (Figures 1A and B, 4A and B). Previously published experiments already suggested that mPer1 is a direct target of glucocorticoid signaling (Balsalobre et al., 2000a,b). In keeping with this conjecture, mPer1 contains two almost perfect GRE consensus sequences of the type AGAACAN3TGTTCT in the 5′-flanking region (AGAACACGATGTTCc centered around –1088) and in the first intron (gGAACATCC TGTTCT centered around +490), respectively, with respect to the transcription start site determined by Hida et al. (2000). Moreover, dexamethasone rapidly induces a surge of Per1 mRNA accumulation in rat-1 tissue culture cells or peripheral cell types of intact mice (Balsalobre et al., 2000a,b). We thus consider it likely that the altered expression of mPer1 after switching mice to daytime feeding participates in the glucocorticoid-dependent kinetics of food-induced phase resettings in liver and kidney. We also wish to emphasize, however, that after 1 week of daytime feeding, corticosterone accumulates in the blood in a biphasic temporal pattern, the phase of the second peak of which resembles the circadian pattern of glucocorticoid secretion in animals fed ad libitum. Thus, once the new steady-state phase is reached after an extended duration of daytime feeding, glucocorticoid signaling does not appear to interfere with the food-imposed reversed phase of circadian gene expression in peripheral tissues.
Restricted feeding not only affects the phasing of circadian gene expression, but it can also entrain food-anticipatory activity (see Davidson and Stephan, 1999 and references therein). Thus, shortly before the daily meal is offered to laboratory rodents, they display increased locomotor activity and body temperature. Interestingly, food-anticipatory behavior does not depend on either a functional SCN pacemaker or an intact pituitary gland. Whether circadian gene expression in other brain regions or in peripheral tissues is involved in driving the feeding-entrainable oscillator remains to be examined.
How does feeding time reset the phase of oscillators in organs such as liver and kidney? As mentioned in the Introduction, a plethora of chemical signals can induce circadian gene expression in tissue culture cells, and at least some of them may be involved in food-induced phase resetting. Additional bloodborne signals (e.g. insulin, glucagon and leptin), whose secretion is known to depend on food metabolites, may also play a role. However, a recent report by Rutter et al. (2001) opens up the possibility that feeding may also influence circadian gene expression more directly. At least in vitro, the DNA binding of the Clock–BMAL1 or NPAS2–BMAL1 heterodimers, which constitute the positive limbs of the central circadian feedback loop, is highly sensitive to the ratio of NAD cofactors. The reduced electron carriers NADH or NADPH strongly stimulate the occupancy of E-box motifs by Clock–Bmal1 heterodimers, while the oxidized versions NAD+ or NADP+ inhibit this protein– DNA interaction. As the ratio of reduced to oxidized NAD cofactors is likely to depend on the availability of fuels such as glucose, the metabolic state of cells may directly influence molecular oscillators in liver and other peripheral tissues through changes in redox potential. If applicable, this mechanism would have to be specific to peripheral oscillators, since feeding time does not impinge on the phase entrainment of the central SCN pacemaker (Damiola et al., 2000; Stokkan et al., 2001).
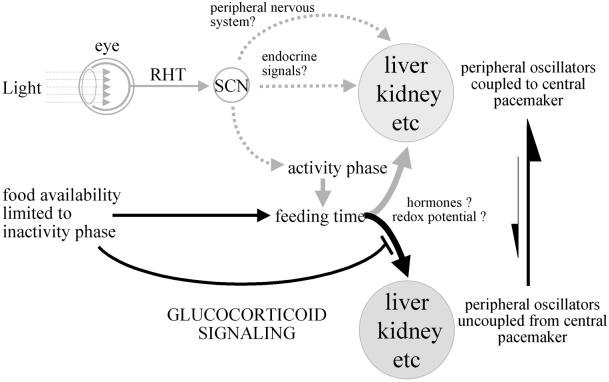
The scheme depicted in Figure 7 summarizes how the SCN pacemaker, feeding time and glucocorticoid signaling may control the phase entrainment of peripheral oscillators. The elucidation of the signals involved in the complex interactions between central and peripheral clocks, and of the mechanisms that govern the interplay between metabolism and circadian oscillators, has just begun. We hope that further studies addressing these issues will be greatly facilitated by the possibility of investigating circadian gene expression in tissue culture cells that are readily amenable to genetic and biochemical manipulation.
Fig. 7. The phase entrainment of circadian oscillators in peripheral organs. The scheme displays a hypothetical model of the synchronization of peripheral oscillators by the central SCN pacemaker and feeding time. The SCN is entrained by the photoperiod via synaptic connections with the retina (retino–hypothalamic tract, RHT). In turn, the SCN master clock entrains circadian gene expression in peripheral tissues (e.g. liver, kidney) through direct neuronal and humoral pathways, or indirectly by determining the activity phase and thus feeding time. When food, the dominant Zeitgeber for (at least some) peripheral oscillators, is only available during the resting phase (light phase in nocturnal rodents), the phases of these clocks are inverted. This process is slowed down by glucocorticoid signaling, probably through the abundant secretion of corticosterones during the dark phase in animals switched to daytime feeding. In contrast to the slow-phase adaptation of peripheral gene expression accompanying the switch from night-time to daytime feeding, during which peripheral oscillators become uncoupled from the central pacemaker, the switch back from daytime to night-time feeding causes an almost instantaneous phase inversion. The molecular mechanisms by which food resets the phase of peripheral oscillators are not known, but they may involve changes in the redox potential [NAD(P)H/NAD(P)+ ratio] or in hormones whose secretion is provoked or suppressed by food metabolites.
Materials and methods
Animal care and handling
All experiments were performed with mice between 10 and 16 weeks of age. Adrenalectomized and sham-operated C57BL/6J mice were purchased from IFFA CREDO. GRAlfpCre and their respective wild-type control mice were established at the Deutsches Krebsforschungzentrum (Heidelberg, Germany) (Kellendonk et al., 2000). GRAlfpCre mice have a mixed genetic background (129-C57BL/6-FVB/N). The animals were kept under a 12 h light–dark regimen (lights on at ZT0). Mice fed during the day received food from ZT0 to ZT12, whereas those fed during the night received food from ZT12 to ZT0.
RNase protection experiments
Mouse tissues were removed within 4 min after decapitation, frozen in liquid N2 and stored at –70°C until use. The extraction of whole-cell RNA and its analysis by RNase protection assays were performed as described (Schmidt and Schibler, 1995). The Dbp antisense RNA probe is complementary to rat Dbp mRNA (+1126 to +1221) (Fonjallaz et al., 1996). The Tbp probe is complementary to mouse Tbp mRNA (+36 to +135) (Schmidt and Schibler, 1995). The Per3 and Cry1 probes (kindly provided by A.Balsalobre) are complementary to mouse Per3 mRNA (+367 to +572) and mouse Cry1 mRNA (+83 to +312), respectively (Balsalobre et al., 2000a). The Per2 probe is complementary to mouse Per2 mRNA (+165 to +287) (Balsalobre et al., 1998). The Per1 probe is complementary to mouse Per1 mRNA (+ 2397 to + 2523) (a generous gift from J.Ripperger).
In all cases, the plasmids were linearized with a suitable restriction enzyme, and the antisense RNA probes were prepared by in vitro transcription of the linearized templates with T7 or T3 RNA polymerase using [α-32P]UTP. Autoradiography was performed with an intensifying screen (Fuji) at –70°C for 1–5 days.
In situ hybridization to coronal brain sections
Immediately after removal, brains were frozen in isopentane (4 min at –20°C) and stored at –70°C until use. Serial coronal brain cryosections of 12 µm above the optical chiasma were prepared using standard procedures. In situ hybridizations with sections though the central SCN were performed as described previously (Nef et al., 1996). The mPer1 and mPer2 riboprobes used in these experiments were prepared from the templates pKS-mPer1-Fl and pKS-mPer2-nqFl (a generous gift from S.Brown), using RNA polymerases T3 and T7 for the antisense and sense strands, respectively. pKS-mPer1-Fl and pKS-mPer2-nqFl were obtained by cloning the inserts of the plasmids pcDNA3.1-P1 and pcDNA3.1-P2, respectively, into the plasmid vector pBS-KS. To this end, the inserts of pcDNA3.1-P1 and pcDNA3.1-P2 were excised with XbaI and ClaI–NotI, respectively. The plasmids pcDNA3.1-P1 and pcDNA3.1-P2 (Jin et al., 1999) were kindly provided by Dr Steven Reppert.
Determination of corticosterone levels
Mice were killed within 10 s after entering the animal facility (to avoid interference of stress with corticosterone levels) and, after decapitation, their blood was collected. After coagulation (10 min at room temperature and 10 min on ice), samples were centrifuged at 3000 r.p.m. (920 g) for 20 min and the sera were stored at –70°C until use. Corticosterone measurements were carried out using the corticosterone 125I RIA kit from ICN Biomedicals, Inc. (Costa Mesa, CA) according to the manufacturer’s instructions.
Acknowledgments
Acknowledgements
We are grateful to Steven Reppert for his gift of the mPer1 and mPer2 cDNA plasmids, Steven Brown and Juergen Ripperger for their valuable discussions, and Nicolas Roggli for expert preparation of the illustrations. This work was supported by the Swiss National Science Foundation, the State of Geneva, the Bonizzi-Theler Stiftung and the Louis-Jeantet Foundation for Medicine (U.S.), the Deutsche Forschungsgemeinschaft and the European Union (G.S.) and the Roche Research Foundation (F.D.).
References
- Akashi M. and Nishida,E. (2000) Involvement of the MAP kinase cascade in resetting of the mammalian circadian clock. Genes Dev., 14, 645–649. [PMC free article] [PubMed] [Google Scholar]
- Albrecht U., Sun,Z.S., Eichele,G. and Lee,C.C. (1997) A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell, 91, 1055–1064. [DOI] [PubMed] [Google Scholar]
- Balsalobre A., Damiola,F. and Schibler,U. (1998) A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell, 93, 929–937. [DOI] [PubMed] [Google Scholar]
- Balsalobre A., Brown,S.A., Marcacci,L., Tronche,F., Kellendonk,C., Reichardt,H.M., Schutz,G. and Schibler,U. (2000a) Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science, 289, 2344–2347. [DOI] [PubMed] [Google Scholar]
- Balsalobre A., Marcacci,L. and Schibler,U. (2000b) Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr. Biol., 10, 1291–1294. [DOI] [PubMed] [Google Scholar]
- Brown S.A. and Schibler,U. (1999) The ins and outs of circadian timekeeping. Curr. Opin. Genet. Dev., 9, 588–594. [DOI] [PubMed] [Google Scholar]
- Bunger M.K., Wilsbacher,L.D., Moran,S.M., Clendenin,C., Radcliffe,L.A., Hogenesch,J.B., Simon,M.C., Takahashi,J.S. and Bradfield,C.A. (2000) Mop3 is an essential component of the master circadian pacemaker in mammals. Cell, 103, 1009–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo A.J., Duke,P.G. and Dunn,J.D. (1980) Episodic corticosterone secretion in the female rat. Horm. Res., 13, 40–47. [DOI] [PubMed] [Google Scholar]
- Cheifetz P.N. (1971) The daily rhythm of the secretion of corticotrophin and corticosterone in rats and mice. J. Endocrinol., 49, xi–xii. [PubMed] [Google Scholar]
- Dallman M.F. et al. (1999) Starvation: early signals, sensors and sequelae. Endocrinology, 140, 4015–4023. [DOI] [PubMed] [Google Scholar]
- Damiola F., Le Minh,N., Preitner,N., Kornmann,B., Fleury-Olela,F. and Schibler,U. (2000) Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev., 14, 2950–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson A.J. and Stephan,F.K. (1999) Feeding-entrained circadian rhythms in hypophysectomized rats with suprachiasmatic nucleus lesions. Am. J. Physiol., 277, R1376–R1384. [DOI] [PubMed] [Google Scholar]
- Ding J.M., Chen,D., Weber,E.T., Faiman,L.E., Rea,M.A. and Gillette,M.U. (1994) Resetting the biological clock: mediation of nocturnal circadian shifts by glutamate and NO. Science, 266, 1713–1717. [DOI] [PubMed] [Google Scholar]
- Ebling F.J. (1996) The role of glutamate in the photic regulation of the suprachiasmatic nucleus. Prog. Neurobiol., 50, 109–132. [DOI] [PubMed] [Google Scholar]
- Fonjallaz P., Ossipow,V., Wanner,G. and Schibler,U. (1996) The two PAR leucine zipper proteins, TEF and DBP, display similar circadian and tissue-specific expression, but have different target promoter preferences. EMBO J., 15, 351–362. [PMC free article] [PubMed] [Google Scholar]
- Hida A., Koike,N., Hirose,M., Hattori,M., Sakaki,Y. and Tei,H. (2000) The human and mouse Period1 genes: five well-conserved E-boxes additively contribute to the enhancement of mPer1 transcription. Genomics, 65, 224–233. [DOI] [PubMed] [Google Scholar]
- Jin X., Shearman,L.P., Weaver,D.R., Zylka,M.J., de Vries,G.J. and Reppert,S.M. (1999) A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell, 96, 57–68. [DOI] [PubMed] [Google Scholar]
- Keesler G.A., Camacho,F., Guo,Y., Virshup,D., Mondadori,C. and Yao,Z. (2000) Phosphorylation and destabilization of human period I clock protein by human casein kinase Iε. Neuroreport, 11, 951–955. [DOI] [PubMed] [Google Scholar]
- Kellendonk C., Opherk,C., Anlag,K., Schutz,G. and Tronche,F. (2000) Hepatocyte-specific expression of Cre recombinase. Genesis, 26, 151–153. [DOI] [PubMed] [Google Scholar]
- King D.P. et al. (1997) Positional cloning of the mouse circadian clock gene. Cell, 89, 641–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K., Zylka,M.J., Sriram,S., Shearman,L.P., Weaver,D.R., Jin,X., Maywood,E.S., Hastings,M.H. and Reppert,S.M. (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell, 98, 193–205. [DOI] [PubMed] [Google Scholar]
- Lejeune-Lenain C., Van Cauter,E., Desir,D., Beyloos,M. and Franckson,J.R. (1987) Control of circadian and episodic variations of adrenal androgens secretion in man. J. Endocrinol. Invest., 10, 267–276. [DOI] [PubMed] [Google Scholar]
- Lowrey P.L., Shimomura,K., Antoch,M.P., Yamazaki,S., Zemenides,P.D., Ralph,M.R., Menaker,M. and Takahashi,J.S. (2000) Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science, 288, 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara P., Seo,S.P., Rudic,R.D., Sehgal,A., Chakravarti,D. and FitzGerald,G.A. (2001) Regulation of CLOCK and MOP4 by nuclear hormone receptors in the vasculature: a humoral mechanism to reset a peripheral clock. Cell, 105, 877–889. [DOI] [PubMed] [Google Scholar]
- Morris M.E., Viswanathan,N., Kuhlman,S., Davis,F.C. and Weitz,C.J. (1998) A screen for genes induced in the suprachiasmatic nucleus by light. Science, 279, 1544–1547. [DOI] [PubMed] [Google Scholar]
- Nef S., Allaman,I., Fiumelli,H., De Castro,E. and Nef,P. (1996) Olfaction in birds: differential embryonic expression of nine putative odorant receptor genes in the avian olfactory system. Mech. Dev., 55, 65–77. [DOI] [PubMed] [Google Scholar]
- Reppert S.M. and Weaver,D.R. (2001) Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol., 63, 647–676. [DOI] [PubMed] [Google Scholar]
- Ripperger J.A. and Schibler,U. (2001) Circadian regulation of gene expression in animals. Curr. Opin. Cell Biol., 13, 357–362. [DOI] [PubMed] [Google Scholar]
- Ripperger J.A., Shearman,L.P., Reppert,S.M. and Schibler,U. (2000) CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev., 14, 679–689. [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld P., Van Eekelen,J.A., Levine,S. and De Kloet,E.R. (1988) Ontogeny of the type 2 glucocorticoid receptor in discrete rat brain regions: an immunocytochemical study. Brain Res., 470, 119–127. [DOI] [PubMed] [Google Scholar]
- Rutter J., Reick,M., Wu,L.C. and McKnight,S.L. (2001) Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science, 293, 510–514. [DOI] [PubMed] [Google Scholar]
- Schmidt E.E. and Schibler,U. (1995) High accumulation of components of the RNA polymerase II transcription machinery in rodent spermatids. Development, 121, 2373–2383. [DOI] [PubMed] [Google Scholar]
- Shearman L.P., Zylka,M.J., Weaver,D.R., Kolakowski,L.F.,Jr and Reppert,S.M. (1997) Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron, 19, 1261–1269. [DOI] [PubMed] [Google Scholar]
- Shearman L.P. et al. (2000) Interacting molecular loops in the mammalian circadian clock. Science, 288, 1013–1019. [DOI] [PubMed] [Google Scholar]
- Shigeyoshi Y. et al. (1997) Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell, 91, 1043–1053. [DOI] [PubMed] [Google Scholar]
- Shiraishi I., Honma,K., Honma,S. and Hiroshige,T. (1984) Ethosecreto gram: relation of behavior to plasma corticosterone in freely moving rats. Am. J. Physiol., 247, R40–R45. [DOI] [PubMed] [Google Scholar]
- Shirakawa T. and Moore,R.Y. (1994) Glutamate shifts the phase of the circadian neuronal firing rhythm in the rat suprachiasmatic nucleus in vitro. Neurosci. Lett., 178, 47–50. [DOI] [PubMed] [Google Scholar]
- Stokkan K.A., Yamazaki,S., Tei,H., Sakaki,Y. and Menaker,M. (2001) Entrainment of the circadian clock in the liver by feeding. Science, 291, 490–493. [DOI] [PubMed] [Google Scholar]
- Sun Z.S., Albrecht,U., Zhuchenko,O., Bailey,J., Eichele,G. and Lee,C.C. (1997) RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell, 90, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Tei H., Okamura,H., Shigeyoshi,Y., Fukuhara,C., Ozawa,R., Hirose,M. and Sakaki,Y. (1997) Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature, 389, 512–516. [DOI] [PubMed] [Google Scholar]
- Toh K.L., Jones,C.R., He,Y., Eide,E.J., Hinz,W.A., Virshup,D.M., Ptacek,L.J. and Fu,Y.H. (2001) An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science, 291, 1040–1043. [DOI] [PubMed] [Google Scholar]
- Tronche F., Kellendonk,C., Reichardt,H.M. and Schutz,G. (1998) Genetic dissection of glucocorticoid receptor function in mice. Curr. Opin. Genet. Dev., 8, 532–538. [DOI] [PubMed] [Google Scholar]
- van der Horst G.T. et al. (1999) Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature, 398, 627–630. [DOI] [PubMed] [Google Scholar]
- Yagita K. and Okamura,H. (2000) Forskolin induces circadian gene expression of rPer1, rPer2 and dbp in mammalian rat-1 fibroblasts. FEBS Lett., 465, 79–82. [DOI] [PubMed] [Google Scholar]
- Yagita K., Tamanini,F., van der Horst,G.T. and Okamura,H. (2001) Molecular mechanisms of the biological clock in cultured fibroblasts. Science, 292, 278–281. [DOI] [PubMed] [Google Scholar]
- Yamazaki S. et al. (2000) Resetting central and peripheral circadian oscillators in transgenic rats. Science, 288, 682–685. [DOI] [PubMed] [Google Scholar]
- Zheng B., Larkin,D.W., Albrecht,U., Sun,Z.S., Sage,M., Eichele,G., Lee,C.C. and Bradley,A. (1999) The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature, 400, 169–173. [DOI] [PubMed] [Google Scholar]